Application of an Unstructured Grid-Based Water Quality Model to Chesapeake Bay and Its Adjacent Coastal Ocean
Abstract
:1. Introduction
2. Material and Methods
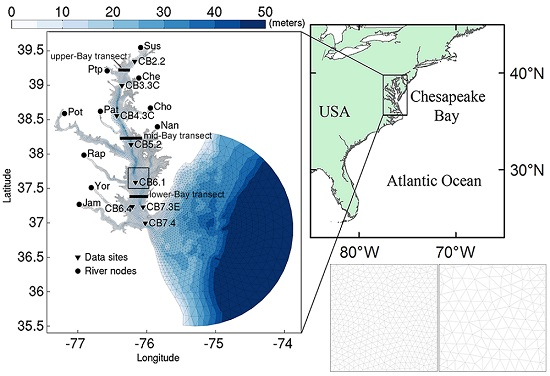
2.1. Study Site
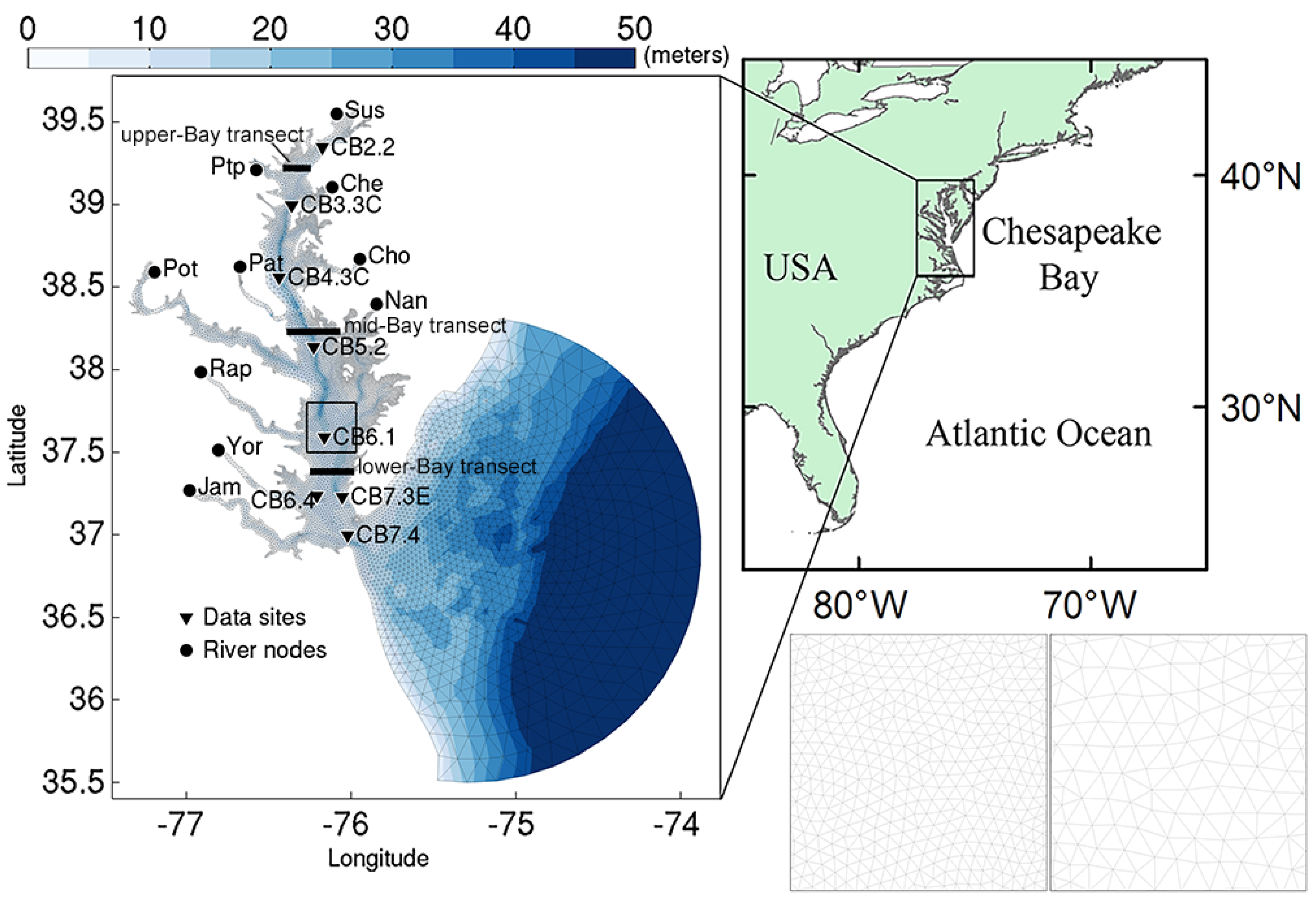
2.2. Model Description
2.3. Model Settings
2.4. Design of Numerical Experiments
3. Sensitivity Experiments
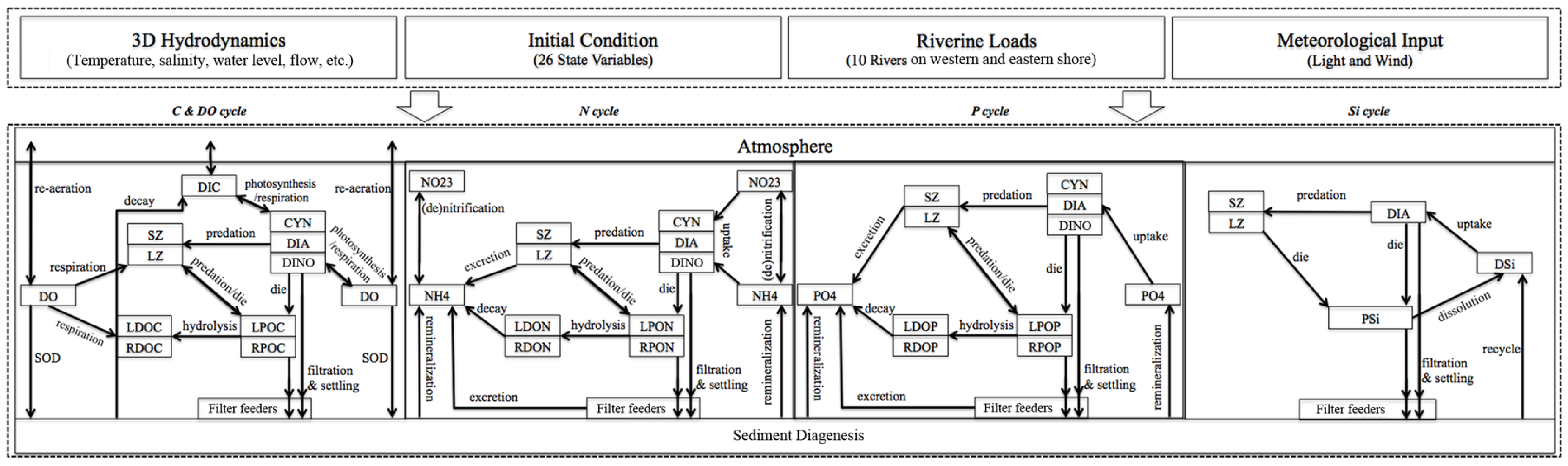
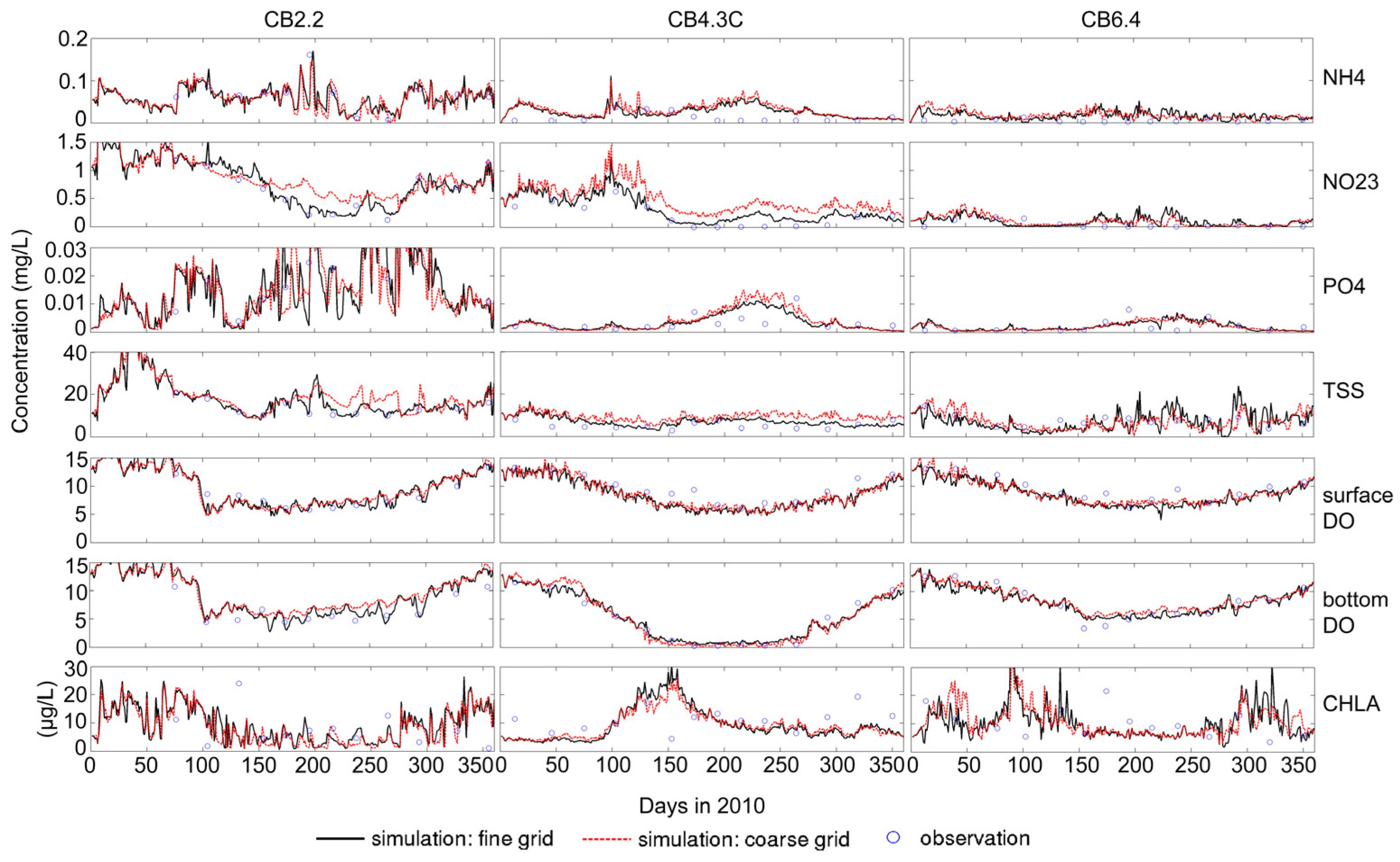
3.1. Sensitivity of Main Water Quality Variables to Grid Resolution
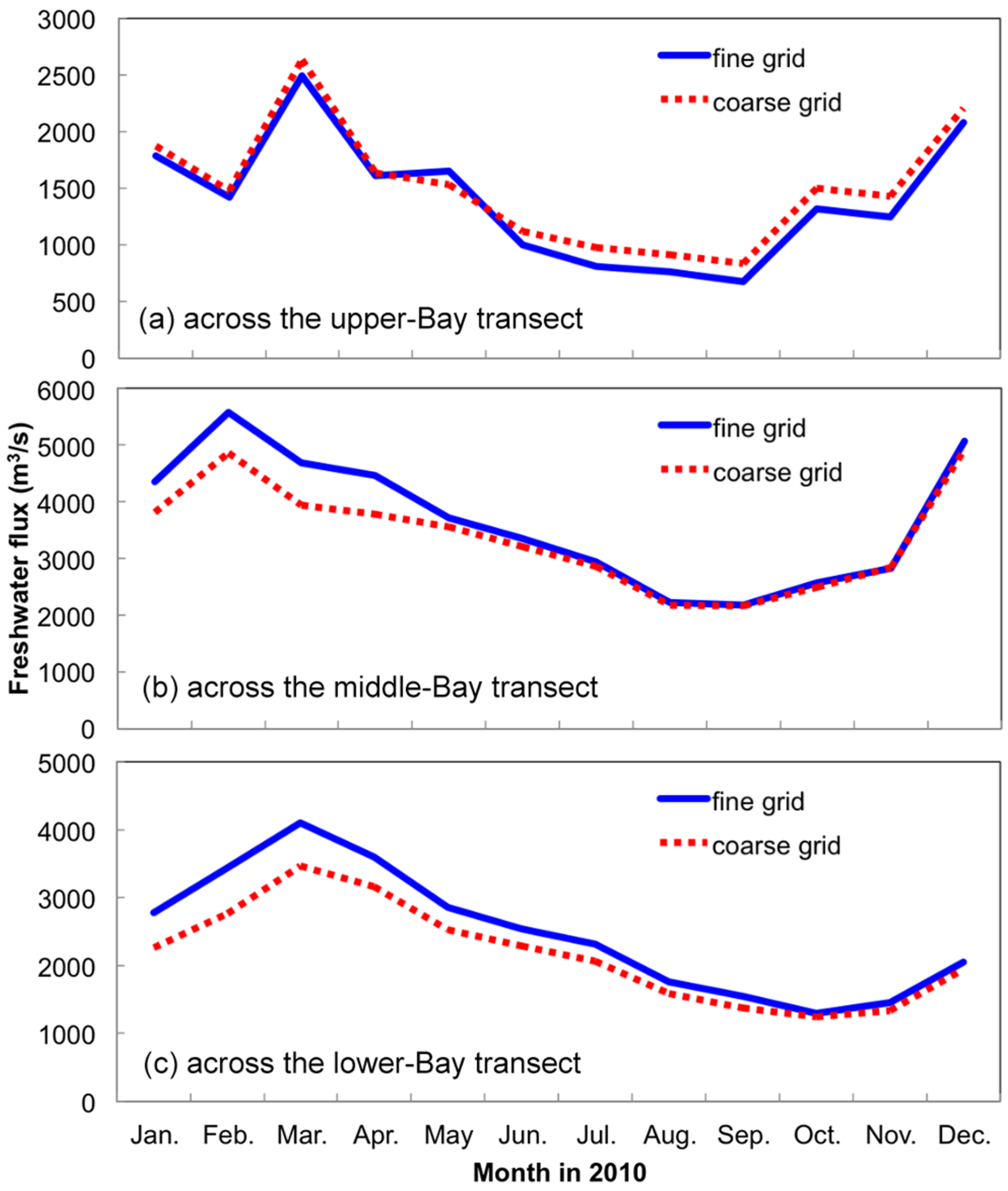
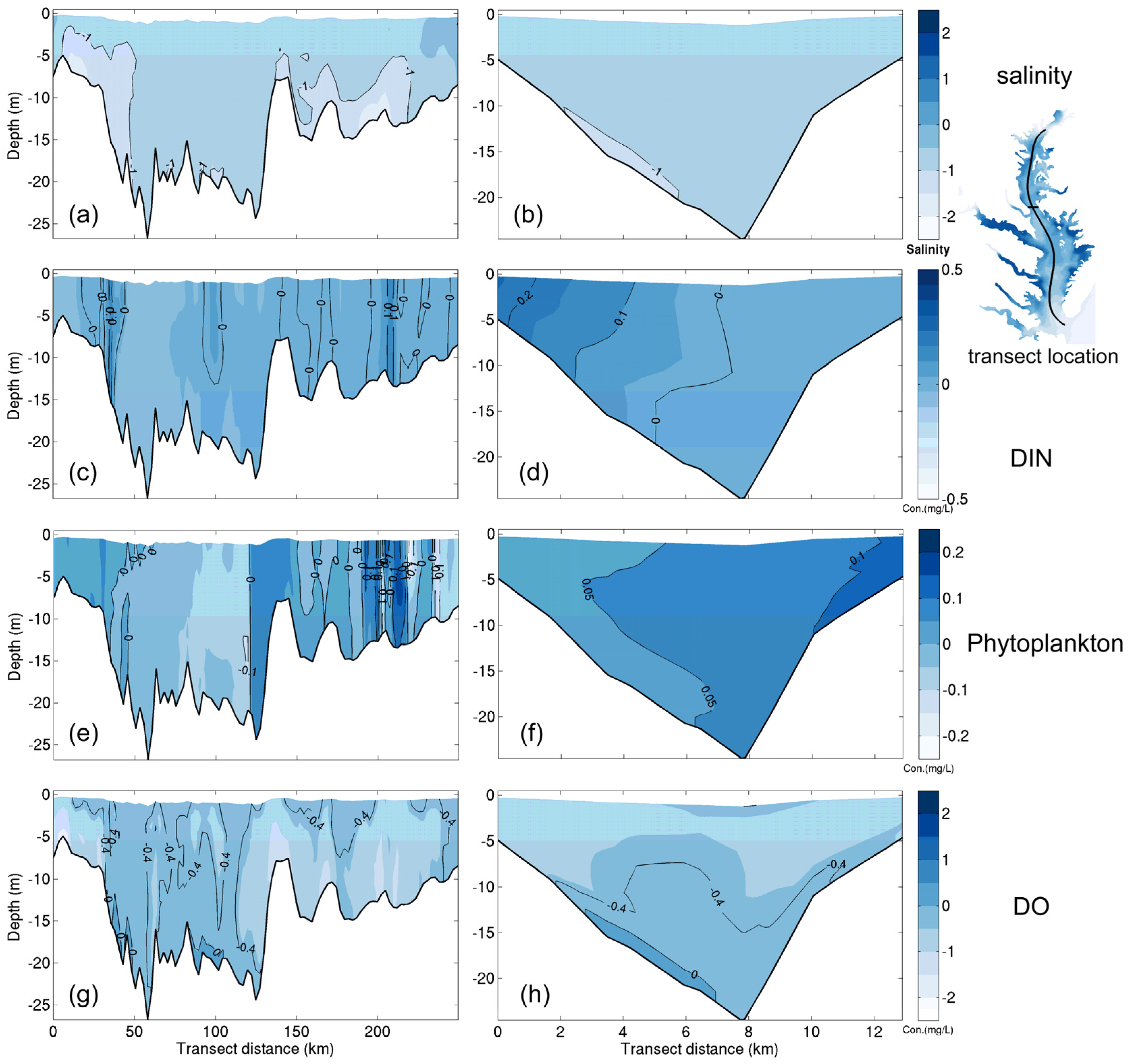
3.1.1. Effect of Horizontal Resolution
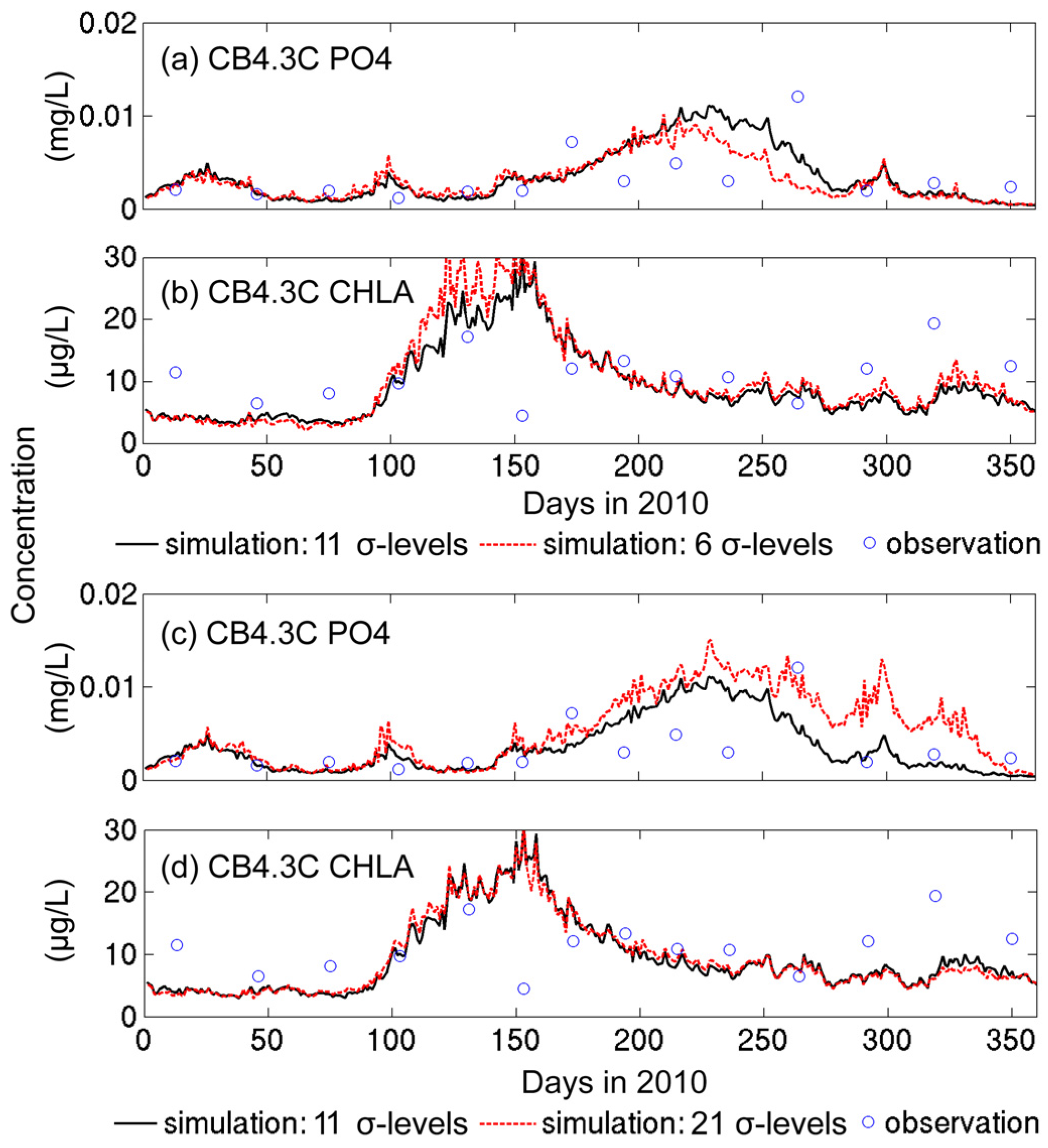
3.1.2. Sensitivity to Vertical Resolution
3.2. Uncertainties Associated with the Hydrodynamic Simulation
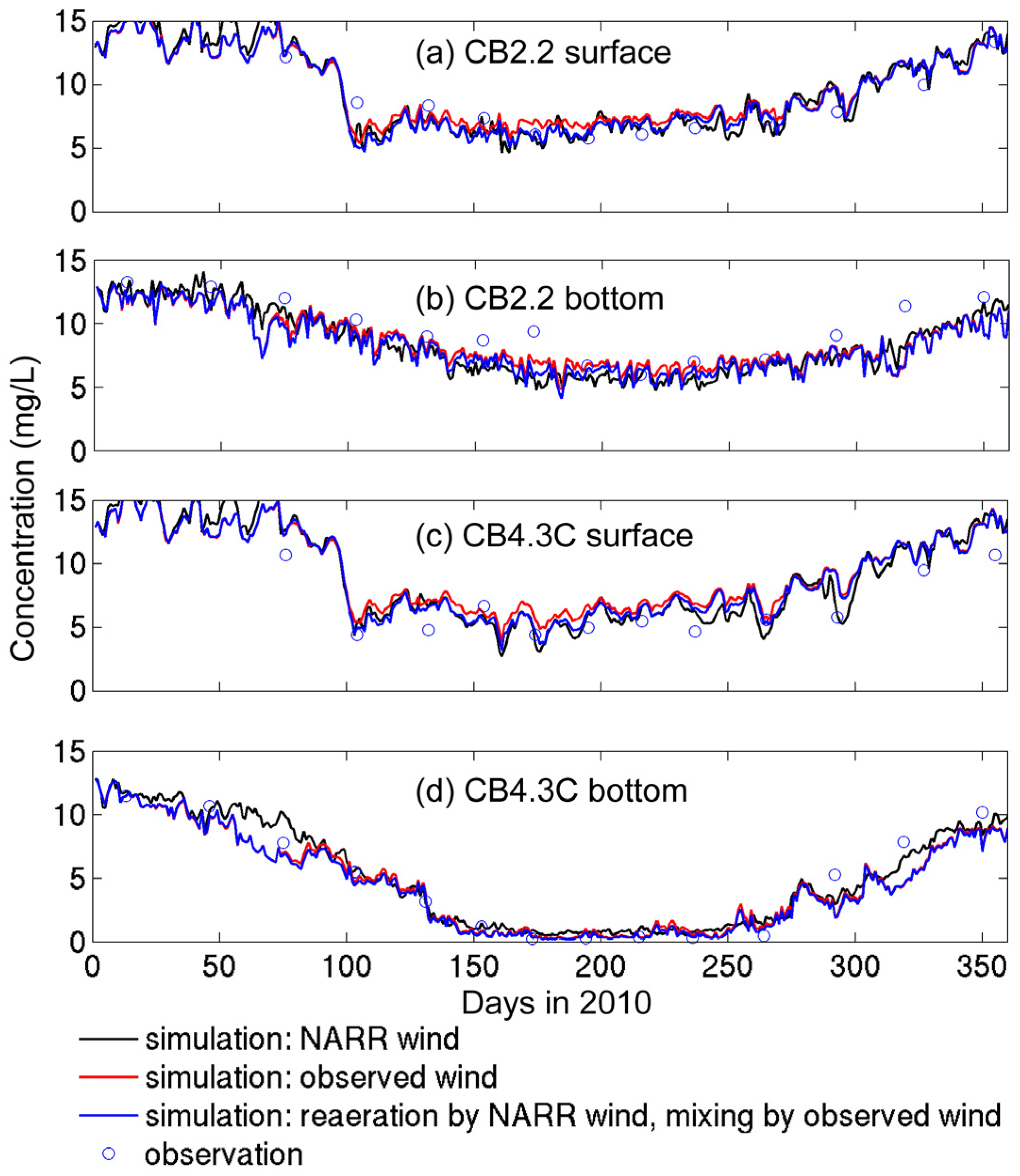
3.2.1. Sensitivity to Different Wind Datasets
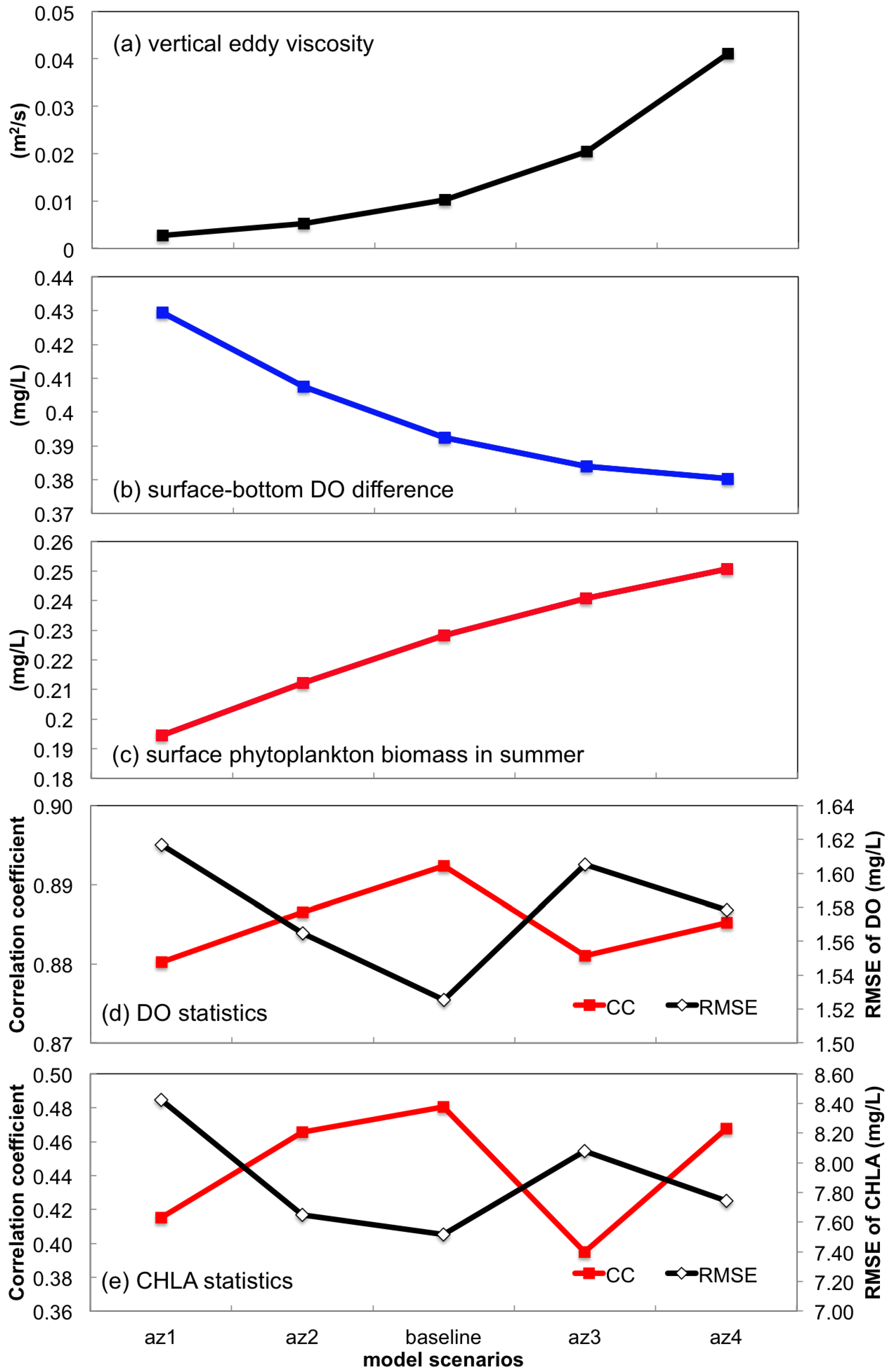
3.2.2. Sensitivity to Vertical Eddy Viscosity
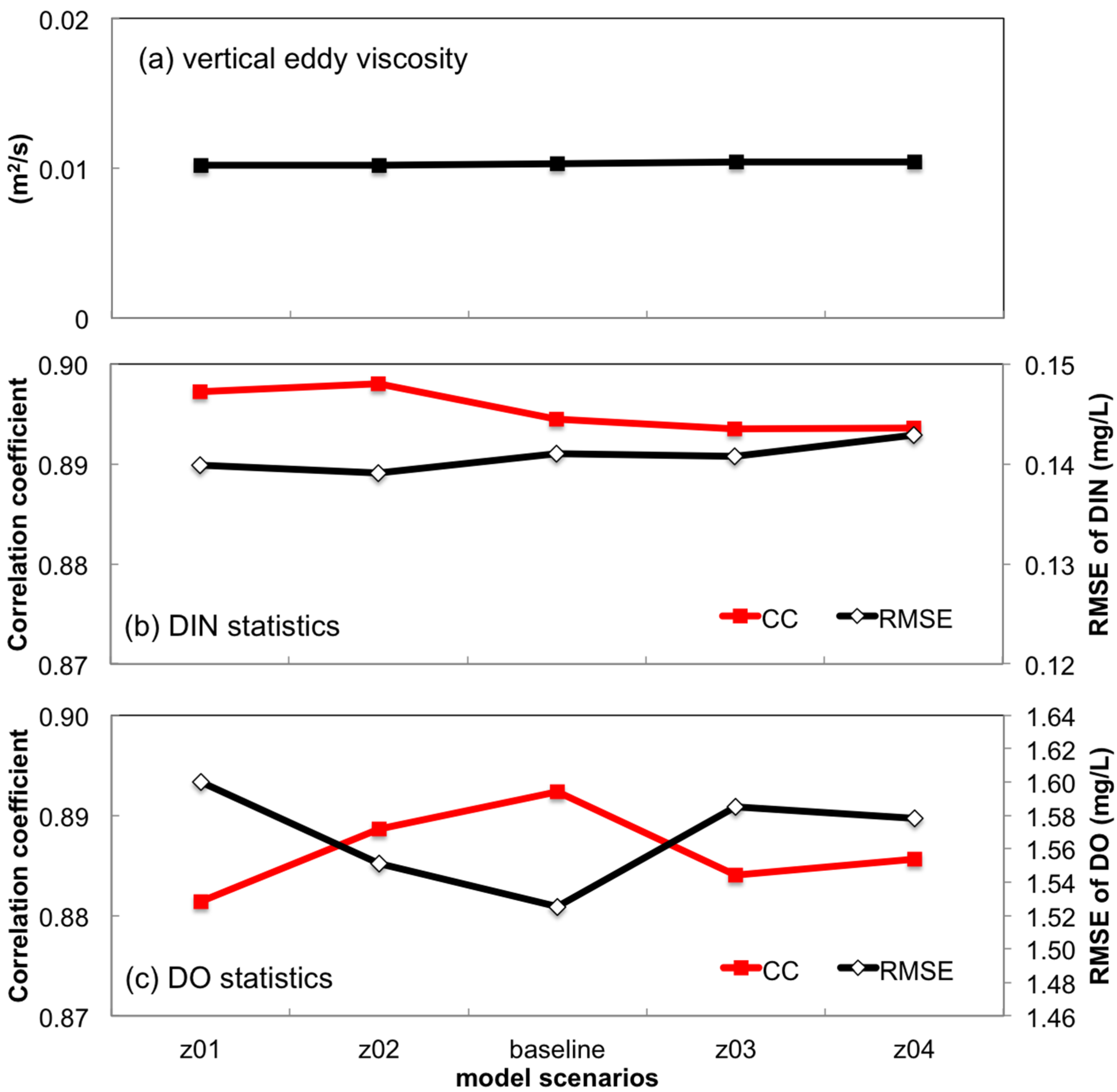
3.2.3. Sensitivity to Bottom Roughness Length Scale
3.3. Uncertainties Associated with the Model Inputs
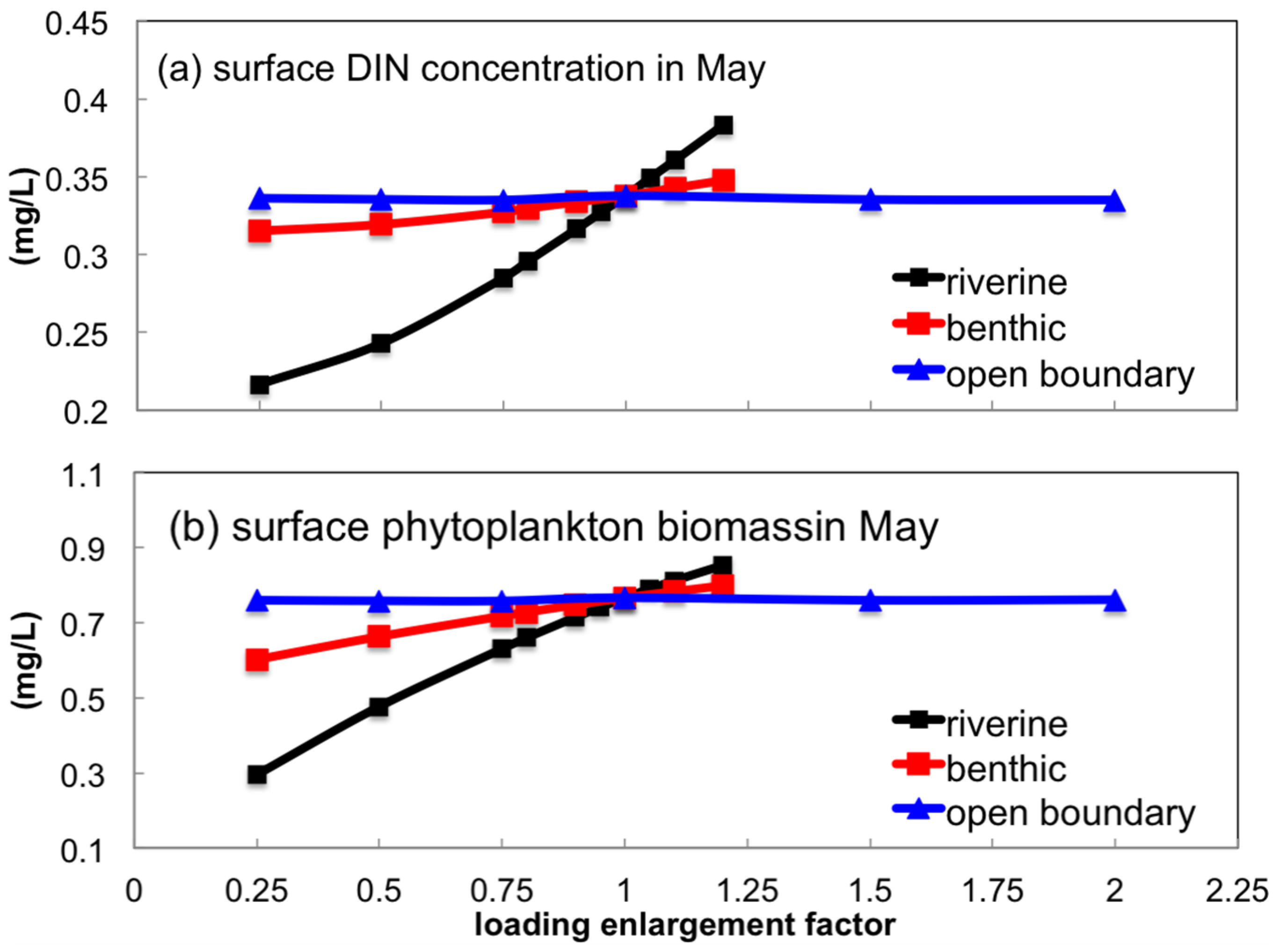
3.3.1. Sensitivity to Boundary Nutrient Loading
3.3.2. Sensitivity of Predation Terms to Phytoplankton Simulation
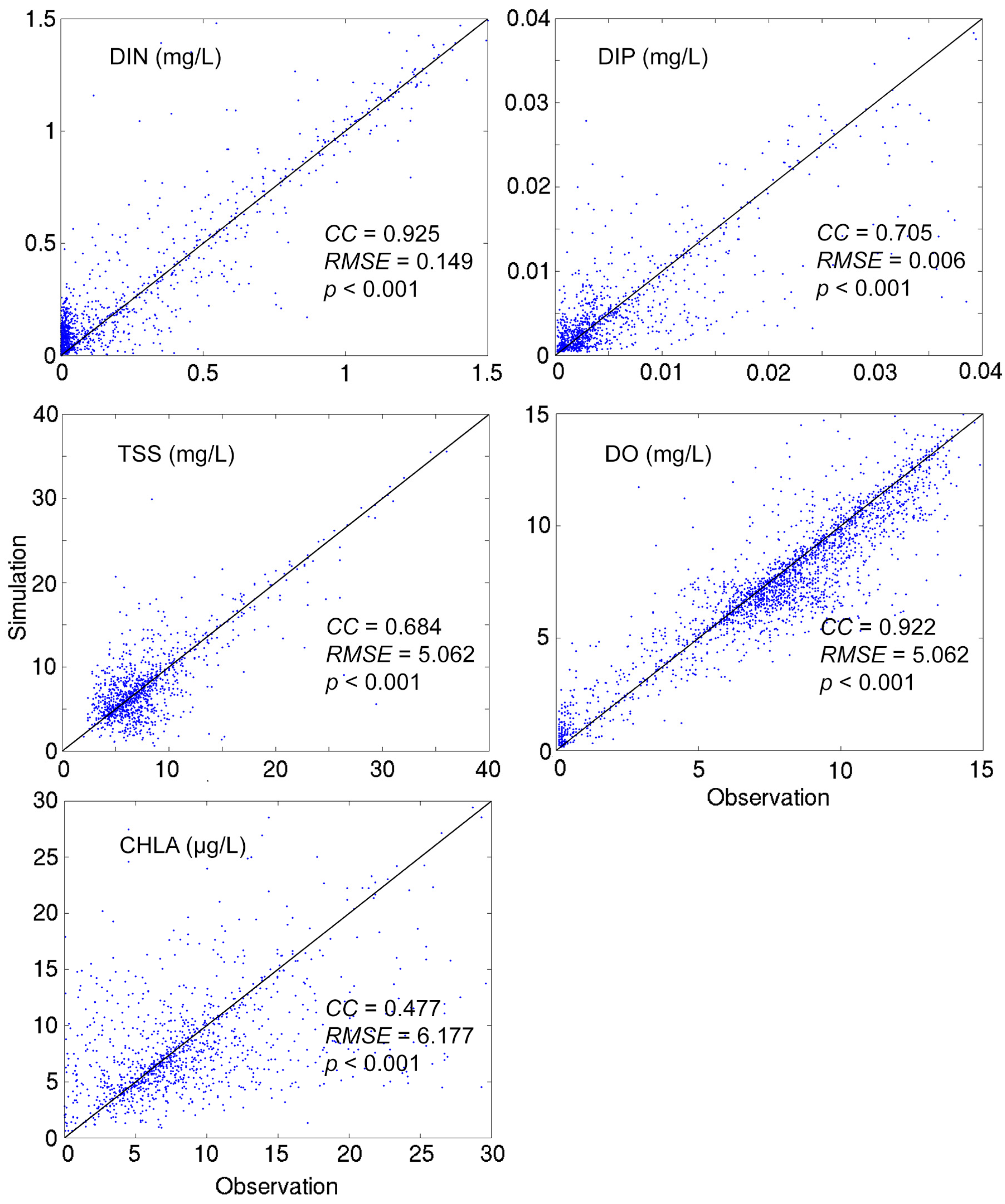
4. Model Calibration and Validation
4.1. A 10-Year (2003–2012) Model Simulation
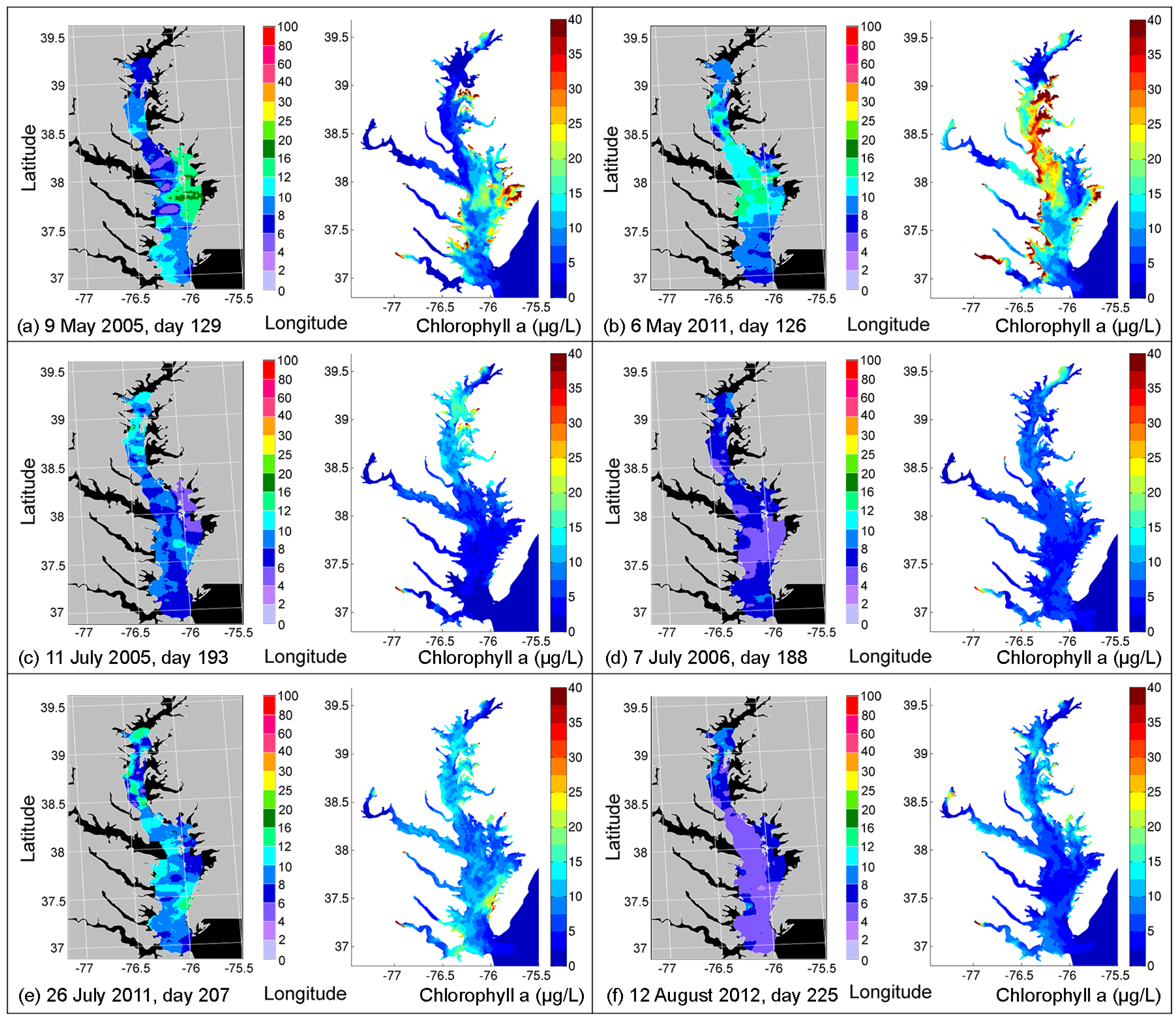
4.2. Comparison of Simulated CHLA with Remote Sensing Images
5. Conclusions
- (1)
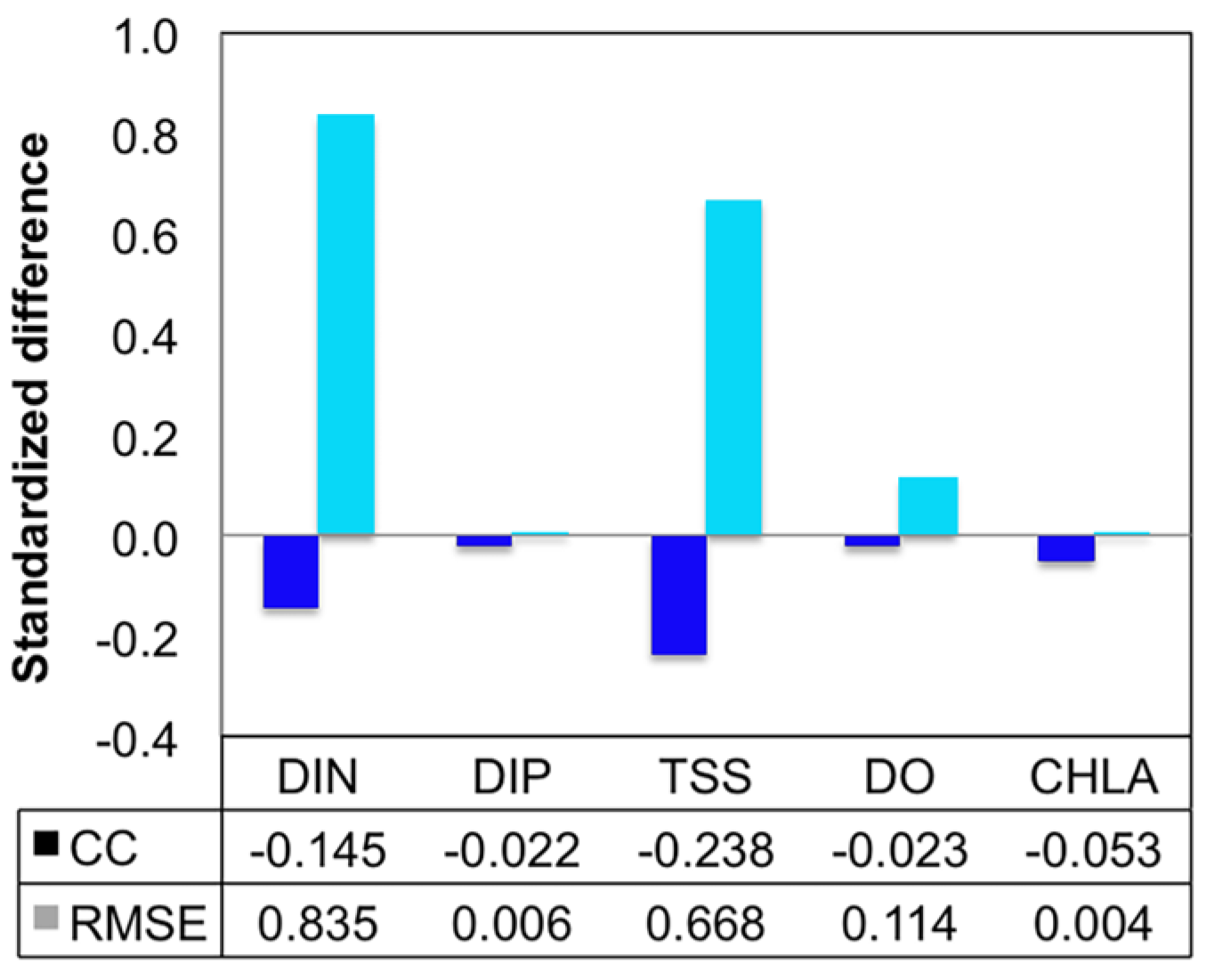
