In Vivo Level of Poly(ADP-ribose)
Abstract
:1. Introduction
1.1. What Is PolyADP-Ribosylation?
1.2. Proposed Functions of PolyADP-Ribosylation
1.3. Importance to Understand the In Vivo Level of PAR that is Rapidly Turning-Over under Various Biological Conditions
1.4. Measurement of the Amount of PAR In Vivo (i.e., in Cells or Tissues)
1.5. Prospect: In Vivo PAR Level to Understand More Biological Functions and Its Application for Clinic
2. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sugimura, T. Poly(adenosine diphosphate ribose). Prog. Nucleic Acid Res. Mol. Biol. 1973, 13, 127–151. [Google Scholar] [PubMed]
- Miwa, M.; Masutani, M. PolyADP-ribosylation and cancer. Cancer Sci. 2007, 98, 1528–1535. [Google Scholar] [CrossRef] [PubMed]
- Hottiger, M.O.; Hassa, P.O.; Lüscher, B.; Schüler, H.; Koch-Nolte, F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010, 35, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Chambon, P.; Weill, J.; Doly, J.; Strosser, M.; Mandel, P. On the formation of a novel adenylic compound by enzymatic extracts of liver nuclei. Biochem. Biophys Res. Commun. 1966, 25, 634–643. [Google Scholar] [CrossRef]
- Sugimura, T.; Fujimura, S.; Hasegawa, S.; Kawamura, Y. Polymerization of the adenosine 5′-diphosphate ribose moiety of NAD by rat liver nuclear enzyme. Biochim. Biophys. Acta 1967, 138, 438–441. [Google Scholar] [CrossRef]
- Nishizuka, Y.; Ueda, K.; Nakazawa, K.; Hayaishi, O. Studies on the polymer of adenosine diphosphate ribose. I. Enzymic formation from nicotinamide adenine dinuclotide in mammalian nuclei. J. Biol. Chem. 1967, 242, 3164–3171. [Google Scholar] [PubMed]
- Morrison, A.R.; Moss, J.; Stevens, L.A.; Evans, J.E.; Farrell, C.; Merithew, E.; Lambright, D.G.; Greiner, D.L.; Mordes, J.P.; Rossini, A.A.; et al. ART2, a T cell surface mono-ADP-ribosyltransferase, generates extracellular poly(ADP-ribose). J. Biol. Chem. 2006, 281, 33363–33372. [Google Scholar] [CrossRef] [PubMed]
- Miwa, M.; Sugimura, T. Splitting of the ribose-ribose linkage of poly(adenosine diphosphate-robose) by a calf thymus extract. J. Biol. Chem. 1971, 246, 6362–6364. [Google Scholar] [PubMed]
- Ueda, K.; Oka, J.; Narumiya, S.; Miyakawa, N.; Hayaishi, O. Poly ADP-ribose glycohydrolase from rat liver nuclei, a novel enzyme degrading the polymer. Biochem. Biophys Res. Commun. 1972, 46, 516–523. [Google Scholar] [CrossRef]
- Oka, S.; Kato, J.; Moss, J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2006, 281, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Oka, J.; Ueda, K.; Hayaishi, O.; Komura, H.; Nakanishi, K. ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J. Biol. Chem. 1984, 259, 986–995. [Google Scholar] [PubMed]
- Sharifi, R.; Morra, R.; Appel, C.D.; Tallis, M.; Chioza, B.; Jankevicius, G.; Simpson, M.A.; Matic, I.; Ozkan, E.; Golia, B.; et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013, 32, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, F.; Feijs, K.L.; Frugier, E.; Bonalli, M.; Forst, A.H.; Imhof, R.; Winkler, H.C.; Fischer, D.; Caflisch, A.; Hassa, P.O.; et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013, 20, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Jankevicius, G.; Hassler, M.; Golia, B.; Rybin, V.; Zacharias, M.; Timinszky, G.; Ladurner, A.G. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013, 20, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Poltronieri, P.; Cerekovic, N. Roles of nicotinamide adenine dinucleotide (NAD+) in biological systems. Challenges 2018, 9, 3. [Google Scholar] [CrossRef]
- Ueda, K.; Hayaishi, O. ADP-ribosylation. Annu. Rev. Biochem. 1985, 54, 73–100. [Google Scholar] [CrossRef] [PubMed]
