Non-Invasive Prenatal Diagnosis in the Management of Preimplantation Genetic Diagnosis Pregnancies
Abstract
:1. Introduction
2. Experimental Section
2.1. Fetal Sex Determination in Maternal Blood
2.2. Study of the Paternal Alleles in Maternal Plasma
2.2.1. Marfan Syndrome
2.2.1.1. Direct Analysis of the Paternal Mutation
2.2.1.2. Haplotype Analysis
2.2.2. Huntington Disease (HD)
| Disease/Case | Paternal Mutation/STR | Oligos Name | Oligos Sequence (5′ → 3′) |
|---|---|---|---|
| Marfan Syndrome | c.7636G>A (FBN1 gene) | FBN1-F | 5′-GCCCCCACTGCTTCTCA-3′ |
| FBN1-R | 5′-CCTCCACAAGGATTCACCAG-3′ | ||
| FBN1-S | 5′-CATTTGCCAGAACACTCCT-3′ | ||
| MTS2 [14] | MTS2-F | 5′-GTAGTTGTTATCTTGCAGA-3′ | |
| MTS2-R | 5′-CTGCCCTCTAGGACTCTAAG-3′ | ||
| MTS4 [14] | MTS4-F | 5′-GATGTCCCTATTGCCATCACCAC-3′ | |
| MTS4-R | 5′-CCTGTGCAGGGTAAGACAAG-3 | ||
| Huntington disease | CAG repeats (HTT gene) | HTT-F | 5′-ATGGCGACCCTGGAAAAGCTGATGAA-3′ |
| HTT-R | 5′-GGCGGTGGCGGCTGTTGCTGC-3′ | ||
| I1CAHD | I1CAHD-F | 5′-TATGCCACTACACTACAACCTGGGC-3′ | |
| I1CAHD-F | 5′-ACCAGCATGTGGTATTGTCAAAGTG-3′ | ||
| D4S126 | D4S126-F | 5′-GGATCCTGTCACTGTACT-3′ | |
| D4S126-R | 5′-GTTTCTTTGCTTAACCAGTTTGACCATGAGG-3′ | ||
| D4S127 | D4S127-F | 5′-CCTCTGTTTGCAATCCATTT-3′ | |
| D4S127-R | 5′-GTCCCTTGCATGCCCTGGCT-3′ |
3. Results and Discussion
3.1. Fetal Sex Determination in Maternal Blood
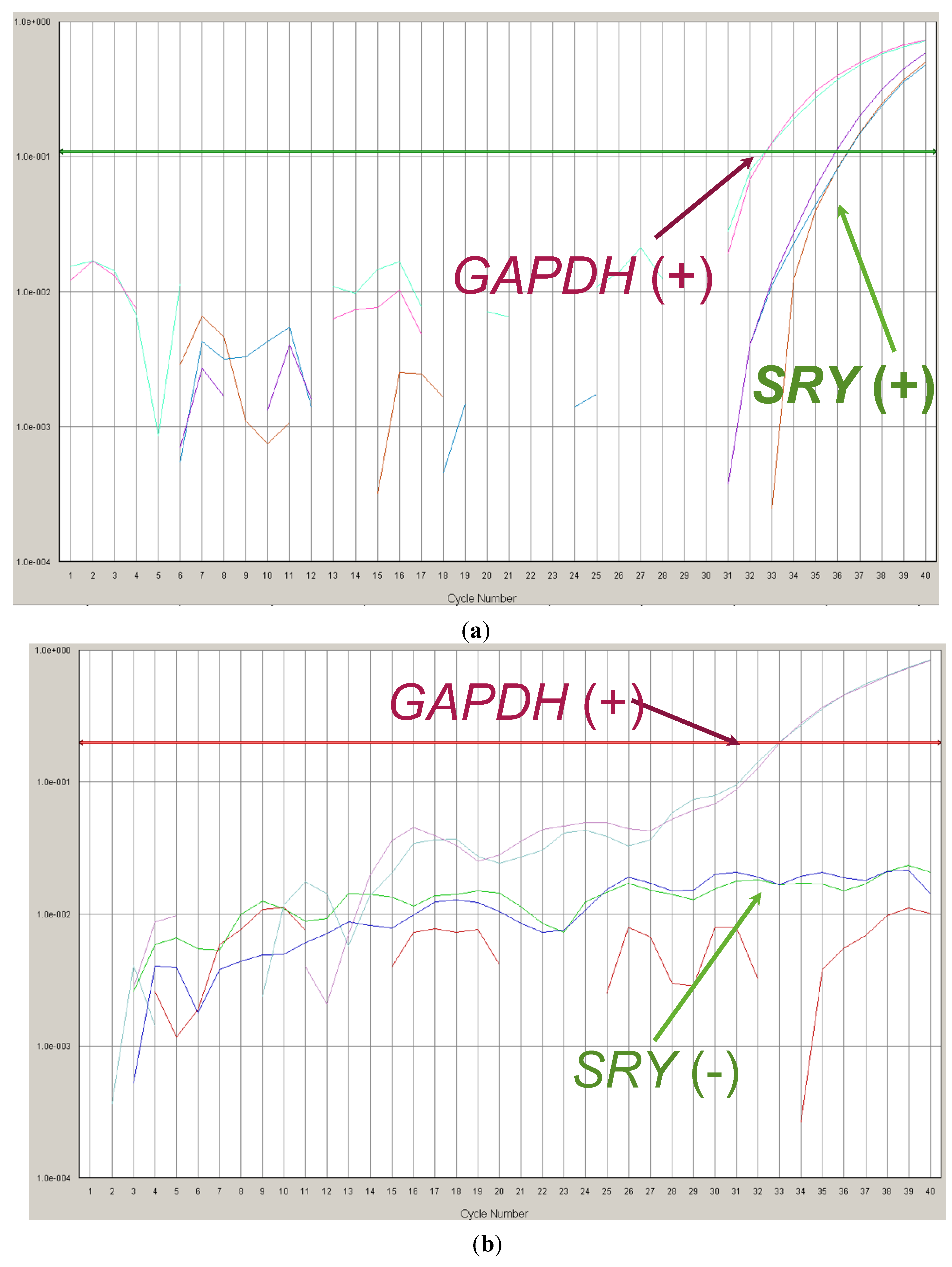
3.2. Study of Paternal Alleles/Mutations in Maternal Plasma
3.2.1. Marfan Syndrome
3.2.2. Huntington Disease
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Harper, J.C.; Wilton, L.; Traeger-Synodinos, J.; Goossens, V.; Moutou, C.; SenGupta, S.B.; Pehlivan Budak, T.; Renwick, P.; de Rycke, M.; Geraedts, J.P.; et al. The ESHRE PGD Consortium: 10 years of data collection. Hum. Reprod. Update 2012, 18, 234–247. [Google Scholar] [CrossRef]
- Lun, F.M.; Chiu, R.W.; Allen Chan, K.C.; Yeung Leung, T.; Kin Lau, T.; Dennis Lo, Y.M. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin. Chem. 2008, 54, 1664–1672. [Google Scholar] [CrossRef]
- Lam, K.W.; Jiang, P.; Liao, G.J.; Chan, K.C.; Leung, T.Y.; Chiu, R.W.; Lo, Y.M. Noninvasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: Application to β-thalassemia. Clin. Chem. 2012, 58, 1467–1475. [Google Scholar] [CrossRef]