- Grid refinement improved the model performance of most variables, particularly for DIN and TSS. The finer grid favored modeling the realistic material transport from the Susquehanna River to the bay mouth, especially in the mid-bay portion. Eleven sigma levels were applied in consideration of the balance between computational accuracy and efficiency. The unstructured grid-based water quality models made it feasible to reach high resolution in biologically-active regions (e.g., littoral zones and the main channel) without significantly adding to the overall computational burden.
- (2)
- The effects of wind source on DO simulation were compared between the NARR modeled and observed winds. Both winds represented directional asymmetry in summer (southerly winds strongest and westerly weakest), while the observed winds showed lower frequency in southerly winds. The DO simulation forced by these two wind sources had good agreement with empirical data, except that the surface DO was overestimated under the observed winds. Due to stronger mixing from the more frequent southerly winds, the NARR winds were preferred in our water quality model.
- (3)
- Turbulent mixing and bottom stress were two potential sources of uncertainties in water quality simulation. Appropriate representation of vertical eddy viscosity was propitious to model the vertical oxygen ventilation and the new primary production fueled with recycled nutrients. Bottom friction exerted a moderate impact only on the horizontal oxygen mixing. In terms of water quality simulation, the vertical mixing process was more influential than bottom roughness.
- (4)
- Uncertainties in the riverine source could exert a relatively larger influence on the inner-bay nutrient concentration and phytoplankton biomass during the spring bloom than those in benthic nutrient flux and open boundary loading. Zooplankton predation on phytoplankton was more significant than that of the filtration of suspension feeders.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gillmore, J.; Glendening, P.; Ridge, T.; Williams, A.; Browner, C.; Bolling, B. Chesapeake 2000 Agreement; United States Environmental Protection Agency Chesapeake Bay Program: Annapolis, MD, USA, 2000. [Google Scholar]