- Althaus, F.R.; Richter, C. ADP-ribosylation of proteins. Enzymology and biological significance. Mol. Biol. Biochem. Biophys 1987, 37, 1–237. [Google Scholar] [PubMed]
- Schreiber, V.; Dantzer, F.; Ame, J.C.; de Murcia, G. Poly(ADP-ribose): Novel functions for an old molecule. Nat. Rev. Mol. Cell. Biol. 2006, 7, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Kraus, W.L. PARPs and ADP-Ribosylation: 50 Years … and Counting. Mol. Cell 2015, 58, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Hottiger, M.O. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu. Rev. Biochem. 2015, 4, 227–263. [Google Scholar] [CrossRef] [PubMed]
- Barkauskaite, E.; Jankevicius, G.; Ahel, I. Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation. Mol. Cell. 2015, 58, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Crawford, K.; Bonfiglio, J.J.; Mikoč, A.; Matic, I.; Ahel, I. Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 64–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Q.; Stingl, L.; Morrison, C.; Jantsch, M.; Los, M.; Schulze-Osthoff, K.; Wagner, E.F. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997, 11, 2347–2358. [Google Scholar] [CrossRef] [PubMed]
- De Murcia, J.M.; Niedergang, C.; Trucco, C.; Ricoul, M.; Dutrillaux, B.; Mark, M.; Oliver, F.J.; Masson, M.; Dierich, A.; LeMeur, M.; et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. USA 1997, 94, 7303–7307. [Google Scholar] [CrossRef] [PubMed]
- Masutani, M.; Suzuki, H.; Kamada, N.; Watanabe, M.; Ueda, O.; Nozaki, T.; Jishage, K.; Watanabe, T.; Sugimoto, T.; Nakagama, H.; et al. Poly(ADP-ribose) polymerase gene disruption conferred mice resistant to streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. USA 1999, 96, 2301–2304. [Google Scholar] [CrossRef] [PubMed]
- Hanai, S.; Kanai, M.; Ohashi, S.; Okamoto, K.; Yamada, M.; Takahashi, H.; Miwa, M. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2004, 101, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Koh, D.W.; Lawler, A.M.; Poitras, M.F.; Sasaki, M.; Wattler, S.; Nehls, M.C.; Stöger, T.; Poirier, G.G.; Dawson, V.L.; Dawson, T.M. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. USA 2004, 101, 17699–17704. [Google Scholar] [CrossRef] [PubMed]
- Durkacz, B.W.; Omidiji, O.; Gray, D.A.; Shall, S. (ADP-ribose)n participates in DNA excision repair. Nature 1980, 283, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Smulson, M.; Stark, P.; Gazzoli, M.; Roberts, J. Release of template restriction for DNA synthesis by poly ADP(ribose) polymerase during the HeLa cell cycle. Exp. Cell Res. 1975, 90, 175–182. [Google Scholar] [CrossRef]
- Simbulan-Rosenthal, C.M.; Rosenthal, D.S.; Smulson, M.E. Purification and characterization of poly(ADP-ribosyl)ated DNA replication/repair complexes. Methods Mol. Biol. 2011, 780, 165–190. [Google Scholar] [PubMed]
- Tulin, A.; Spradling, A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 2003, 299, 560–562. [Google Scholar] [CrossRef] [PubMed]
- Kraus, W.L. Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 2008, 20, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Aubin, R.J.; Fréchette, A.; de Murcia, G.; Mandel, P.; Lord, A.; Grondin, G.; Poirier, G.G. Correlation between endogenous nucleosomal hyper(ADP-ribosyl)ation of histone H1 and the induction of chromatin relaxation. EMBO J. 1983, 2, 1685–1693. [Google Scholar] [PubMed]
- Realini, C.A.; Althaus, F.R. Histone shuttling by poly(ADP-ribosylation). J. Biol. Chem. 1992, 267, 18858–18865. [Google Scholar] [PubMed]
- Utakoji, T.; Hosoda, K.; Umezawa, K.; Sawamura, M.; Matsushima, T.; Miwa, M.; Sugimura, T. Induction of sister chromatid exchanges by nicotinamide in Chinese hamster lung fibroblasts and human lymphoblastoid cells. Biochem. Biophys. Res. Commun. 1979, 90, 1147–1152. [Google Scholar] [CrossRef]
