Host Transcriptional Response to Ebola Virus Infection
Abstract
:1. Ebola Virus Disease
2. Transcriptional Response to Ebola Virus Disease In Vitro
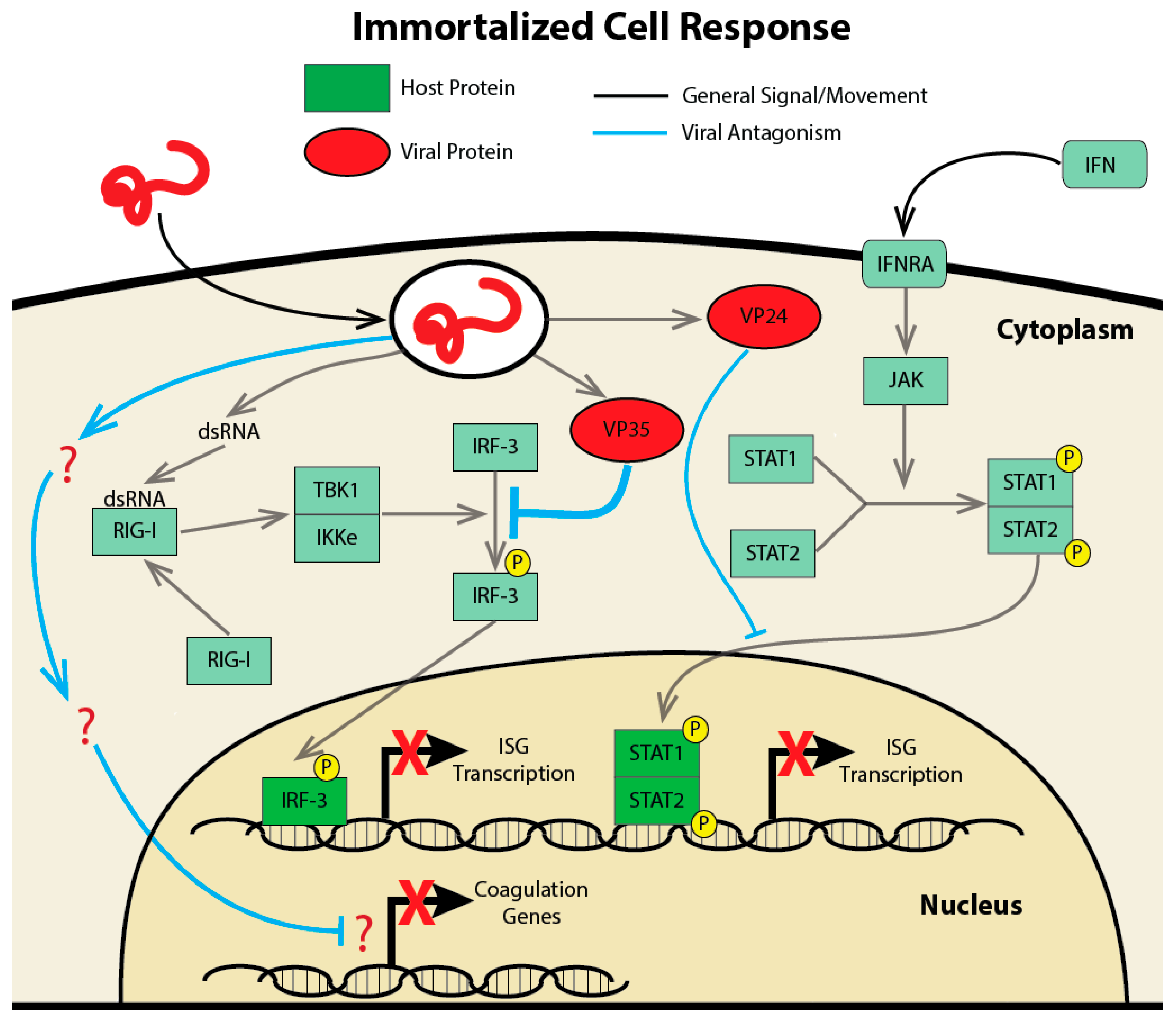
2.1. Studying the Transcriptional Response in Immortalized Cell Lines
2.2. Transcriptional Analysis of a Potential Reservoir
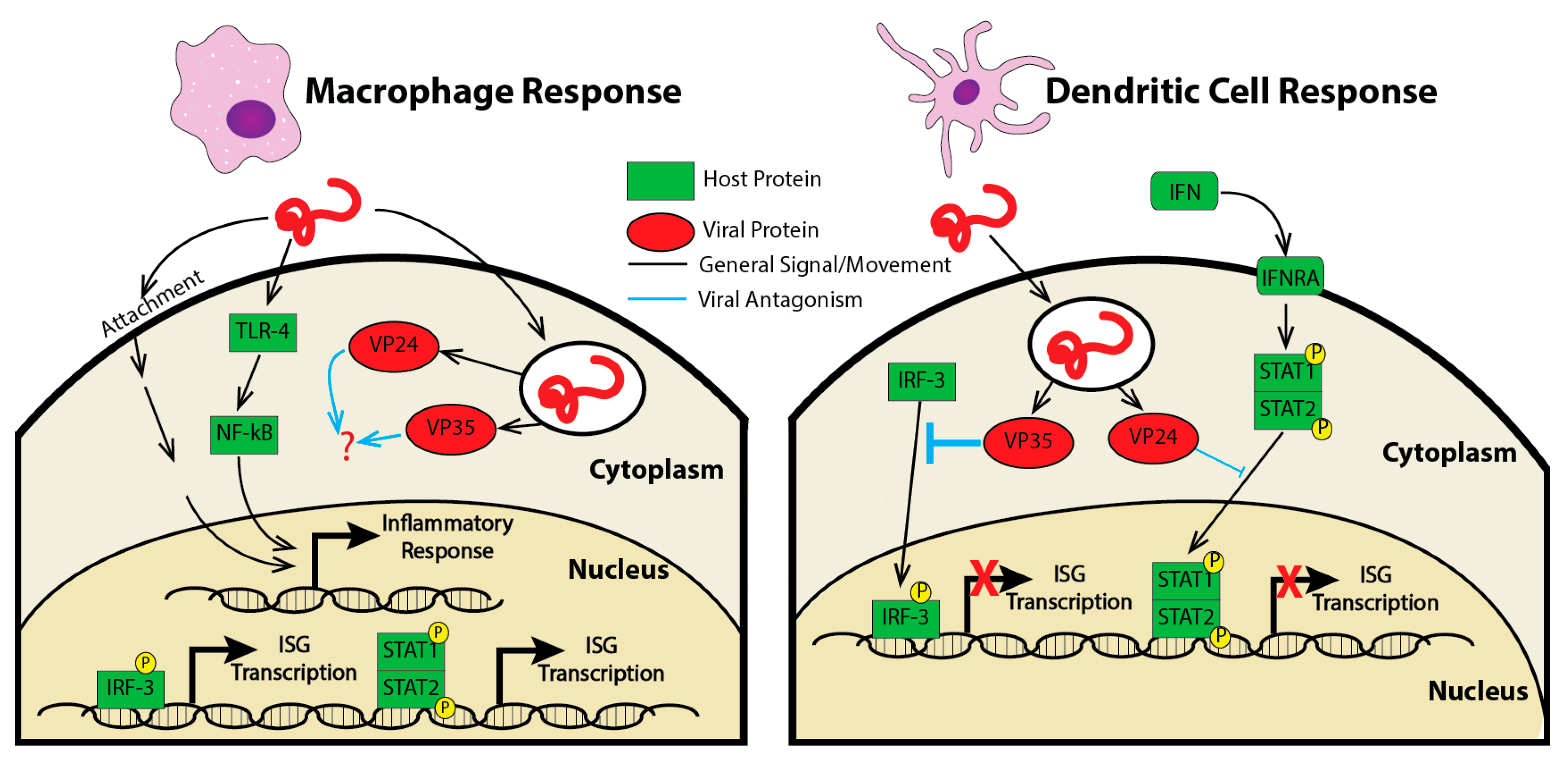
2.3. Studying the Transcriptoinal Response in Primary Cells