- Kemp, W.M.; Boynton, W.R.; Adolf, J.E.; Boesch, D.F.; Boicourt, W.C.; Brush, G.; Cornwell, J.C.; Fisher, T.R.; Glibert, P.M.; Hagy, J.D.; et al. Eutrophication of Chesapeake Bay: Historical trends and ecological interactions. Mar. Ecol. Prog. Ser. 2005, 303, 1–29. [Google Scholar] [CrossRef]
- Hagy, J.D.; Boynton, W.R.; Keefe, C.W.; Wood, K.V. Hypoxia in Chesapeake Bay, 1950–2001: Long-term change in relation to nutrient loading and river flow. Estuaries 2004, 27, 634–658. [Google Scholar] [CrossRef]
- Murphy, R.R.; Kemp, W.M.; Ball, W.P. Long-term trends in Chesapeake Bay seasonal hypoxia, stratification, and nutrient loading. Estuar. Coasts 2011, 34, 1293–1309. [Google Scholar] [CrossRef]
- Harding, L.W.; Mallonee, M.E.; Perry, E.S. Toward a predictive understanding of primary productivity in a temperate, partially stratified estuary. Estuar. Coast. Shelf Sci. 2002, 55, 437–463. [Google Scholar] [CrossRef]
- Marshall, H.G.; Lane, M.F.; Nesius, K.K.; Burchardt, L. Assessment and significance of phytoplankton species composition within Chesapeake Bay and Virginia tributaries through a long-term monitoring program. Environ. Monit. Assess. 2009, 150, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.M.; Kemp, W.M. Seasonal and regional variations in plankton community production and respiration for Chesapeake Bay. Mar. Ecol. Prog. Ser. 1995, 116, 217–231. [Google Scholar] [CrossRef]
- Harding, L.W. Long-term trends in the distribution of phytoplankton in Chesapeake Bay: Roles of light, nutrients and streamflow. Mar. Ecol. Prog. Ser. 1994, 104, 267. [Google Scholar] [CrossRef]