- Oikawa, A.; Tohda, H.; Kanai, M.; Miwa, M.; Sugimura, T. Inhibitors of poly(adenosine diphosphate ribose) polymerase induce sister chromatid exchanges. Biochem. Biophys. Res. Commun. 1980, 97, 1311–1316. [Google Scholar] [CrossRef]
- Kanai, M.; Tong, W.M.; Sugihara, E.; Wang, Z.Q.; Fukasawa, K.; Miwa, M. Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-Ribosyl)ation in regulation of centrosome function. Mol. Cell. Biol. 2003, 23, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Giriat, I.; Schmitt, A.; de Lange, T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 1998, 282, 1484–1487. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Armon, M.; Visochek, L.; Rozensal, D.; Kalal, A.; Geistrikh, I.; Klein, R.; Bendetz-Nezer, S.; Yao, Z.; Seger, R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: A link to histone acetylation. Mol. Cell 2007, 25, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M.; Hanashiro, K.; Kim, S.H.; Hanai, S.; Boulares, A.H.; Miwa, M.; Fukasawa, K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol. 2007, 9, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Stilmann, M.; Hinz, M.; Arslan, S.C.; Zimmer, A.; Schreiber, V.; Scheidereit, C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol. Cell 2009, 36, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kim, N.S.; Haince, J.F.; Kang, H.C.; David, K.K.; Andrabi, S.A.; Poirier, G.G.; Dawson, V.L.; Dawson, T.M. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci. Signal. 2011, 4, ra20. [Google Scholar] [CrossRef] [PubMed]
- Masutani, M.; Fujimori, H. Poly(ADP-ribosyl)ation in carcinogenesis. Mol. Asp. Med. 2013, 34, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Thomas, C.; Tulin, N.; Lodhi, N.; Boamah, E.; Kolenko, V.; Tulin, A.V. Charon Mediates Immune Deficiency-Driven PARP-1-Dependent Immune Responses in Drosophila. J. Immunol. 2016, 197, 2382–2389. [Google Scholar] [CrossRef] [PubMed]
- Szabó, C.; Dawson, V.L. Role of poly(ADP-ribose) synthetase in inflammation and ischaemia-reperfusion. Trends Pharmacol. Sci. 1998, 19, 287–298. [Google Scholar] [CrossRef]
- Ji, J.; Kinders, R.J.; Zhang, Y.; Rubinstein, L.; Kummar, S.; Parchment, R.E.; Tomaszewski, J.E.; Doroshow, J.H. Modeling pharmacodynamic response to the poly(ADP-Ribose) polymerase inhibitor ABT-888 in human peripheral blood mononuclear cells. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Hashida, T.; Yoshihara, H.; Tanaka, Y.; Ohgushi, H. Enzyme-bound early product of purified poly(ADP-ribose) polymerase. Biochem. Biophys. Res. Commun. 1977, 78, 1281–1288. [Google Scholar] [CrossRef]
- Ogata, N.; Ueda, K.; Hayaishi, O. ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site. J. Biol. Chem. 1980, 255, 7610–7615. [Google Scholar] [PubMed]
- Ogata, N.; Ueda, K.; Kagamiyama, H.; Hayaishi, O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J. Biol. Chem. 1980, 255, 7616–7620. [Google Scholar] [PubMed]
- Suzuki, H.; Quesada, P.; Farina, B.; Leone, E. In vitro poly(ADP-ribosyl)ation of seminal ribonuclease. J. Biol. Chem. 1986, 261, 6048–6055. [Google Scholar] [PubMed]
- Wesierska-Gadek, J.; Schmid, G.; Cerni, C. ADP-ribosylation of wild-type p53 in vitro: Binding of p53 protein to specific p53 consensus sequence prevents its modification. Biochem. Biophys. Res. Commun. 1996, 224, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Miwa, M.; Saikawa, N.; Yamaizumi, Z.; Nishimura, S.; Sugimura, T. Structure of poly(adenosine diphosphate ribose): Identification of 2′-[1′′-ribosyl-2′′-(or 3′′-)(1′′′-ribosyl)]adenosine-5′,5′′,5′′′-tris(phosphate) as a branch linkage. Proc. Natl. Acad. Sci. USA 1979, 76, 595–599. [Google Scholar] [CrossRef] [PubMed]