- Lo, Y.M. Non-invasive prenatal diagnosis by massively parallel sequencing of maternal plasma DNA. Open Biol. 2012, 2. [Google Scholar] [CrossRef]
- Hyett, J.A.; Gardener, G.; Stojilkovic-Mikic, T.; Finning, K.M.; Martin, P.G.; Rodeck, C.H.; Chitty, L.S. Reduction in diagnostic and therapeutic interventions by non-invasive determination of fetal sex in early pregnancy. Prenat. Diagn. 2005, 25, 1111–1116. [Google Scholar] [CrossRef]
- Minon, J.M.; Schaaps, J.P.; Retz, M.C.; Dricot, J.F.; Foidart, J.M.; Senterre, J.M. Prenatal determination of fetal RHD in maternal plasma: Two-years experience of routine clinical use. J. Gynecol. Obstet. Biol. Reprod. 2005, 34, 448–453. [Google Scholar] [CrossRef]
- Minon, J.M.; Gerard, C.; Senterre, J.M.; Schaaps, J.P.; Foidart, J.M. Routine fetal RHD genotyping with maternal plasma: A four-year experience in Belgium. Transfusion 2008, 48, 373–381. [Google Scholar]
- Bustamante-Aragones, A.; Rodriguez de Alba, M.; Gonzalez-Gonzalez, C.; Trujillo-Tiebas, M.J.; Diego-Alvarez, D.; Vallespin, E.; Plaza, J.; Ayuso, C.; Ramos, C. Foetal sex determination in maternal blood from the seventh week of gestation and its role in diagnosing haemophilia in the foetuses of female carriers. Haemophilia 2008, 14, 593–598. [Google Scholar]
- Harton, G.L.; de Rycke, M.; Fiorentino, F.; Moutou, C.; SenGupta, S.; Traeger-Synodinos, J.; Harper, J.C.; European Society for Human Reproduction and Embryology (ESHRE) PGD Consortium. ESHRE PGD consortium best practice guidelines for amplification-based PGD. Hum. Reprod. 2011, 26, 33–40. [Google Scholar]
- Gonzalez-Gonzalez, M.C.; Trujillo, M.J.; Rodriguez de Alba, M.; Garcia-Hoyos, M.; Lorda-Sanchez, I.; Diaz-Recasens, J.; Ayuso, C.; Ramos, C. Huntington disease-unaffected fetus diagnosed from maternal plasma using QF-PCR. Prenat. Diagn. 2003, 23, 232–234. [Google Scholar]
- Gonzalez-Gonzalez, M.C.; Trujillo, M.J.; Rodrí de Alba, M.; guez de Alba, M.; Ramos, C. Early Huntington disease prenatal diagnosis by maternal semiquantitative fluorescent-PCR. Neurology 2003, 60, 1214–1215. [Google Scholar]
