Structural and Immunological Characterization of Novel Recombinant MOMP-Based Chlamydial Antigens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analyses
2.2. Cloning Strategy
2.3. Recombinant Expression and Purification of PorB/VDs
2.4. Molecular Modeling
2.5. Structural Analyses
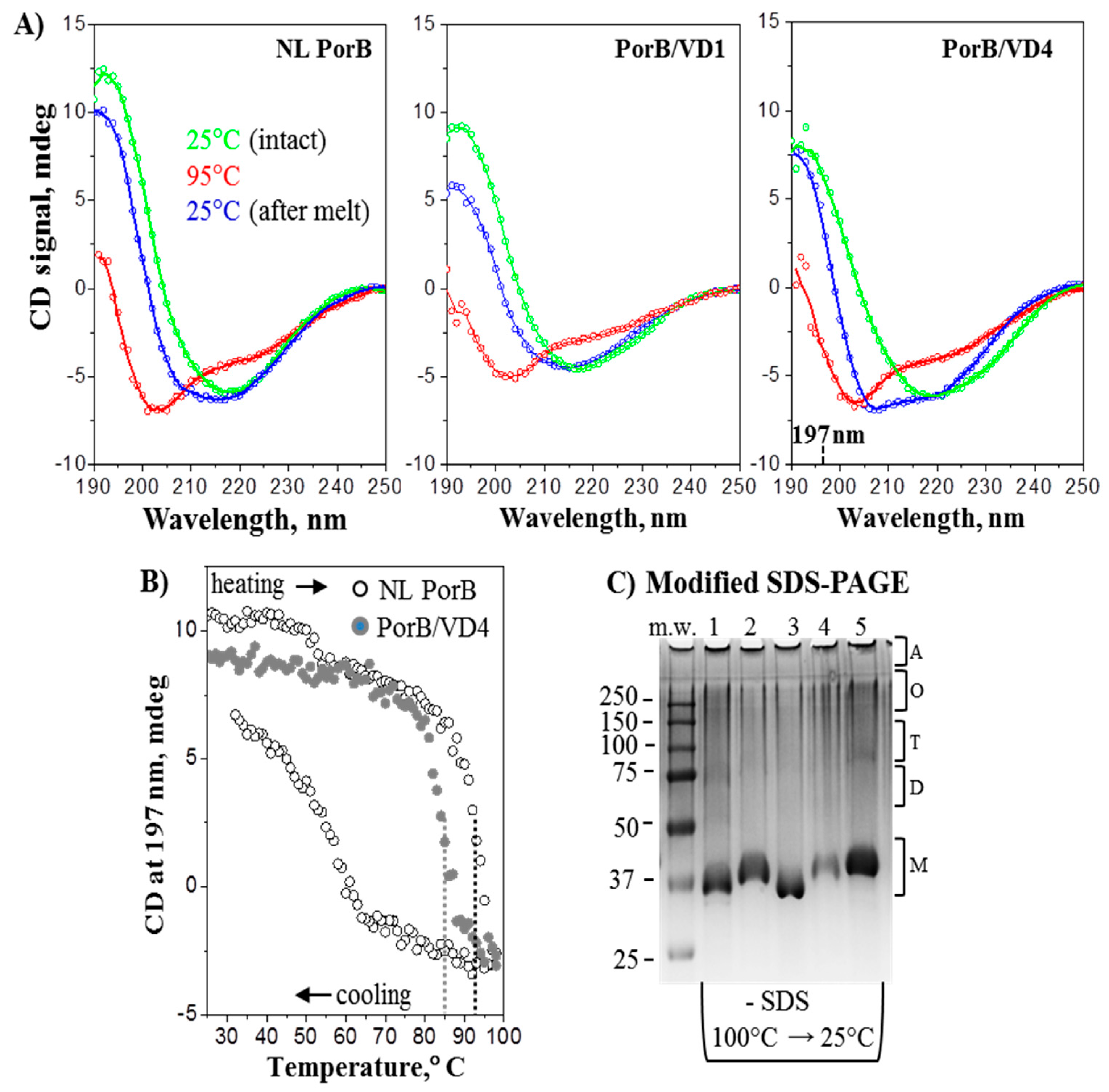
2.5.1. Electrophoresis
2.5.2. Circular Dichroism Spectroscopy
2.6. Cell Cultures and Stimulations
2.7. Vaccine Preparation and Immunization of Mice
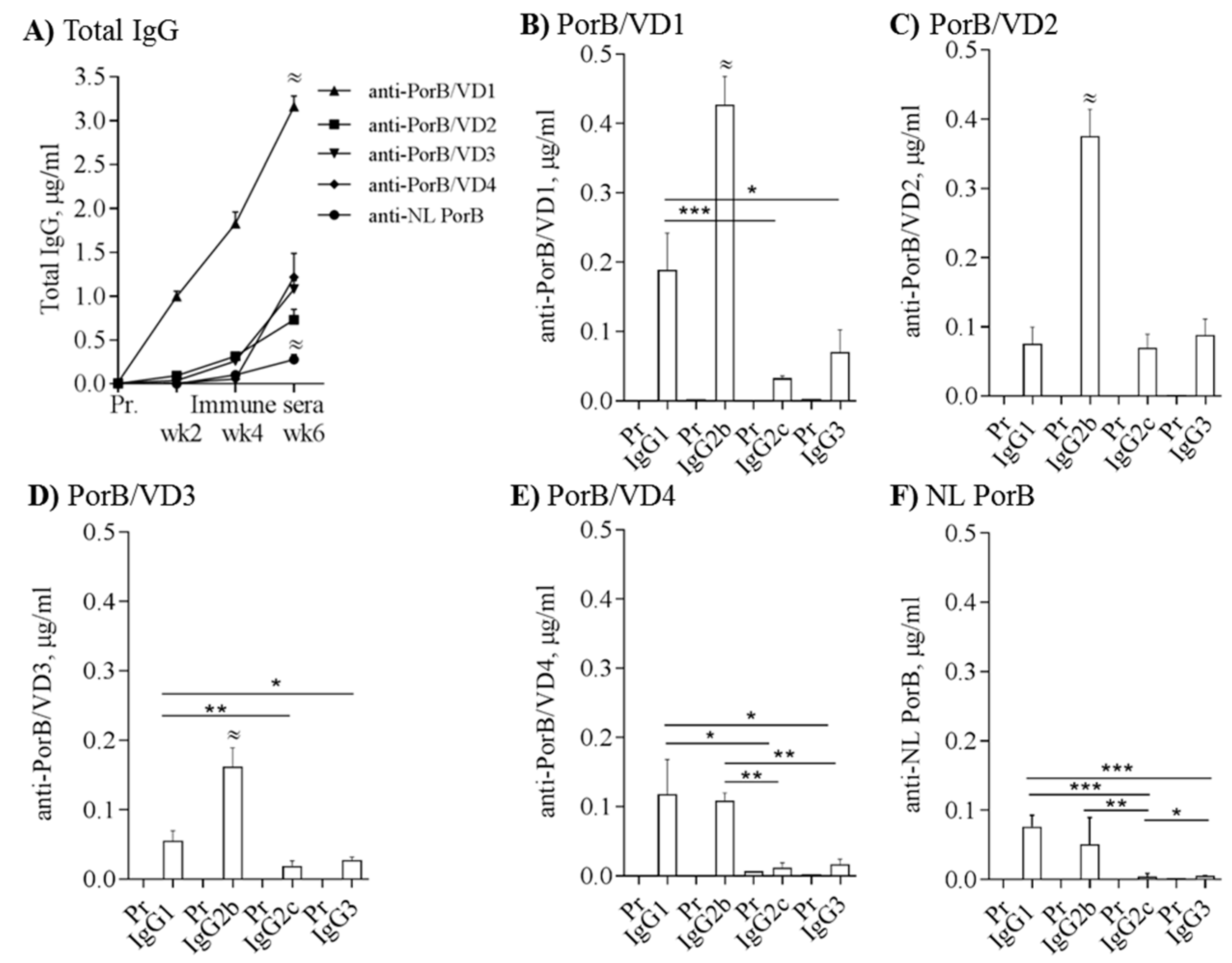
2.8. Antibody ELISA
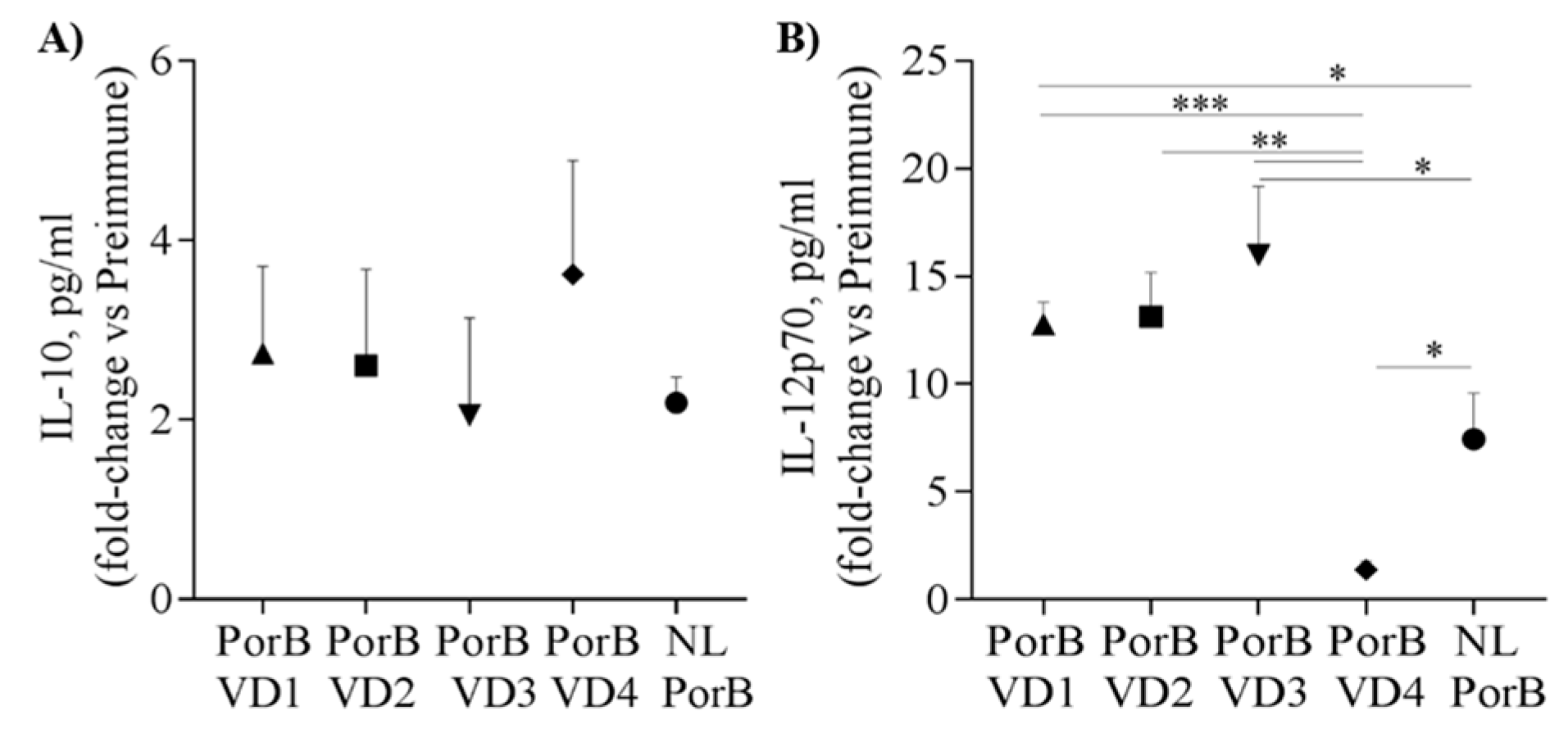
2.9. Cytokine ELISA
2.10. Statistical Analyses
3. Results
3.1. MOMP and PorB Sequence Analyses
3.2. Molecular Representation
3.3. Cloning and Expression of Recombinant PorB/VDs
3.4. Structural Analyses
3.5. PorB/VDs Induce TLR2 Signaling and Are Free of LPS
3.6. Immunogenicity of PorB/VDs
3.7. Serum Cytokines
3.8. Cross-Reactivity and Cross-Immunogenicity
4. Discussion
5. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A


References
- Schachter, J. Chlamydial infections (first of three parts). N. Engl. J. Med. 1978, 298, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Chlamydia screening among sexually active young female enrollees of health plans–United States, 2000–2007. MMWR Morb. Mortal. Wkly. Rep. 2009, 58, 362–365. [Google Scholar]
- Torrone, E.; Papp, J.; Weinstock, H. Prevalence of Chlamydia trachomatis genital infection among persons aged 14–39 years—United States, 2007–2012. MMWR Morb. Mortal. Wkly. Rep. 2014, 63, 834–838. [Google Scholar] [PubMed]
- Newman, L.; Rowley, J.; Vander, H.S.; Wijesooriya, N.S.; Unemo, M.; Low, N.; Stevens, G.; Gottlieb, S.; Kiarie, J.; Temmerman, M. Global Estimates of the Prevalence and Incidence of Four Curable Sexually Transmitted Infections in 2012 Based on Systematic Review and Global Reporting. PLoS ONE 2015, 10, e0143304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.P.; Grayston, J.T. Classification of trachoma virus strains by protection of mice from toxic death. J. Immunol. 1963, 90, 849–856. [Google Scholar] [PubMed]
- Wang, S.P.; Kuo, C.C.; Barnes, R.C.; Stephens, R.S.; Grayston, J.T. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J. Infect. Dis. 1985, 152, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Brunham, R.C.; Pourbohloul, B.; Mak, S.; White, R.; Rekart, M.L. The unexpected impact of a Chlamydia trachomatis infection control program on susceptibility to reinfection. J. Infect. Dis. 2005, 192, 1836–1844. [Google Scholar] [CrossRef] [PubMed]