- De Murcia, G.; Jongstra-Bilen, J.; Ittel, M.E.; Mandel, P.; Delain, E. Poly(ADP-ribose) polymerase auto-modification and interaction with DNA: Electron microscopic visualization. EMBO J. 1983, 2, 543–548. [Google Scholar] [PubMed]
- Hayashi, K.; Tanaka, M.; Shimada, T.; Miwa, M.; Sugimura, T. Size and shape of poly(ADP-ribose): Examination by gel filtration, gel electrophoresis and electron microscopy. Biochem. Biophys. Res. Commun. 1983, 112, 102–107. [Google Scholar] [CrossRef]
- Panzeter, P.L.; Realini, C.A.; Althaus, F.R. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry 1992, 31, 1379–1385. [Google Scholar] [CrossRef] [PubMed]
- Malanga, M.; Althaus, F.R. Noncovalent protein interaction with poly(ADP-ribose). Methods Mol. Biol. 2011, 780, 67–82. [Google Scholar] [PubMed]
- Li, M.; Yu, X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 2013, 23, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Zaja, R.; Mikoč, A.; Barkauskaite, E.; Ahel, I. Molecular Insights into Poly(ADP-ribose) Recognition and Processing. Biomolecules 2012, 3, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Barkauskaite, E.; Jankevicius, G.; Ladurner, A.G.; Ahel, I.; Timinszky, G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 2013, 280, 3491–3507. [Google Scholar] [CrossRef] [PubMed]
- Daniels, C.M.; Ong, S.E.; Leung, A.K. The Promise of Proteomics for the Study of ADP-Ribosylation. Mol. Cell 2015, 58, 911–924. [Google Scholar] [CrossRef] [PubMed]
- Pleschke, J.M.; Kleczkowska, H.E.; Strohm, M.; Althaus, F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000, 275, 40974–40980. [Google Scholar] [CrossRef] [PubMed]
- Miwa, M.; Ida, C.; Yamashita, S.; Tanaka, M.; Fujisawa, J. Poly(ADP-ribose): Structure, Physicochemical Properties and Quantification In Vivo, with Special Reference to Poly(ADP-ribose) Binding Protein Modules. Curr. Protein Pept. Sci. 2016, 17, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Doly, J.; Mandel, P. Mise en évidence de la biosynthèse in vivo d’un polymère composé, le polyadénosine diphosphoribose dans les noyaux de foie de poulet. C. R. Acad. Sci. 1967, 264, 2687–2690. [Google Scholar]
- Ueda, K.; Omachi, A.; Kawaichi, M.; Hayaishi, O. Natural occurrence of poly(ADP-ribosyl) histones in rat liver. Proc. Natl. Acad. Sci. USA 1975, 72, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Kawaminami, Y.; Miwa, M.; Matsushima, T.; Sugimura, T. Naturally-occurring antibodies to poly(ADP-ribose) in patients with systemic lupus erythematosus. Nature 1977, 265, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Stocken, L.A. Identification of poly (ADP-ribose) covalently bound to histone F1 in vivo. Biochem. Biophys. Res. Commun. 1973, 54, 297–300. [Google Scholar] [CrossRef]
- Stone, P.R.; Bredehorst, R.; Kittler, M.; Lengyel, H.; Hilz, H. Quantitative determination of poly(adenosine diphosphate ribose) in different hepatic tissues by an isotope dilution procedure. Hoppe Seylers Z. Physiol. Chem. 1976, 357, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Bredehorst, R.; Wielckens, K.; Adamietz, P.; Steinhagen-Thiessen, E.; Hilz, H. Mono(ADP-ribosyl)ation and poly(ADP-ribosyl)ation of proteins in developing liver and in hepatomas: Relation of conjugate subfractions to metabolic competence and proliferation rates. Eur. J. Biochem. 1981, 120, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Wielckens, K.; Bredehorst, R.; Adamietz, P.; Hilz, H. Protein-bound polymeric and monomeric ADP-ribose residues in hepatic tissues. Comparative analyses using a new procedure for the quantification of poly(ADP-ribose). Eur. J. Biochem. 1981, 117, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Miwa, M.; Matsushima, T.; Sugimura, T. Anti-poly(ADP-ribose) antibody and its specificity. J. Biochem. 1975, 77, 5–6. [Google Scholar] [CrossRef]