3. Transcriptional Response to Ebola Virus Disease In Vivo
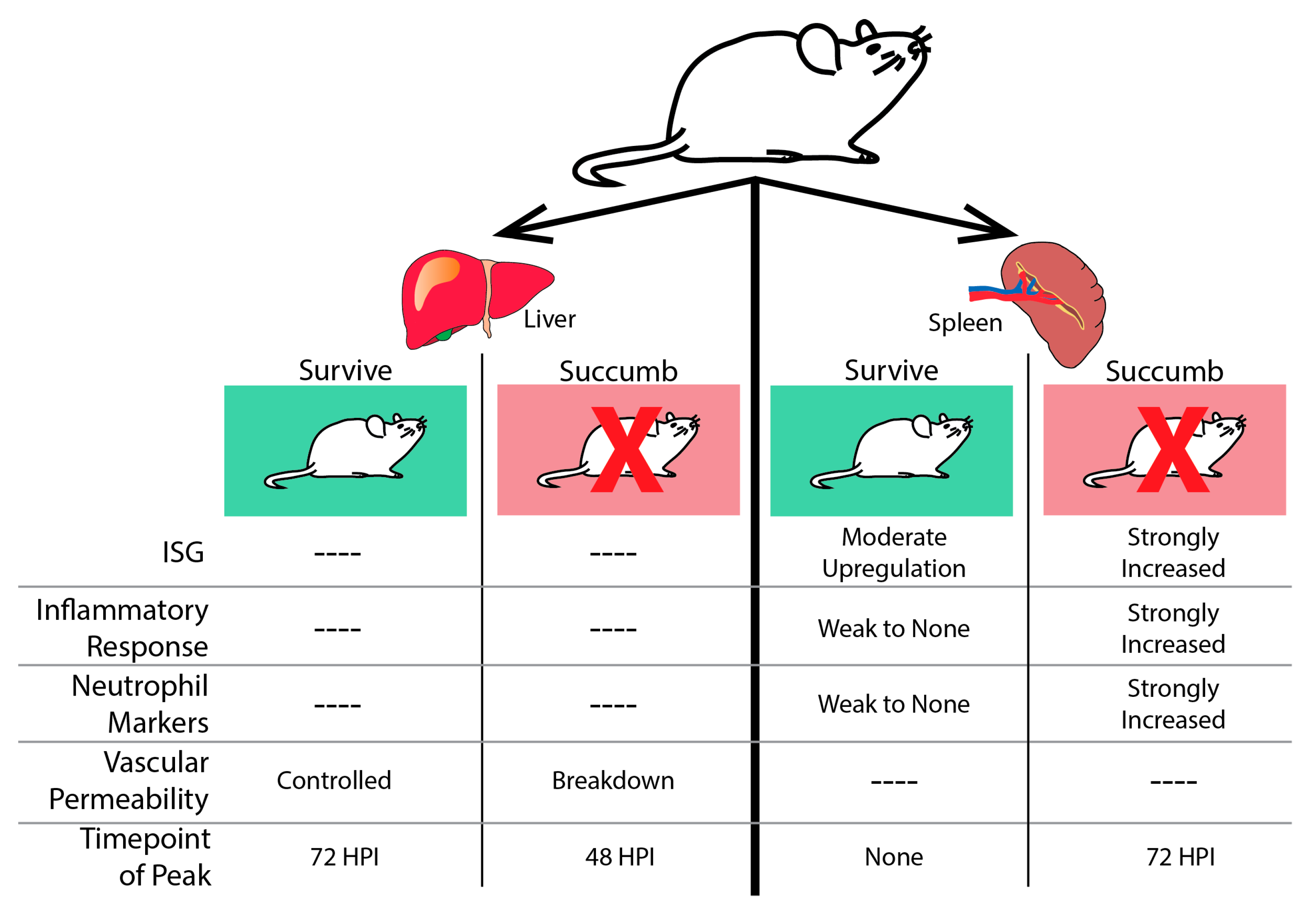
3.1. Rodent Models and Transcriptional Response
3.2. Non-Human Primate Models and Transcriptional Response
4. Transcriptional Response to Ebola Virus Disease in Humans In Vivo
5. Comparison of Various Platforms
6. Conclusions and Future Directions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Feldmann, H.; Geisbert, T.W. Ebola haemorrhagic fever. Lancet 2011. [Google Scholar] [CrossRef]
- Laupland, K.B.; Valiquette, L. Ebola virus disease. Can. J. Infect. Dis. Med. Microbiol. 2014, 25, 128–129. [Google Scholar] [CrossRef] [PubMed]
- CDC. 2014 Ebola Outbreak in West Africa: Case Counts. Centers for Disease Control and Prevention. Available online: http://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/case-counts.html (accessed on 1 June 2017).