- Prasad, M.B.K.; Sapiano, M.R.; Anderson, C.R.; Long, W.; Murtugudde, R. Long-term variability of nutrients and chlorophyll in the Chesapeake Bay: A retrospective analysis, 1985–2008. Estuar. Coasts 2010, 33, 1128–1143. [Google Scholar] [CrossRef]
- Son, S.; Wang, M.; Harding, L.W. Satellite-measured net primary production in the Chesapeake Bay. Remote Sens. Environ. 2014, 144, 109–119. [Google Scholar] [CrossRef]
- Cerco, C.F.; Kim, C.-S.; Noel, M.R. The 2010 Chesapeake Bay Eutrophication Model; U.S. Army Corps of Engineers Waterways Experiment Station: Vicksburg, MS, USA, 2010. [Google Scholar]
- Feng, Y.; Friedrichs, M.A.; Wilkin, J.; Tian, H.; Yang, Q.; Hofmann, E.E.; Wiggert, J.D.; Hood, R.R. Chesapeake Bay nitrogen fluxes derived from a land-estuarine ocean biogeochemical modeling system: Model description, evaluation and nitrogen budgets. J. Geophys. Res. Biogeosci. 2015, 120, 1666–1695. [Google Scholar] [CrossRef]
- Pennock, J.R. Chlorophyll distributions in the Delaware estuary: Regulation by light-limitation. Estuar. Coast. Shelf Sci. 1985, 21, 711–725. [Google Scholar] [CrossRef]
- Jiang, L.; Xia, M.; Ludsin, S.A.; Rutherford, E.S.; Mason, D.M.; Jarrin, J.M.; Pangle, K.L. Biophysical modeling assessment of the drivers for plankton dynamics in dreissenid-colonized western Lake Erie. Ecol. Model. 2015, 308, 18–33. [Google Scholar] [CrossRef]
- Testa, J.M.; Li, Y.; Lee, Y.J.; Li, M.; Brady, D.C.; di Toro, D.M.; Kemp, W.M.; Fitzpatrick, J.J. Quantifying the effects of nutrient loading on dissolved O2 cycling and hypoxia in Chesapeake Bay using a coupled hydrodynamic-biogeochemical model. J. Mar. Syst. 2014, 139, 139–158. [Google Scholar] [CrossRef]