- Kidwell, W.R.; Mage, M.G. Changes in poly(adenosine diphosphate-ribose) and poly(adenosine diphosphate-ribose) polymerase in synchronous HeLa cells. Biochemistry 1976, 15, 1213–1217. [Google Scholar] [CrossRef] [PubMed]
- Martello, R.; Mangerich, A.; Sass, S.; Dedon, P.C.; Bürkle, A. Quantification of cellular poly(ADP-ribosyl)ation by stable isotope dilution mass spectrometry reveals tissue- and drug-dependent stress response dynamics. ACS Chem. Biol. 2013, 8, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Ida, C.; Yamashita, S.; Tsukada, M.; Sato, T.; Eguchi, T.; Tanaka, M.; Ogata, S.; Fujii, T.; Nishi, Y.; Ikegami, S.; et al. An enzyme-linked immunosorbent assay-based system for determining the physiological level of poly(ADP-ribose) in cultured cells. Anal. Biochem. 2016, 494, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Sakura, H.; Miwa, M.; Tanaka, M.; Kanai, Y.; Shimada, T.; Matsushima, T.; Sugimura, T. Natural occurence of a biopolymer, poly (adenosine diphosphate ribose). Nucleic Acids Res. 1977, 4, 2903–2915. [Google Scholar] [CrossRef] [PubMed]
- Juarez-Salinas, H.; Sims, J.L.; Jacobson, M.K. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature 1979, 282, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Juarez-Salinas, H.; Levi, V.; Jacobson, E.L.; Jacobson, M.K. Poly(ADP-ribose) has a branched structure in vivo. J. Biol. Chem. 1982, 257, 607–609. [Google Scholar] [PubMed]
- Jacobson, E.L.; Antol, K.M.; Juarez-Salinas, H.; Jacobson, M.K. Poly(ADP-ribose) metabolism in ultraviolet irradiated human fibroblasts. J. Biol. Chem. 1983, 258, 103–107. [Google Scholar] [PubMed]
- Juarez-Salinas, H.; Duran-Torres, G.; Jacobson, M.K. Alteration of poly(ADP-ribose) metabolism by hyperthermia. Biochem. Biophys. Res. Commun. 1984, 122, 1381–1388. [Google Scholar] [CrossRef]
- Shah, G.M.; Poirier, D.; Duchaine, C.G.; Desnoyers, S.; Lagueux, J.; Verreault, A.; Hoflack, J.C.; Kirkland, J.B.; Poirier, G.G. Methods for biochemical study of poly(ADP-ribose) metabolism in vitro and in vivo. Anal. Biochem. 1995, 227, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Malanga, M.; Bachmann, S.; Panzeter, P.L.; Zweifel, B.; Althaus, F.R. Poly(ADP-ribose) quantification at the femtomole level in mammalian cells. Anal. Biochem. 1995, 228, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Kinders, R.J.; Hollingshead, M.; Khin, S.; Rubinstein, L.; Tomaszewski, J.E.; Doroshow, J.H.; Parchment, R.E. National Cancer Institute Phase 0 Clinical Trials Team. Preclinical modeling of a phase 0 clinical trial: Qualification of a pharmacodynamic assay of poly (ADP-ribose) polymerase in tumor biopsies of mouse xenografts. Clin. Cancer Res. 2008, 14, 6877–6885. [Google Scholar] [CrossRef] [PubMed]
- Kawamitsu, H.; Hoshino, H.; Okada, H.; Miwa, M.; Momoi, H.; Sugimura, T. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry 1984, 23, 3771–3777. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Tanaka, M.; Sato, T.; Ida, C.; Ohta, N.; Hamada, T.; Uetsuki, T.; Nishi, Y.; Moss, J.; Miwa, M. Effect of mild temperature shift on poly(ADP-ribose) and γH2AX levels in cultured cells. Biochem. Biophys. Res. Commun. 2016, 476, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.D.; Gagné, J.P.; Poirier, G.G.; Goodlett, D.R. Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2013, 12, 1868–1880. [Google Scholar] [CrossRef] [PubMed]