- Geisbert, T.W.; Hensley, L.E.; Larsen, T.; Young, H.A.; Reed, D.S.; Geisbert, J.B.; Scott, D.P.; Kagan, E.; Jahrling, P.B.; Davis, K.J. Pathogenesis of Ebola Hemorrhagic Fever in Cynomolgus Macaques. Am. J. Pathol. 2003, 163, 2347–2370. [Google Scholar] [CrossRef]
- Lüdtke, A.; Ruibal, P.; Wozniak, D.M.; Pallasch, E.; Wurr, S.; Bockholt, S.; Gómez-Medina, S.; Qiu, X.; Kobinger, G.P.; Rodríguez, E.; et al. Ebola virus infection kinetics in chimeric mice reveal a key role of T cells as barriers for virus dissemination. Sci. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Vernet, M.-A.; Reynard, S.; Fizet, A.; Schaeffer, J.; Pannetier, D.; Guedj, J.; Rives, M.; Georges, N.; Garcia-Bonnet, N.; Sylla, A.I.; et al. Clinical, virological, and biological parameters associated with outcomes of Ebola virus infection in Macenta, Guinea. JCI Insight 2017, 2, e88864. [Google Scholar] [CrossRef] [PubMed]
- Bosio, C.M.; Aman, M.J.; Grogan, C.; Hogan, R.; Ruthel, G.; Negley, D.; Mohamadzadeh, M.; Bavari, S.; Schmaljohn, A. Ebola and Marburg Viruses Replicate in Monocyte-Derived Dendritic Cells without Inducing the Production of Cytokines and Full Maturation. J. Infect. Dis. 2003, 188, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.; Geisbert, T.W. Ebola virus: The role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int. J. Biochem. Cell Biol. 2005, 37, 1560–1566. [Google Scholar] [CrossRef] [PubMed]
- Ruibal, P.; Oestereich, L.; Lüdtke, A.; Becker-Ziaja, B.; Wozniak, D.M.; Kerber, R.; Korva, M.; Cabeza-Cabrerizo, M.; Bore, J.A.; Koundouno, F.R.; et al. Unique human immune signature of Ebola virus disease in Guinea. Nature 2016, 533, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Bradfute, S.B.; Braun, D.R.; Shamblin, J.D.; Geisbert, J.B.; Paragas, J.; Garrison, A.; Hensley, L.E.; Geisbert, T.W. Lymphocyte Death in a Mouse Model of Ebola Virus Infection EBOV infection in humans results in mortality rates. J. Infect. Dis. 2007, 196, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Reed, D.S.; Hensley, L.E.; Geisbert, J.B.; Jahrling, P.B.; Geisbert, T.W. Depletion of peripheral blood T lymphocytes and NK cells during the course of ebola hemorrhagic Fever in cynomolgus macaques. Viral Immunol. 2004, 17, 390–400. [Google Scholar] [CrossRef] [PubMed]
- McElroy, A.K.; Erickson, B.R.; Flietstra, T.D.; Rollin, P.E.; Nichol, S.T.; Towner, J.S.; Spiropoulou, C.F. Ebola Hemorrhagic Fever: Novel Biomarker Correlates of Clinical Outcome. J. Infect. Dis. 2014, 210, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Baize, S.; Leroy, E.M.; Georges, A.J.; Georges-Courbot, M.-C.; Capron, M.; Bedjabaga, I.; Lansoud-Soukate, J.; Mavoungou, E. Inflammatory responses in Ebola virus-infected patients. Clin. Exp. Immunol. 2002, 128, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Bixler, S.L.; Goff, A.J. The Role of Cytokines and Chemokines in Filovirus Infection. Viruses 2015, 7, 5489–5507. [Google Scholar] [CrossRef] [PubMed]