- Gonzalez-Gonzalez, M.C.; Garcia-Hoyos, M.; Trujillo-Tiebas, M.J.; Bustamante-Aragones, A.; Rodriguez de Alba, M.; Diego Alvarez, D.; Diaz-Recasens, J.; Ayuso, C.; Ramos, C. Improvement in strategies for the non-invasive prenatal diagnosis of Huntington disease. J. Assist. Reprod. Genet. 2008, 25, 477–481. [Google Scholar]
- Bustamante-Aragones, A.; Trujillo-Tiebas, M.J.; Gallego-Merlo, J.; Rodriguez de Alba, M.; Gonzalez-Gonzalez, C.; Cantalapiedra, D.; Ayuso, C.; Ramos, C. Prenatal diagnosis of Huntington disease in maternal plasma: Direct and indirect study. Eur. J. Neurol. 2008, 15, 13338–1344. [Google Scholar]
- Judge, D.P.; Biery, N.J.; Dietz, H.C. Characterization of microsatellite markers flanking FBN1: Utility in the diagnostic evaluation for Marfan syndrome. Am. J. Med. Genet. 2001, 99, 39–47. [Google Scholar] [CrossRef]
- Barrett, A.N.; McDonnell, T.C.; Chan, K.C.; Chitty, L.S. A digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clin. Chem. 2012, 58, 1026–1032. [Google Scholar] [CrossRef]
- Saito, H.; Sekizawa, A.; Morimoto, T.; Suzuki, M.; Yanaihara, T. Prenatal DNA diagnosis of a single-gene disorder from maternal plasma. Lancet 2000, 356. [Google Scholar] [CrossRef]
- Chiu, R.W.; Lau, T.K.; Leung, T.N.; Chow, K.C.; Chui, D.H.; Lo, Y.M. Prenatal exclusion of beta thalassaemia major by examination of maternal plasma. Lancet 2002, 360, 998–1000. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, M.C.; Garcia-Hoyos, M.; Trujillo, M.J.; Rodriguez de Alba, M.; Lorda-Sanchez, I.; Diaz-Recasens, J.; Gallardo, E.; Ayuso, C.; Ramos, C. Prenatal detection of a cystic fibrosis mutation in fetal DNA from maternal plasma. Prenat. Diagn. 2002, 22, 946–948. [Google Scholar]
- Li, Y.; Holzgreve, W.; Page-Christiaens, G.C.; Gille, J.J.; Hahn, S. Improved prenatal detection of a fetal point mutation for achondroplasia by the use of size-fractionated circulatory DNA in maternal plasma—Case report. Prenat. Diagn. 2004, 24, 896–898. [Google Scholar] [CrossRef]
- Bustamante-Aragones, A.; Gallego-Merlo, J.; Trujillo-Tiebas, M.J.; de Alba, M.R.; Gonzalez-Gonzalez, C.; Glover, G.; Diego-Alvarez, D.; Ayuso, C.; Ramos, C. New strategy for the prenatal detection/exclusion of paternal cystic fibrosis mutations in maternal plasma. J. Cyst. Fibros. 2008, 7, 505–510. [Google Scholar]