- Malanga, M.; Althaus, F.R. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 2005, 83, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Gagné, J.P.; Isabelle, M.; Lo, K.S.; Bourassa, S.; Hendzel, M.J.; Dawson, V.L.; Dawson, T.M.; Poirier, G.G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008, 36, 6959–6976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.J.; Szabo, C. Therapeutic applications of PARP inhibitors: Anticancer therapy and beyond. Mol. Asp. Med. 2013, 34, 1217–1256. [Google Scholar] [CrossRef] [PubMed]
- Kummar, S.; Ji, J.; Morgan, R.; Lenz, H.J.; Puhalla, S.L.; Belani, C.P.; Gandara, D.R.; Allen, D.; Kiesel, B.; Beumer, J.H.; et al. A phase I study of veliparib in combination with metronomic cyclophosphamide in adults with refractory solid tumors and lymphomas. Clin. Cancer Res. 2012, 18, 1726–1734. [Google Scholar] [CrossRef] [PubMed]
- Kummar, S.; Wade, J.L.; Oza, A.M.; Sullivan, D.; Chen, A.P.; Gandara, D.R.; Ji, J.; Kinders, R.J.; Wang, L.; Allen, D.; et al. Randomized phase II trial of cyclophosphamide and the oral poly (ADP-ribose) polymerase inhibitor veliparib in patients with recurrent, advanced triple-negative breast cancer. Investig. New Drugs 2016, 34, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Drew, Y.; Ledermann, J.; Hall, G.; Rea, D.; Glasspool, R.; Highley, M.; Jayson, G.; Sludden, J.; Murray, J.; Jamieson, D.; et al. Phase 2 multicentre trial investigating intermittent and continuous dosing schedules of the poly(ADP-ribose) polymerase inhibitor rucaparib in germline BRCA mutation carriers with advanced ovarian and breast cancer. Br. J. Cancer 2016, 114, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Michels, J.; Vitale, I.; Galluzzi, L.; Adam, J.; Olaussen, K.A.; Kepp, O.; Senovilla, L.; Talhaoui, I.; Guegan, J.; Enot, D.P.; et al. Cisplatin resistance associated with PARP hyperactivation. Cancer Res. 2013, 73, 2271–2280. [Google Scholar] [CrossRef] [PubMed]
- Berger, N.A.; Besson, V.C.; Boulares, A.H.; Bürkle, A.; Chiarugi, A.; Clark, R.S.; Curtin, N.J.; Cuzzocrea, S.; Dawson, T.M.; Dawson, V.L.; et al. Opportunities for the repurposing of PARP inhibitors for the therapy of non-oncological diseases. Br. J. Pharmacol. 2018, 175, 192–222. [Google Scholar] [CrossRef] [PubMed]

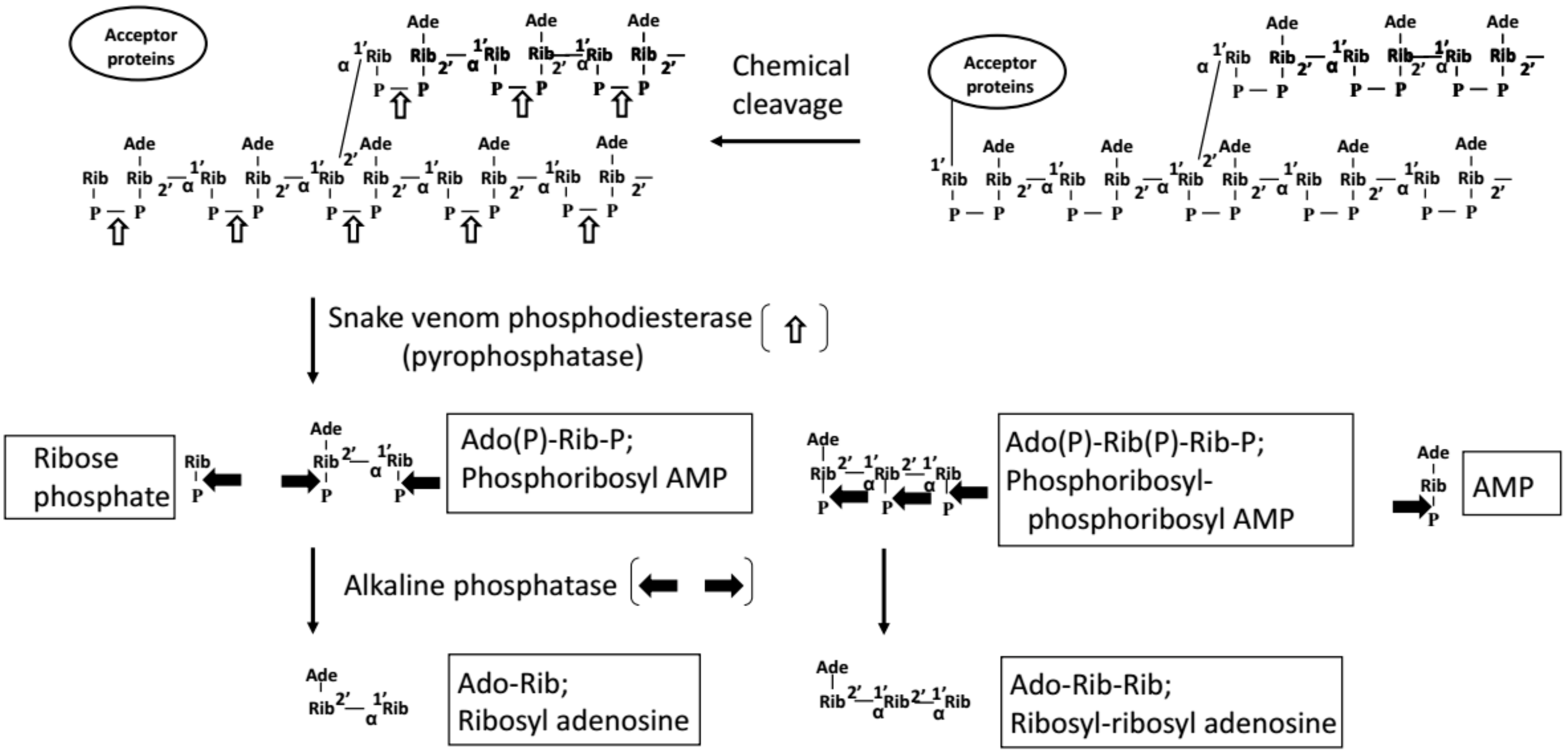
| Name of Methods | Target | Reagents | Tissues or Cells | Fixation | Extraction, Purification | Sensitivity * | Level of PAR * | Comments by Authors | References |
|---|---|---|---|---|---|---|---|---|---|
| Chemical analysis | Histone F1 | Rat liver nuclei | Histone F1 extracted with 5% HClO4 and precipitated with 20% TCA | DEAE-cellulose column chromatography to separate from non-histone proteins | 0.89–1.73 nmol/mg F1 histone protein | Smith and Stocken (1973) [66] | |||
| Isotope dilution | Ado(P)-Rib-P | [3H]PAR as indicator, prepared using sonicated Ehrlich carcinoma cell lysate as enzyme source | Rat liver (adult and neonatal), Zajdela hepatoma | Freeze-clamped (rat liver) | Homogenized in 0.25 N KOH, sonicated, 1 M NH2OH; venom PDE; anion exchange column chromatography; paper chromatography; phosphatase; paper chromatography | 5.59 (adult), 6.32 (neonatal), 1.24 (hepatoma) nmol/mg DNA | Stone et al. (1976) [67] | ||
| RIA | PAR | [14C]PAR, polyclonal antibody against PAR | HeLa cells | 0.1 N NaOH, 2 h, 37 °C; 4 °C overnight | DNase I, micrococcal nuclease; Pronase; phenol extraction | 3.7 pmol/106 HeLa cells; small peak of PAR at S phase and large peak at G2 phase | Kidwell and Mage (1976) [71] | ||