- Xia, M.; Craig, P.M.; Schaeffer, B.; Stoddard, A.; Liu, Z.; Peng, M.; Zhang, H.; Wallen, C.M.; Bailey, N.; Mandrup-Poulsen, J. Influence of physical forcing on bottom-water dissolved oxygen within Caloosahatchee River Estuary, Florida. J. Environ. Eng. 2010, 136, 1032–1044. [Google Scholar] [CrossRef]
- Xia, M.; Craig, P.M.; Wallen, C.M.; Stoddard, A.; Mandrup-Poulsen, J.; Peng, M.; Schaeffer, B.; Liu, Z. Numerical simulation of salinity and dissolved oxygen at Perdido Bay and adjacent coastal ocean. J. Coast. Res. 2011, 27, 73–86. [Google Scholar] [CrossRef]
- Xia, M.; Jiang, L. Influence of wind and river discharge on the hypoxia in a shallow bay. Ocean Dyn. 2015, 65, 665–678. [Google Scholar] [CrossRef]
- Fitzpatrick, J.J. Assessing skill of estuarine and coastal eutrophication models for water quality managers. J. Mar. Syst. 2009, 76, 195–211. [Google Scholar] [CrossRef]
- Ganju, N.K.; Brush, M.J.; Rashleigh, B.; Aretxabaleta, A.L.; del Barrio, P.; Grear, J.S.; Harris, L.A.; Lake, S.J.; McCardell, G.; O’Donnell, J.; et al. Progress and challenges in coupled hydrodynamic-ecological estuarine modeling. Estuar. Coasts 2015, 39, 311–332. [Google Scholar] [CrossRef]
- Khangaonkar, T.; Sackmann, B.; Long, W.; Mohamedali, T.; Roberts, M. Simulation of annual biogeochemical cycles of nutrient balance, phytoplankton bloom(s), and DO in Puget Sound using an unstructured grid model. Ocean Dyn. 2012, 62, 1353–1379. [Google Scholar] [CrossRef]
- Ganju, N.K.; Sherwood, C.R. Effect of roughness formulation on the performance of a coupled wave, hydrodynamic, and sediment transport model. Ocean Model. 2010, 33, 299–313. [Google Scholar] [CrossRef]