- Gotz, H.; Lindback, J.; Ripa, T.; Arneborn, M.; Ramsted, K.; Ekdahl, K. Is the increase in notifications of Chlamydia trachomatis infections in Sweden the result of changes in prevalence, sampling frequency or diagnostic methods? Scand. J. Infect. Dis. 2002, 34, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Darville, T. Pelvic inflammatory disease: Identifying research gaps--proceedings of a workshop sponsored by Department of Health and Human Services/National Institutes of Health/National Institute of Allergy and Infectious Diseases, November 3–4, 2011. Sex Transm. Dis. 2013, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Wiesenfeld, H.C. Screening for Chlamydia trachomatis Infections in Women. N. Engl. J. Med. 2017, 376, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Plummer, F.A.; Simonsen, J.N.; Cameron, D.W.; Ndinya-Achola, J.O.; Kreiss, J.K.; Gakinya, M.N.; Waiyaki, P.; Cheang, M.; Piot, P.; Ronald, A.R. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J. Infect. Dis. 1991, 163, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Paba, P.; Bonifacio, D.; Di, B.L.; Ombres, D.; Favalli, C.; Syrjanen, K.; Ciotti, M. Co-expression of HSV2 and Chlamydia trachomatis in HPV-positive cervical cancer and cervical intraepithelial neoplasia lesions is associated with aberrations in key intracellular pathways. Intervirology 2008, 51, 230–234. [Google Scholar] [CrossRef] [PubMed]
- De la Maza, L.M.; Zhong, G.; Brunham, R.C. Update on Chlamydia trachomatis Vaccinology. Clin. Vaccine Immunol. 2017, 24. [Google Scholar] [CrossRef] [PubMed]
- Owusu-Edusei, K., Jr.; Chesson, H.W.; Gift, T.L.; Brunham, R.C.; Bolan, G. Cost-effectiveness of Chlamydia vaccination programs for young women. Emerg. Infect. Dis. 2015, 21, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Bulir, D.; Kaushic, C.; Mahony, J. Considerations for the rational design of a Chlamydia vaccine. Hum. Vaccine Immunother. 2017, 13, 831–835. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, S.L.; Johnston, C. Future prospects for new vaccines against sexually transmitted infections. Curr. Opin. Infect. Dis. 2017, 30, 77–86. [Google Scholar] [CrossRef] [PubMed]
- CDC. Sexually transmitted diseases surveillance 2013. In Prevention; U.S. Department of Health and Human Services: Atlanta, GA, USA, 2014; pp. 1–176. [Google Scholar]
- Plotkin, S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010, 17, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Li, L.X.; McSorley, S.J. B cells enhance antigen-specific CD4 T cell priming and prevent bacteria dissemination following Chlamydia muridarum genital tract infection. PLoS Pathog. 2013, 9, e1003707. [Google Scholar] [CrossRef] [PubMed]
- Farris, C.M.; Morrison, S.G.; Morrison, R.P. CD4+ T cells and antibody are required for optimal major outer membrane protein vaccine-induced immunity to Chlamydia muridarum genital infection. Infect. Immun. 2010, 78, 4374–4383. [Google Scholar] [CrossRef] [PubMed]
- Morrison, S.G.; Morrison, R.P. A predominant role for antibody in acquired immunity to chlamydial genital tract reinfection. J. Immunol. 2005, 175, 7536–7542. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.P.; Grayston, J.T. Pannus with experimental trachoma and inclusion conjunctivitis agent infection of Taiwan monkeys. Am. J. Ophthalmol. 1967, 63, 1133–1145. [Google Scholar] [CrossRef]