- Wauquier, N.; Becquart, P.; Padilla, C.; Baize, S.; Leroy, E.M. Human Fatal Zaire Ebola Virus Infection Is Associated with an Aberrant Innate Immunity and with Massive Lymphocyte Apoptosis. PLoS Negl. Trop. Dis. 2010, 4, e837. [Google Scholar] [CrossRef] [PubMed]
- Rougeron, V.; Feldmann, H.; Grard, G.; Becker, S.; Leroy, E.M. Ebola and Marburg haemorrhagic fever. J. Clin. Virol. 2015, 64, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhou, Y.; Yang, Z. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol. Immunol. 2016, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Edwards, M.R.; Liu, G.; Mire, C.E.; Sureshchandra, S.; Luthra, P.; Yen, B.; Shabman, R.S.; Leung, D.W.; Messaoudi, I.; Geisbert, T.W.; et al. Differential Regulation of Interferon Responses by Ebola and Marburg Virus VP35 Proteins. Cell Rep. 2016, 14, 1632–1640. [Google Scholar] [CrossRef] [PubMed]
- Basler, C.F.; Wang, X.; Mühlberger, E.; Volchkov, V.; Paragas, J.; Klenk, H.D.; García-Sastre, A.; Palese, P. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 2000, 97, 12289–12294. [Google Scholar] [CrossRef] [PubMed]
- Leung, L.W.; Park, M.-S.; Martinez, O.; Valmas, C.; López, C.B.; Basler, C.F. Ebolavirus VP35 suppresses IFN production from conventional but not plasmacytoid dendritic cells. Immunol. Cell Biol. 2011, 89, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Reid, S.P.; Valmas, C.; Martinez, O.; Sanchez, F.M.; Basler, C.F. Ebola Virus VP24 Proteins Inhibit the Interaction of NPI-1 Subfamily Karyopherin Proteins with Activated STAT1. J. Virol. 2007, 81, 13469–13477. [Google Scholar] [CrossRef] [PubMed]
- Martines, R.B.; Ng, D.L.; Greer, P.W.; Rollin, P.E.; Zaki, S.R. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J. Pathol. 2015, 235, 153–174. [Google Scholar] [CrossRef] [PubMed]
- Basler, C.F.; Mikulasova, A.; Martinez-Sobrido, L.; Paragas, J.; Mühlberger, E.; Bray, M.; Klenk, H.-D.; Palese, P.; García-Sastre, A. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 2003, 77, 7945–7956. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, W.B.; Loo, Y.-M.; Gale, M.; Hartman, A.L.; Kimberlin, C.R.; Martínez-Sobrido, L.; Saphire, E.O.; Basler, C.F. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 2006, 80, 5168–5178. [Google Scholar] [CrossRef] [PubMed]
- Kash, J.C.; Muhlberger, E.; Carter, V.; Grosch, M.; Perwitasari, O.; Proll, S.C.; Thomas, M.J.; Weber, F.; Klenk, H.-D.; Katze, M.G. Global Suppression of the Host Antiviral Response by Ebola- and Marburgviruses: Increased Antagonism of the Type I Interferon Response Is Associated with Enhanced Virulence. J. Virol. 2006, 80, 3009–3020. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.; Josset, L.; Tchitchek, N.; Chang, J.; Belser, J.A.; Swayne, D.E.; Pantin-Jackwood, M.J.; Tumpey, T.M.; Katze, M.G. H7N9 and other pathogenic avian influenza viruses elicit a three-pronged transcriptomic signature that is reminiscent of 1918 influenza virus and is associated with lethal outcome in mice. J. Virol. 2014, 88, 10556–10568. [Google Scholar] [CrossRef] [PubMed]