- Irby, I.D.; Friedrichs, M.A.M.; Friedrichs, C.T.; Bever, A.J.; Hood, R.R.; Lanerolle, L.W.J.; Scully, M.E.; Sellner, K.; Shen, J.; Testa, J.; et al. Challenges associated with modeling low-oxygen waters in Chesapeake Bay: A multiple model comparison. Biogeosciences 2016, 13, 2011–2028. [Google Scholar] [CrossRef]
- Kim, T.; Khangaonkar, T. An offline unstructured biogeochemical model (UBM) for complex estuarine and coastal environments. Environ. Model. Softw. 2012, 31, 47–63. [Google Scholar] [CrossRef]
- Shen, J.; Wang, H.V. Determining the age of water and long-term transport timescale of the Chesapeake Bay. Estuar. Coast. Shelf Sci. 2007, 74, 585–598. [Google Scholar] [CrossRef]
- Fisher, T.R.; Harding, L.W.; Stanley, D.W.; Ward, L.G. Phytoplankton, nutrients, and turbidity in the Chesapeake, Delaware, and Hudson estuaries. Estuar. Coast. Shelf Sci. 1988, 27, 61–93. [Google Scholar] [CrossRef]
- Wang, D.-P. Wind-driven circulation in the Chesapeake Bay, winter 1975. J. Phys. Oceanogr. 1979, 9, 564–572. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Chesapeake Bay Total Maximum Daily Load for Nitrogen, Phosphorus, and Sediment; Chesapeake Bay Program Office: Annapolis, MD, USA, 2010.
- Testa, J.M.; Kemp, W.M. Hypoxia-induced shifts in nitrogen and phosphorus cycling in Chesapeake Bay. Limnol. Oceanogr. 2012, 57, 835–850. [Google Scholar] [CrossRef]
- Li, J.; Glibert, P.M.; Gao, Y. Temporal and spatial changes in Chesapeake Bay water quality and relationships to Prorocentrum minimum, Karlodinium veneficum, and CyanoHAB events, 1991–2008. Harmful Algae 2015, 42, 1–14. [Google Scholar] [CrossRef]
- Jiang, L.; Xia, M. Dynamics of the Chesapeake Bay outflow plume: Realistic plume simulation and its seasonal and interannual variability. J. Geophys. Res. Oceans 2016, 121, 1424–1445. [Google Scholar] [CrossRef]
- Di Toro, D.M.; Fitzpatrick, J.J. Chesapeake Bay Sediment Flux Model; HydroQual, Inc.: Mahwah, NJ, USA, 1993. [Google Scholar]