- Morrison, R.P.; Lyng, K.; Caldwell, H.D. Chlamydial disease pathogenesis. Ocular hypersensitivity elicited by a genus-specific 57-kD protein. J. Exp. Med. 1989, 169, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Brunham, R.C.; Rey-Ladino, J. Immunology of Chlamydia infection: Implications for a Chlamydia trachomatis vaccine. Nat. Rev. Immunol. 2005, 5, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, H.D.; Kromhout, J.; Schachter, J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 1981, 31, 1161–1176. [Google Scholar] [PubMed]
- Rodriguez-Maranon, M.J.; Bush, R.M.; Peterson, E.M.; Schirmer, T.; de la Maza, L.M. Prediction of the membrane-spanning beta-strands of the major outer membrane protein of Chlamydia. Protein Sci. 2002, 11, 1854–1861. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Pal, S.; Sarcon, A.K.; Kim, S.; Sugawara, E.; Nikaido, H.; Cocco, M.J.; Peterson, E.M.; de la Maza, L.M. Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis. J. Bacteriol. 2007, 189, 6222–6235. [Google Scholar] [CrossRef] [PubMed]
- Feher, V.A.; Randall, A.; Baldi, P.; Bush, R.M.; de la Maza, L.M.; Amaro, R.E. A 3-dimensional trimeric beta-barrel model for Chlamydia MOMP contains conserved and novel elements of Gram-negative bacterial porins. PLoS ONE 2013, 8, e68934. [Google Scholar] [CrossRef] [PubMed]
- Fitch, F.W.; Lancki, D.W.; Gajewski, T.F. T-cell-mediated immune regulation: Help and suppression. In Fundamental Immunology, 3rd ed.; Paul, W.E., Ed.; Raven Press, Ltd.: New York, NY, USA, 1993; pp. 733–762. [Google Scholar]
- Baehr, W.; Zhang, Y.X.; Joseph, T.; Su, H.; Nano, F.E.; Everett, K.D.; Caldwell, H.D. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. USA 1988, 85, 4000–4004. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, L.; Angevine, M.; Kim, S.K.; Watkins, D.; DeMars, R. T-cell epitopes in variable segments of Chlamydia trachomatis major outer membrane protein elicit serovar-specific immune responses in infected humans. Infect. Immun. 2000, 68, 1719–1723. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.M.; Cheng, X.; Markoff, B.A.; Fielder, T.J.; de la Maza, L.M. Functional and structural mapping of Chlamydia trachomatis species-specific major outer membrane protein epitopes by use of neutralizing monoclonal antibodies. Infect. Immun. 1991, 59, 4147–4153. [Google Scholar] [PubMed]
- Pal, S.; Theodor, I.; Peterson, E.M.; de la Maza, L.M. Monoclonal immunoglobulin A antibody to the major outer membrane protein of the Chlamydia trachomatis mouse pneumonitis biovar protects mice against a chlamydial genital challenge. Vaccine 1997, 15, 575–582. [Google Scholar] [CrossRef]