- Bustamante-Aragones, A.; Vallespin, E.; Rodriguez de Alba, M.; Trujillo-Tiebas, M.J.; Gonzalez-Gonzalez, C.; Diego-Alvarez, D.; Riveiro-Alvarez, R.; Lorda-Sanchez, I.; Ayuso, C.; Ramos, C. Early noninvasive prenatal detection of a fetal CRB1 mutation causing Leber congenital amaurosis. Mol. Vis. 2008, 14, 1388–1394. [Google Scholar]
- Chan, K.; Yam, I.; Leung, K.Y.; Tang, M.; Chan, T.K.; Chan, V. Detection of paternal alleles in maternal plasma for non-invasive prenatal diagnosis of beta-thalassemia: A feasibility study in southern Chinese. Eur. J. Obstet. Gynecol. Reprod. Biol. 2010, 150, 28–33. [Google Scholar] [CrossRef]
- Perlado-Marina, S.; Bustamante-Aragonés, A.; Trujillo-Tiebas, M.J.; Gallego, J.; Ramos, C.; Lorda, I.; Horcajada, L.; Plaza, P.; Rodriguez de Alba, M. NIPD of monogenic disorders in the clinical practice: Experience in 301 cases. In Proceedings of the Circulating Nucleic Acids in Plasma and Serum (CNAPS-VIII), Baltimore, MD, USA, 7–8 November 2013.
- Chan, K.C.; Zhang, J.; Hui, A.B.; Wong, N.; Lau, T.K.; Leung, T.N.; Lo, K.W.; Huang, D.W.; Lo, Y.M. Size distributions of maternal and fetal DNA in maternal plasma. Clin. Chem. 2004, 50, 88–92. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bustamante-Aragones, A.; Perlado-Marina, S.; Trujillo-Tiebas, M.J.; Gallego-Merlo, J.; Lorda-Sanchez, I.; Rodríguez-Ramirez, L.; Linares, C.; Hernandez, C.; De Alba, M.R. Non-Invasive Prenatal Diagnosis in the Management of Preimplantation Genetic Diagnosis Pregnancies. J. Clin. Med. 2014, 3, 913-922. https://doi.org/10.3390/jcm3030913
Bustamante-Aragones A, Perlado-Marina S, Trujillo-Tiebas MJ, Gallego-Merlo J, Lorda-Sanchez I, Rodríguez-Ramirez L, Linares C, Hernandez C, De Alba MR. Non-Invasive Prenatal Diagnosis in the Management of Preimplantation Genetic Diagnosis Pregnancies. Journal of Clinical Medicine. 2014; 3(3):913-922. https://doi.org/10.3390/jcm3030913
Chicago/Turabian StyleBustamante-Aragones, Ana, Sara Perlado-Marina, Maria José Trujillo-Tiebas, Jesús Gallego-Merlo, Isabel Lorda-Sanchez, Luz Rodríguez-Ramirez, Concepcion Linares, Corazón Hernandez, and Marta Rodriguez De Alba. 2014. "Non-Invasive Prenatal Diagnosis in the Management of Preimplantation Genetic Diagnosis Pregnancies" Journal of Clinical Medicine 3, no. 3: 913-922. https://doi.org/10.3390/jcm3030913