- Meyers, M.B.; di Toro, D.M.; Lowe, S.A. Coupling suspension feeders to the Chesapeake Bay Eutrophication Model. Water Qual. Ecosyst. Model. 2000, 1, 123–140. [Google Scholar] [CrossRef]
- Cerco, C.F.; Cole, T. User’s Guide to the CE-QUAL-ICM Three-Dimensional Eutrophication Model: Release, version 1.0 Report; US Army Engineer Waterways Experiment Station: Vicksburg, MS, USA, 1995. [Google Scholar]
- Malone, T.C.; Crocker, L.H.; Pike, S.E.; Wendler, B.W. Influences of river flow on the dynamics of phytoplankton production in a partially stratified estuary. Mar. Ecol. Prog. Ser. 1988, 48, 235–249. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Chesapeake Bay Program water quality data. Available online: http://www.chesapeakebay.net/data (accessed on 18 August 2016).
- National Atmospheric Deposition Program data. Available online: http://nadp.sws.uiuc.edu/data/ (accessed on 18 August 2016).
- World Ocean Atlas 2005 data. Available online: http://www.nodc.noaa.gov/OC5/WOA05/pr_woa05.html (accessed on 18 August 2016).
- National Center for Environmental Prediction. North America Regional Reanalysis data. Available online: http://www.esrl.noaa.gov/psd/data/gridded/data.narr.html (accessed on 18 August 2016).
- Chesapeake Bay Remote Sensing Program data. Available online: http://www.cbrsp.org/index.html (accessed on 18 August 2016).
- U.S. Geological Survey data. Available online: http://md.water.usgs.gov/waterdata/chesinflow/wy/ (accessed on 18 August 2016).
- National Data Buoy Center data. Available online: http://www.ndbc.noaa.gov (accessed on 18 August 2016).
- National Centers for Environmental Information data. Available online: http://www.ncdc.noaa.gov (accessed on 18 August 2016).