- Stephens, R.S.; Wagar, E.A.; Schoolnik, G.K. High-resolution mapping of serovar-specific and common antigenic determinants of the major outer membrane protein of Chlamydia trachomatis. J. Exp. Med. 1988, 167, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.W.; Follmann, F.; Erneholm, K.; Rosenkrands, I.; Andersen, P. Protection Against Chlamydia trachomatis Infection and Upper Genital Tract Pathological Changes by Vaccine-Promoted Neutralizing Antibodies Directed to the VD4 of the Major Outer Membrane Protein. J. Infect. Dis. 2015, 212, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Nigg, C. An unidentified virus which produces pneumonia and systemic infection in mice. Science 1942, 95, 49–50. [Google Scholar] [CrossRef] [PubMed]
- De la Maza, L.M.; Pal, S.; Khamesipour, A.; Peterson, E.M. Intravaginal inoculation of mice with the Chlamydia trachomatis mouse pneumonitis biovar results in infertility. Infect. Immun. 1994, 62, 2094–2097. [Google Scholar] [PubMed]
- Carmichael, J.R.; Pal, S.; Tifrea, D.; de la Maza, L.M. Induction of protection against vaginal shedding and infertility by a recombinant Chlamydia vaccine. Vaccine 2011, 29, 5276–5283. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Peterson, E.M.; de la Maza, L.M. New murine model for the study of Chlamydia trachomatis genitourinary tract infections in males. Infect. Immun. 2004, 72, 4210–4216. [Google Scholar] [CrossRef] [PubMed]
- Farris, C.M.; Morrison, R.P. Vaccination against Chlamydia genital infection utilizing the murine C. muridarum model. Infect. Immun. 2011, 79, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Pal, S.; Tifrea, D.; Jia, Z.; de la Maza, L.M. A vaccine formulated with a combination of TLR-2 and TLR-9 adjuvants and the recombinant major outer membrane protein elicits a robust immune response and significant protection against a Chlamydia muridarum challenge. Microbes Infect. 2014, 16, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Tifrea, D.F.; Pal, S.; Toussi, D.N.; Massari, P.; de la Maza, L.M. Vaccination with major outer membrane protein proteosomes elicits protection in mice against a Chlamydia respiratory challenge. Microbes Infect. 2013, 15, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Kari, L.; Whitmire, W.M.; Crane, D.D.; Reveneau, N.; Carlson, J.H.; Goheen, M.M.; Peterson, E.M.; Pal, S.; de la Maza, L.M.; Caldwell, H.D. Chlamydia trachomatis native major outer membrane protein induces partial protection in nonhuman primates: Implication for a trachoma transmission-blocking vaccine. J. Immunol. 2009, 182, 8063–8070. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Peterson, E.M.; Rappuoli, R.; Ratti, G.; de la Maza, L.M. Immunization with the Chlamydia trachomatis major outer membrane protein, using adjuvants developed for human vaccines, can induce partial protection in a mouse model against a genital challenge. Vaccine 2006, 24, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Hou, B.; Wang, B.; Lin, X.; Gong, W.; Dong, H.; Zhu, S.; Chen, S.; Xue, X.; Zhao, K.N.; et al. A multi-epitope vaccine based on Chlamydia trachomatis major outer membrane protein induces specific immunity in mice. Acta