| RIA | PAR | [14C]PAR, polyclonal antibody against PAR | Calf tissues | Frozen after slaughterhouse | Homogenized to isolate nuclei and ethanol precipitation; or homogenized in 5% TCA without isolating nuclei; Pronase; phenol extraction; DNase I, RNase, nuclease P1; Pronase; phenol extraction; hydroxyapatite column chromatography | 10 pmol of PAR | 37 pmol/mg DNA or 740 pmol/g calf thymus | Chain length distribution shown with hydroxyapatite column chromatography | Sakura et al. (1977) [74] |
| Fluorescence method | Etheno Ado-Rib | Fluorescent labeling of Ado-Rib | SV40-transformed 3T3 cells | 20% TCA | 0.1 M potassium phosphate (pH 5) in 6 M guanidine hydrochloride, sonication; dihydroxyboryl-Sepharose column chromatography; alkaline phosphatase and venom PDE; chloroacetaldehyde; HPLC | 5 pmol of Ado-Rib | 0.05 pmol/106 SV40 virus-transformed 3T3 cells, increased to 7.50 pmol/106 cells after 20 min of MNNG | Juarez-Salinas et al. (1979) [75] | |
| RIA: Method A | Ado(P)-Rib-P | [3H]PAR as tracer prepared using Ehrlich carcinoma cell nuclei, [3H]Ado(P)-Rib-P; antibody against Ado(P)-Rib-P | Rat liver (adult) | Freeze-clamped and frozen | Homogenized in 20% TCA; dissolved in 6 M guanidine hydrochloride and morpholine buffer; 0.3 M NaOH, 1 h, 56 °C; boronate column chromatography; venom PDE; HPLC | 1 pmol Ado(P)-Rib-P | 88.6 ± 20.7 and 72.6 pmol/g tissue | Wielkens et al. (1981) [69] | |
| RIA: Method B | Ado(P)-Rib-P | Same as above | Same as above | Same as above | Homogenized in 20% TCA; dissolved in cold water; 0.33 M NaOH, 3 h, 56 °C; alkaline phosphatase; proteinase K; heat inactivation; venom PDE; 5% TCA; anion exchange column chromatography, HPLC | 1 pmol Ado(P)-Rib-P | 89.0 ± 10.7 and 128.1 ± 7.3 pmol/g tissue | Wielkens et al. (1981) [69] | |
| RIA | Ado(P)-Rib-P | RIA Method A (Wielkens et al. 1981) [69] | Rat liver (adult and neonatal) and hepatoma | Freeze-clamped and frozen (rat liver) or washed and frozen in liquid nitrogen (hepatoma cells) | RIA Method A (Wielkens et al., 1981) [69] | 1 pmol Ado(P)-Rib-P | 32 ± 9, 10 ± 2, 39 ± 5 pmol/mg DNA of neonatal (1 day), neonatal (17 days) and adult (>150 days) rat liver, respectively. 61 ± 11, 25 ± 3 and 60 ± 17 pmol/mg DNA for AH130, AH 7974 and Reuber H35 hepatomas, respectively | Bredehorst et al. (1981) [68] | |
| Fluorescence method | Etheno Ado-Rib and etheno Ado-Rib-Rib | Fluorescent labeling of specific degradation product, Ado-Rib and Ado-Rib-Rib | Rat liver, kidney and spleen | 20% TCA | Juarez-Salinas et al. (1979) [75] | 14, 18, and 8.0 pmol of Ado-Rib/mg DNA of liver, kidney and spleen of rat, respectively; Ado-Rib-Rib constituted 0.8-1.6% of total PAR | Juarez-Salinas et al. (1982) [76] | ||
| Fluorescence method | Etheno Ado-Rib | Fluorescent labeling of specific degradation product, Ado-Rib | Normal human diploid fibroblasts (CF-3) | 20% TCA | MOPS/KOH (pH 8.8) in 6 M guanidine hydrochloride, sonication; dihydroxyboryl BioRex 70 resin; alkaline phosphatase and venom PDE; fluorescent etheno derivatization; dihydroxyboronate column chromatography; HPLC | 0.12 pmol Ado-Rib/106 cells; increased to 2.0 pmol/106 cells after UV | Jacobson et al. (1983) [77] | ||
| Fluorescence method | Etheno Ado-Rib | Fluorescent labeling of Ado-Rib | SV40-transformed Balb/3T3 fibroblasts | 20% TCA | Jacobson et al. (1983) [77] | 0.09 or 0.14 pmol Ado-Rib/106 cells: increased by hyperthermia at 43 °C up to 5.25 pmol/106 cells after 8 h | No significant DNA strand breaks upon alkaline sucrose gradient centrifugation | Juarez-Salinas et al. (1984) [78] | |
| PAGE, silver staining and computer-aided scanning densitometry | PAR | PAR standard | Human keratinocyte cell line (HaKaT) | 20% TCA | Proteinase K (DNase I, RNase); 1 M KOH, 37 °C 2 h; dihydroxyboronate column chromatography; 20% PAGE; silver staining; scanned with computing densitometer (ImageQuant 3.15 software) | 0.55 pmol/106 cells; MNNG treatment increased to 11.1 pmol/106 cells | Chain length distribution shown with PAGE | Malanga et al. (1995) [80] | |