- Boynton, W.R.; Garber, J.H.; Summers, R.; Kemp, W.M. Inputs, transformations, and transport of nitrogen and phosphorus in Chesapeake Bay and selected tributaries. Estuaries 1995, 18, 285–314. [Google Scholar] [CrossRef]
- Cerco, C.F.; Noel, M.R. Incremental improvements in Chesapeake Bay environmental model package. J. Environ. Eng. 2005, 131, 745–754. [Google Scholar] [CrossRef]
- Chenillat, F.; Franks, P.J.; Rivière, P.; Capet, X.; Grima, N.; Blanke, B. Plankton dynamics in a cyclonic eddy in the Southern California Current System. J. Geophys. Res. Oceans 2015, 120, 5566–5588. [Google Scholar] [CrossRef]
- Du, J.; Shen, J. Decoupling the influence of biological and physical processes on the dissolved oxygen in the Chesapeake Bay. J. Geophys. Res. Oceans 2015, 120, 78–93. [Google Scholar] [CrossRef]
- Scully, M.E. Physical controls on hypoxia in Chesapeake Bay: A numerical modeling study. J. Geophys. Res. Oceans 2013, 118, 1239–1256. [Google Scholar] [CrossRef]
- Scully, M.E. Wind modulation of dissolved oxygen in Chesapeake Bay. Estuar. Coasts 2010, 33, 1164–1175. [Google Scholar] [CrossRef]
- Belyaev, V.I. Modelling the influence of turbulence on phytoplankton photosynthesis. Ecol. Model. 1992, 60, 11–29. [Google Scholar] [CrossRef]
- Estrada, M.; Berdalet, E. Phytoplankton in a turbulent world. Sci. Mar. 1998, 61 (Suppl. 1), 125–140. [Google Scholar]
- Filippino, K.C.; Bernhardt, P.W.; Mulholland, M.R. Chesapeake Bay plume morphology and the effects on nutrient dynamics and primary productivity in the coastal zone. Estuar. Coasts 2009, 32, 410–424. [Google Scholar] [CrossRef]
- Cerco, C.F.; Noel, M.R. The 2002 Chesapeake Bay Eutrophication Model; U.S. Army Corps of Engineers, Waterways Experiment Station: Vicksburg, MS, USA, 2004. [Google Scholar]
- Robson, B.J. State of the art in modelling of phosphorus in aquatic systems: Review, criticisms and commentary. Environ. Model. Softw. 2014, 61, 339–359. [Google Scholar] [CrossRef]
- Harding, L.W.; Batiuk, R.A.; Fisher, T.R.; Gallegos, C.L.; Malone, T.C.; Miller, W.D.; Mulholland, M.R.; Paerl, H.W.; Perry, E.S.; Tango, P. Scientific bases for numerical chlorophyll criteria in Chesapeake Bay. Estuar. Coasts 2014, 37, 134–148. [Google Scholar] [CrossRef]
- Linker, L.C.; Shenk, G.W.; Wang, P.; Cerco, C.F.; Butt, A.J.; Tango, P.J.; Savidge, R.W. A Comparison of the Chesapeake Bay Estuary Model Calibration with 1985–1994 Observed Data and Method of Application to Water Quality Criteria; Modeling Subcommittee of the Chesapeake Bay Program: Annapolis, MD, USA, 2002. [Google Scholar]
- Adolf, J.E.; Yeager, C.L.; Miller, W.D.; Mallonee, M.E.; Harding, L.W. Environmental forcing of phytoplankton floral composition, biomass, and primary productivity in Chesapeake Bay, USA. Estuar. Coast. Shelf Sci. 2006, 67, 108–122. [Google Scholar] [CrossRef]
- Harding, L.W.; Meeson, B.W.; Fisher, T.R. Phytoplankton production in two east coast estuaries: Photosynthesis-light functions and patterns of carbon assimilation in Chesapeake and Delaware Bays. Estuar. Coast. Shelf Sci. 1986, 23, 773–806. [Google Scholar] [CrossRef]
- Bronk, D.A.; Glibert, P.M.; Malone, T.C.; Banahan, S.; Sahlsten, E. Inorganic and organic nitrogen cycling in Chesapeake Bay: Autotrophic versus heterotrophic processes and relationships to carbon flux. Aquat. Microb. Ecol. 1998, 15, 177–189. [Google Scholar] [CrossRef]
- Roman, M.; Zhang, X.; McGilliard, C.; Boicourt, W. Seasonal and annual variability in the spatial patterns of plankton biomass in Chesapeake Bay. Limnol. Oceanogr. 2005, 50, 480–492. [Google Scholar] [CrossRef]

| Parameters | Value | Literature Range |
|---|---|---|
| Settling velocity W (CYN, m/day) | 0 | 0–0.1 [11,14,34] |
| Settling velocity W (DIA, m/day) | 0.2 | 0–0.5 [11,14,21,34] |
| Settling velocity W (DINO, m/day) | 0.1 | 0–0.2 [21,34] |
| Max photosynthetic rate Pm (CYN, day−1) | 150 | 100–270 [11,14,34] |
| Max photosynthetic rate Pm (DIA, day−1) | 300 | 200–400 [11,14,21,34] |
| Max photosynthetic rate Pm (DINO, day−1) | 200 | 200–350 [21,34] |
| C:CHLA ratio CChl (CYN) | 50 | 30–143 [11,14,34,35] |
| C:CHLA ratio CChl (DIA) | 37 | 30–143 [11,14,21,34,35] |
| C:CHLA ratio CChl (DINO) | 50 | 30–143 [21,34,35] |
| Half-saturation KH-DIN (CYN, mg/L) | 0.02 | 0.01–0.03 [11,14,34] |
| Half-saturation KH-DIN (DIA, mg/L) | 0.025 | 0.003–0.923 [11,14,21,34] |
| Half-saturation KH-DIN (DINO, mg/L) | 0.025 | 0.003–0.923 [21] |
| Half-saturation KH-DIP (all, mg/L) | 0.0025 | 0.001–0.195 [11,14,21,34] |
| Half-saturation KH-DSi (DIA, mg/L) | 0.03 | 0.01–0.05 [11,14,21,34] |
| Photosynthesis Topt (CYN, °C) | 25 | 22–30 [11,14,34] |
| Photosynthesis Topt (DIA, °C) | 16 | 12–35 [11,14,21,34] |
| Photosynthesis Topt (DINO, °C) | 24 | 18–35 [21] |
| Respiration Tref (°C) | 20 | 20 [11,34] |
| Percentage of active respiration Pres | 0.25 | 0.25 [11,14,21,34] |
| Basal metabolic rate M (CYN, day−1) | 0.03 | 0.03–0.05 [11,14,34] |
| Basal metabolic rate M (DIA, day−1) | 0.01 | 0.01–0.1 [11,14,21,34] |
| Basal metabolic rate M (DINO, day−1) | 0.02 | 0.01–0.1 [21,34] |
| Herbivore predation rate F (CYN, day−1) | 0.03 | 0.01–0.05 [14,34] |
| Herbivore predation rate F (DIA, day−1) | 0.1 | 0.05–1 [14,21,34] |
| Herbivore predation rate F (DINO, day−1) | 0.5 | 0.05–1 [21] |
| Max zooplankton predation ration (SZ, day−1) | 2.25 | 0.8–2.25 [11,14] |
| Max zooplankton predation ration (LZ, day−1) | 1.75 | 0.8–1.75 [11,14] |
| Settling velocity of particles (m/day) | 0.25 | 0.03–0.8 [11,14,21,34] |
| Max nitrification rate (g·m−3·day−1) | 0.075 | 0.01–0.75 [11,14,21,34] |
| Optimal nitrification temperature (°C) | 30 | 25–35 [11,14,21,34] |
| Scenarios | Treatment |
|---|---|
| baseline | Described in Section 2 and [31] |
| coarse | Using a low-resolution model grid a (Figure 1) |
| sigma06, sigma20 | Using 6 and 21 uniform sigma levels; i.e., each sigma layer represents 1/5 and 1/20 of the water depth, respectively |
| obs | Using wind data from National Data Buoy Center and National Centers for Environmental Information |
| obs + narr | Using observed wind to drive mixing and NARR wind to drive reaeration |
| az1, az2, az3, az4 | Vertical eddy viscosity (az) computed in the hydrodynamic model was roughly 25%, 50%, 200% and 400% of the baseline scenario. The adjustment of vertical eddy viscosity is achieved by altering the Prandtl number |
| z01, z02, z03, z04 | The bottom roughness length scale (z0) was 25%, 50%, 200% and 300% of the baseline scenario |
| r0.25, r0.50, r0.75, r0.80, r0.90, r0.95, r1.05, r1.10, r1.20 | The riverine nutrient loading was 25%, 50%, 75%, 80%, 90%, 95%, 105%, 110% and 120% of the baseline scenario |
| bf0.25, bf0.50, bf0.75, bf0.8, bf0.9, bf1.1, bf1.2 | The benthic nutrient flux was 80%, 90%, 110% and 120% of the baseline scenario |
| o0.25, o0.50, o0.75, o1.5, o2.0 | The open boundary nutrient concentration was 25%, 50%, 75%, 150% and 200% of the baseline scenario |
| nzp and nsf | Predation of zooplankton (nzp) and suspension feeder (nsf) on phytoplankton was switched off, respectively |
| Grid | Variable | n | CC | p | RMSE |
|---|---|---|---|---|---|
| Fine | DIN | 105 | 0.895 | <0.001 | 0.141 |
| DIP | 105 | 0.410 | <0.001 | 0.016 | |
| TSS | 105 | 0.801 | <0.001 | 2.209 | |
| DO | 210 | 0.892 | <0.001 | 1.525 | |
| CHLA | 104 | 0.480 | <0.001 | 7.516 | |
| Coarse | DIN | 105 | 0.765 | <0.001 | 0.259 |
| DIP | 105 | 0.400 | <0.001 | 0.016 | |
| TSS | 105 | 0.610 | <0.001 | 3.686 | |
| DO | 210 | 0.872 | <0.001 | 1.700 | |
| CHLA | 104 | 0.455 | <0.001 | 7.548 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, M.; Jiang, L. Application of an Unstructured Grid-Based Water Quality Model to Chesapeake Bay and Its Adjacent Coastal Ocean. J. Mar. Sci. Eng. 2016, 4, 52. https://doi.org/10.3390/jmse4030052
Xia M, Jiang L. Application of an Unstructured Grid-Based Water Quality Model to Chesapeake Bay and Its Adjacent Coastal Ocean. Journal of Marine Science and Engineering. 2016; 4(3):52. https://doi.org/10.3390/jmse4030052
Chicago/Turabian StyleXia, Meng, and Long Jiang. 2016. "Application of an Unstructured Grid-Based Water Quality Model to Chesapeake Bay and Its Adjacent Coastal Ocean" Journal of Marine Science and Engineering 4, no. 3: 52. https://doi.org/10.3390/jmse4030052