Biochim. Biophys. Sin. 2014, 46, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.R.; Hulett, K.; Pillai, S.R.; Dennis, V.A.; Oh, M.K.; Scissum-Gunn, K. Mucosal immunization with recombinant MOMP genetically linked with modified cholera toxin confers protection against Chlamydia trachomatis infection. Vaccine 2006, 24, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Parnell, M.; Caldwell, H.D. Protective efficacy of a parenterally administered MOMP-derived synthetic oligopeptide vaccine in a murine model of Chlamydia trachomatis genital tract infection: Serum neutralizing IgG antibodies do not protect against chlamydial genital tract infection. Vaccine 1995, 13, 1023–1032. [Google Scholar] [CrossRef]
- Pal, S.; Barnhart, K.M.; Wei, Q.; Abai, A.M.; Peterson, E.M.; de la Maza, L.M. Vaccination of mice with DNA plasmids coding for the Chlamydia trachomatis major outer membrane protein elicits an immune response but fails to protect against a genital challenge. Vaccine 1999, 17, 459–465. [Google Scholar] [CrossRef]
- Igietseme, J.U.; Murdin, A. Induction of protective immunity against Chlamydia trachomatis genital infection by a vaccine based on major outer membrane protein-lipophilic immune response-stimulating complexes. Infect. Immun. 2000, 68, 6798–6806. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Feng, Y.; Rao, P.; Xue, X.; Chen, S.; Li, W.; Zhu, G.; Zhang, L. Hepatitis B virus surface antigen as delivery vector can enhance Chlamydia trachomatis MOMP multi-epitope immune response in mice. Appl. Microbiol. Biotechnol. 2014, 98, 4107–4117. [Google Scholar] [CrossRef] [PubMed]
- Fairley, S.J.; Singh, S.R.; Yilma, A.N.; Waffo, A.B.; Subbarayan, P.; Dixit, S.; Taha, M.A.; Cambridge, C.D.; Dennis, V.A. Chlamydia trachomatis recombinant MOMP encapsulated in PLGA nanoparticles triggers primarily T helper 1 cellular and antibody immune responses in mice: A desirable candidate nanovaccine. Int. J. Nanomed. 2013, 8, 2085–2099. [Google Scholar]
- Dixit, S.; Singh, S.R.; Yilma, A.N.; Agee, R.D.; Taha, M.; Dennis, V.A. Poly(lactic acid)-poly(ethylene glycol) nanoparticles provide sustained delivery of a Chlamydia trachomatis recombinant MOMP peptide and potentiate systemic adaptive immune responses in mice. Nanomedicine 2014, 10, 1311–1321. [Google Scholar] [CrossRef] [PubMed]
- Childs, T.S.; Webley, W.C. In vitro assessment of halobacterial gas vesicles as a Chlamydia vaccine display and delivery system. Vaccine 2012, 30, 5942–5948. [Google Scholar] [CrossRef] [PubMed]