| Sandwich ELISA | Protein-bound PAR | PAR standard; mouse monoclonal (10H) and rabbit polyclonal antibody against PAR | Xenograft tumors of human melanoma cell lines (A375 and Colo829) | Fresh frozen | Sonicated in lysis buffer (Biosource) with protease inhibitors; 1% SDS; boiled 5 min; supernatant | 5584 units PAR (large tumors); 4146 units PAR (small tumors) [All units are pg PAR/mL per 100 mg protein]; Higher amount of protein and DNA caused interference | Protein and DNA were not digested. Protein concentration should be diluted to 0.1–1 mg/mL before assay. Effect of dose of PARP inhibitor and time course of PAR levels in xenograft were presented | Kinders et al. (2008) [81] | |
| Sandwich ELISA | Protein-bound PAR | PAR standard; mouse monoclonal (10H) and rabbit polyclonal antibody against PAR | Human PBMC | Fresh frozen | Suspended in cell extraction buffer (Invitrogen) with protease inhibitors; 30 min on ice; 1% SDS; boiled 5 min; supernatant | 0.024 pmol/106 PBMC (healthy volunteer); 0.028 pmol/106 PBMC (patients with cancer); 0.008-0.198 pmol/106 PBMC (intra- and inter-individual variation over 3 weeks in healthy volunteers) | Protein and DNA were not digested | Ji et al. (2011) [46] | |
| Stable isotope dilution LC-MS/MS | PAR | 13C,15N-labeled PAR | PBMC, COPFS cells, HeLa cells, mouse tissues | 20% TCA | 0.5 M KOH, 37 °C, 45 min; 13C, 15N labelled PAR added; DNase I, RNase A; proteinase K; solid phase extraction of PAR; venom PDE, alkaline phosphatase; HPLC; mass spectrometry | 50 fmol of Ado-Rib | 0.03 pmol/106 PBMC: 0.05-0.1 pmol/106 HeLa cells | Considerable heterogeneity in the stress-induced PARylation response in human population | Martello et al. (2013) [72] |
| Sandwich ELISA | PAR | PAR standard; mouse monoclonal (10H) and rabbit polyclonal antibody against PAR | HeLa cells; HEK293T cells; HepG2 cells | 20% TCA | Sonication; 0.1 N NaOH, 37 °C 1 h; DNase I, RNase A, Nuclease P1, Proteinase K; | 15 fmol of PAR | 0.039 pmol/106 HeLa cells; 0.038 pmol/106 HEK293T cells; 0.002 pmol/106 HepG2 cells; increased to 6.49 pmol/106 HeLa cells after MNNG. | Big difference of PAR level depending the isolation procedures and cell types | Ida et al. (2016) [73] |
| Sandwich ELISA | PAR | PAR standard; mouse monoclonal (10H) and rabbit polyclonal antibody against PAR | HeLa cells, CHO-K1 cells | 20% TCA | Ida et al. (2016) [73] | 15 fmol of PAR | 0.025 and 0.074 pmol/106 HeLa cells at 37 °C; 0.023 pmol/106 cells at 33.5 °C; 0.085 and 0.155 pmol/106 cells at 40.5 °C; 0.063 pmol/106 CHO-K1 cells at 37 °C; 0.128 pmol/106 cells at 40.5 °C | Phosphorylation of histone H2AX was found at 40.5 °C and further increased by PARP inhibitor. | Yamashita et al. (2016) [83] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miwa, M.; Ida, C.; Yamashita, S.; Kouyama, K.; Kuroda, Y.; Eguchi, T.; Ohta, N.; Sato, T.; Tsuda, M.; Tanaka, M. In Vivo Level of Poly(ADP-ribose). Challenges 2018, 9, 23. https://doi.org/10.3390/challe9010023
Miwa M, Ida C, Yamashita S, Kouyama K, Kuroda Y, Eguchi T, Ohta N, Sato T, Tsuda M, Tanaka M. In Vivo Level of Poly(ADP-ribose). Challenges. 2018; 9(1):23. https://doi.org/10.3390/challe9010023
Chicago/Turabian StyleMiwa, Masanao, Chieri Ida, Sachiko Yamashita, Kenichi Kouyama, Yasuhito Kuroda, Takayuki Eguchi, Narumi Ohta, Teruaki Sato, Masataka Tsuda, and Masakazu Tanaka. 2018. "In Vivo Level of Poly(ADP-ribose)" Challenges 9, no. 1: 23. https://doi.org/10.3390/challe9010023