- Kalbina, I.; Wallin, A.; Lindh, I.; Engstrom, P.; Andersson, S.; Strid, K. A novel chimeric MOMP antigen expressed in Escherichia coli, Arabidopsis thaliana, and Daucus carota as a potential Chlamydia trachomatis vaccine candidate. Protein Expr. Purif. 2011, 80, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Yu, H.; Wang, X.H.; Li, X.Z.; Zhu, Y.P.; Li, H.X.; Luo, S.J.; Yuan, Z.G. Protective efficacy against Chlamydophila psittaci by oral immunization based on transgenic rice expressing MOMP in mice. Vaccine 2013, 31, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Grund, V.; Durling, L.; Crane, D.; Caldwell, H.D. Dendritic cells pulsed with a recombinant chlamydial major outer membrane protein antigen elicit a CD4(+) type 2 rather than type 1 immune response that is not protective. Infect. Immun. 2002, 70, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Tifrea, D.F.; Ralli-Jain, P.; Pal, S.; de la Maza, L.M. Vaccination with the recombinant major outer membrane protein elicits antibodies to the constant domains and induces cross-serovar protection against intranasal challenge with Chlamydia trachomatis. Infect. Immun. 2013, 81, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Toussi, D.N.; Carraway, M.; Wetzler, L.M.; Lewis, L.A.; Liu, X.; Massari, P. The amino acid sequence of Neisseria lactamica PorB surface-exposed loops influences Toll-like receptor 2-dependent cell activation. Infect. Immun. 2012, 80, 3417–3428. [Google Scholar] [CrossRef] [PubMed]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef] [PubMed]
- Madico, G.; Welsch, J.A.; Lewis, L.A.; McNaughton, A.; Perlman, D.H.; Costello, C.E.; Ngampasutadol, J.; Vogel, U.; Granoff, D.M.; Ram, S. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J. Immunol. 2006, 177, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Madico, G.; Ngampasutadol, J.; Gulati, S.; Vogel, U.; Rice, P.A.; Ram, S. Factor H binding and function in sialylated pathogenic neisseriae is influenced by gonococcal, but not meningococcal, porin. J. Immunol. 2007, 178, 4489–4497. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.L.; Tai, J.Y.; Blake, M.S. Expression of large amounts of neisserial porin proteins in Escherichia coli and refolding of the proteins into native trimers. Infect. Immun. 1994, 62, 2432–2439. [Google Scholar] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.S.; Hildebrand, P.W. NGL Viewer: A web application for molecular visualization. Nucleic Acids Res. 2015, 43, W576–W579. [Google Scholar] [CrossRef] [PubMed]
- Kattner, C.; Toussi, D.N.; Zaucha, J.; Wetzler, L.M.; Ruppel, N.; Zachariae, U.; Massari, P.; Tanabe, M. Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition. J. Struct. Biol. 2014, 185, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Massari, P.; King, C.A.; Macleod, H.; Wetzler, L.M. Improved purification of native meningococcal porin PorB and studies on its structure/function. Protein Expr. Purif. 2005, 44, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Peterson, E.M.; de la Maza, L.M. Vaccination with the Chlamydia trachomatis major outer membrane protein can elicit an immune response as protective as that resulting from inoculation with live bacteria. Infect. Immun. 2005, 73, 8153–8160. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wetzler, L.M.; Massari, P. The PorB porin from commensal Neisseria lactamica induces Th1 and Th2 immune responses to ovalbumin in mice and is a potential immune adjuvant. Vaccine 2008, 26, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.S.; Callaghan, M.J.; Derrick, J.P.; Maiden, M.C. Variation in the Neisseria lactamica porin, and its relationship to meningococcal PorB. Microbiology 2008, 154, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Derrick, J.P.; Urwin, R.; Suker, J.; Feavers, I.M.; Maiden, M.C. Structural and evolutionary inference from molecular variation in neisseria porins. Infect. Immun. 1999, 67, 2406–2413. [Google Scholar] [PubMed]
- Minetti, C.A.; Blake, M.S.; Remeta, D.P. Characterization of the structure, function, and conformational stability of PorB class 3 protein from Neisseria meningitidis. A porin with unusual physicochemical properties. J. Biol. Chem. 1998, 273, 25329–25338. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Davis, H.L.; Peterson, E.M.; de la Maza, L.M. Immunization with the Chlamydia trachomatis mouse pneumonitis major outer membrane protein by use of CpG oligodeoxynucleotides as an adjuvant induces a protective immune response against an intranasal chlamydial challenge. Infect. Immun. 2002, 70, 4812–4817. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wetzler, L.M.; Nascimento, L.O.; Massari, P. Human airway epithelial cell responses to Neisseria lactamica and purified porin via Toll-like receptor 2-dependent signaling. Infect. Immun. 2010, 78, 5314–5323. [Google Scholar] [CrossRef] [PubMed]
- Massari, P.; Toussi, D.N.; Tifrea, D.F.; de la Maza, L.M. Toll-Like Receptor 2-Dependent Activity of Native Major Outer Membrane Protein Proteosomes of Chlamydia trachomatis. Infect. Immun. 2013, 81, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Snapper, C.M.; Paul, W.E. B cell stimulatory factor-1 (interleukin 4) prepares resting murine B cells to secrete IgG1 upon subsequent stimulation with bacterial lipopolysaccharide. J. Immunol. 1987, 139, 10–17. [Google Scholar] [PubMed]
- Guy, B.; Krell, T.; Sanchez, V.; Kennel, A.; Manin, C.; Sodoyer, R. Do Th1 or Th2 sequence motifs exist in proteins? Identification of amphipatic immunomodulatory domains in Helicobacter pylori catalase. Immunol. Lett. 2005, 96, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Desclozeaux, M.; Robbins, A.; Jelocnik, M.; Khan, S.A.; Hanger, J.; Gerdts, V.; Potter, A.; Polkinghorne, A.; Timms, P. Immunization of a wild koala population with a recombinant Chlamydia pecorum Major Outer Membrane Protein (MOMP) or Polymorphic Membrane Protein (PMP) based vaccine: New insights into immune response, protection and clearance. PLoS ONE 2017, 12, e0178786. [Google Scholar] [CrossRef] [PubMed]
- Gold, R.; Goldschneider, I.; Lepow, M.L.; Draper, T.F.; Randolph, M. Carriage of Neisseria meningitidis and Neisseria lactamica in infants and children. J. Infect. Dis. 1978, 137, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, K.A.; Stuart, J.M.; Jones, D.M.; Noah, N.D. The Stonehouse survey: Nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiol. Infect. 1987, 99, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Deasy, A.M.; Guccione, E.; Dale, A.P.; Andrews, N.; Evans, C.M.; Bennett, J.S.; Bratcher, H.B.; Maiden, M.C.; Gorringe, A.R.; Read, R.C. Nasal Inoculation of the Commensal Neisseria lactamica Inhibits Carriage of Neisseria meningitidis by Young Adults: A Controlled Human Infection Study. Clin. Infect. Dis. 2015, 60, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.; Winterbotham, M.; Mowe, E.; Gorringe, A.; Tang, C.M. Immunization with live Neisseria lactamica protects mice against meningococcal challenge and can elicit serum bactericidal antibodies. Infect. Immun. 2006, 74, 6348–6355. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, A.T.; Brackenbury, L.S.; Massari, P.; Davenport, V.; Gorringe, A.; Heyderman, R.S.; Williams, N.A. Neisseria lactamica selectively induces mitogenic proliferation of the naive B cell pool via cell surface Ig. J. Immunol. 2010, 185, 3652–3660. [Google Scholar] [CrossRef] [PubMed]





| Ratio | PorB/VD1 | PorB/VD2 | PorB/VD3 | PorB/VD4 | NL PorB |
|---|---|---|---|---|---|
| IgG2b/IgG1 a | 3.5 ± 0.85 | 8 ± 1.6 | 4.1 ± 1.1 | 2 ± 0.69 | 2.48 ± 2.23 |
| IgG2c/IgG1 a | 0.27 ± 0.06 | 1.42 ± 0.41 | 0.46 ± 0.15 | 0.11 ± 0.02 | 0.41 ± 0.4 |
| IgG3/IgG1 a | 0.32 ± 0.08 | 2.38 ± 0.81 | 0.73 ± 0.19 | 0.41 ± 0.26 | 0.14 ± 0.05 |
| Th1:Th2 index b | 0.935 | 2.348 | 1.254 | 0.388 | 0.194 |
| Cytokine | PorB/VD1 | PorB/VD2 | PorB/VD3 | PorB/VD4 | NL PorB |
|---|---|---|---|---|---|
| IL-4 a | 1.19 ± 0.55 | 3.56 ± 0.7 | 3.1 ± 0.72 | 2.29 ± 0.09 | 2.57 ± 0.67 |
| IFN-γ a | 5.44 ± 0.2 | 10.05 ± 4.79 | 3.65 ± 0.25 | 2.08 ± 0.62 | 2.84 ± 1.09 |
| Th1:Th2 index ^ | 2.85 | 2.81 | 1.80 | 0.90 | 1.10 |
| IL-6 a | 5.23 ±0.82 | 16.63 ± 3.47 | 7.6 ± 1.12 | 11.14 ± 2.56 | 4.81 ± 1.7 |
| TNF-α a | 7.87 ± 1.78 | 1.58 ± 0.67 | 5.9 ± 0.68 | 12.75 ± 3.63 | 0.46 ± 0.24 |
| Sera | Cm EBs | rMOMP |
|---|---|---|
| Anti-PorB/VD1 | 12,800 | >12,800 |
| Anti-PorB/VD2 | 100 | <100 |
| Anti-PorB/VD3 | 200 | 3200 |
| Anti-PorB/VD4 | <100 | 100 |
| Anti-NL PorB | <100 | <100 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madico, G.; Gursky, O.; Fairman, J.; Massari, P. Structural and Immunological Characterization of Novel Recombinant MOMP-Based Chlamydial Antigens. Vaccines 2018, 6, 2. https://doi.org/10.3390/vaccines6010002
Madico G, Gursky O, Fairman J, Massari P. Structural and Immunological Characterization of Novel Recombinant MOMP-Based Chlamydial Antigens. Vaccines. 2018; 6(1):2. https://doi.org/10.3390/vaccines6010002
Chicago/Turabian StyleMadico, Guillermo, Olga Gursky, Jeff Fairman, and Paola Massari. 2018. "Structural and Immunological Characterization of Novel Recombinant MOMP-Based Chlamydial Antigens" Vaccines 6, no. 1: 2. https://doi.org/10.3390/vaccines6010002