- Geiss, G.K.; Salvatore, M.; Tumpey, T.M.; Carter, V.S.; Wang, X.; Basler, C.F.; Taubenberger, J.K.; Bumgarner, R.E.; Palese, P.; Katze, M.G.; et al. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 2002, 99, 10736–10741. [Google Scholar] [CrossRef] [PubMed]
- Holzer, M.; Krahling, V.; Amman, F.; Barth, E.; Bernhart, S.H.; Carmelo, V.A.; Collatz, M.; Doose, G.; Eggenhofer, F.; Ewald, J.; et al. Differential transcriptional responses to Ebola and Marburg virus infection in bat and human cells. Sci. Rep. 2016, 6, 34589. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, I.V.; Schwarz, T.M.; Ilinykh, P.A.; Jordan, I.; Ksiazek, T.G.; Sachidanandam, R.; Basler, C.F.; Bukreyev, A. Innate Immune Response of Bat and Human Cells to Filoviruses: Commonalities and Distinctions. J. Virol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hartman, A.L.; Ling, L.; Nichol, S.T.; Hibberd, M.L. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J. Virol. 2008, 82, 5348–5358. [Google Scholar] [CrossRef] [PubMed]
- Hartman, A.L.; Dover, J.E.; Towner, J.S.; Nichol, S.T. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J. Virol. 2006, 80, 6430–6440. [Google Scholar] [CrossRef] [PubMed]
- Bosworth, A.; Dowall, S.D.; Garcia-Dorival, I.; Rickett, N.Y.; Bruce, C.B.; Matthews, D.A.; Fang, Y.; Aljabr, W.; Kenny, J.; Nelson, C.; et al. A comparison of host gene expression signatures associated with infection in vitro by the Makona and Ecran (Mayinga) variants of Ebola virus. Sci. Rep. 2017, 7, 43144. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.R.; Todd, S.; Ashander, L.M.; Charitou, T.; Ma, Y.; Yeh, S.; Crozier, I.; Michael, M.Z.; Appukuttan, B.; Williams, K.A.; et al. Retinal Pigment Epithelial Cells are a Potential Reservoir for Ebola Virus in the Human Eye. Transl. Vis. Sci. Technol. 2017, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Geisbert, T.W.; Young, H.A.; Jahrling, P.B.; Davis, K.J.; Kagan, E.; Hensley, L.E. Mechanisms Underlying Coagulation Abnormalities in Ebola Hemorrhagic Fever: Overexpression of Tissue Factor in Primate Monocytes/Macrophages Is a Key Event. J. Infect. Dis. 2003, 188, 1618–1629. [Google Scholar] [CrossRef] [PubMed]
- McElroy, A.K.; Harmon, J.R.; Flietstra, T.D.; Campbell, S.; Mehta, A.K.; Kraft, C.S.; Lyon, M.G.; Varkey, J.B.; Ribner, B.S.; Kratochvil, C.J.; et al. Kinetic analysis of biomarkers in a cohort of us patients with ebola virus disease. Clin. Infect. Dis. 2016, 63, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Hayman, D.T.S.; Yu, M.; Crameri, G.; Wang, L.-F.; Suu-Ire, R.; Wood, J.L.N.; Cunningham, A.A. Ebola virus antibodies in fruit bats, ghana, west Africa. Emerg. Infect. Dis. 2012, 18, 1207–1209. [Google Scholar] [CrossRef] [PubMed]
- Towner, J.S.; Amman, B.R.; Sealy, T.K.; Carroll, S.A.R.; Comer, J.A.; Kemp, A.; Swanepoel, R.; Paddock, C.D.; Balinandi, S.; Khristova, M.L.; et al. Isolation of Genetically Diverse Marburg Viruses from Egyptian Fruit Bats. PLoS Pathog. 2009, 5, e1000536. [Google Scholar] [CrossRef] [PubMed]
- Ilinykh, P.A.; Lubaki, N.M.; Widen, S.G.; Renn, L.A.; Theisen, T.C.; Rabin, R.L.; Wood, T.G.; Bukreyev, A. Different Temporal Effects of Ebola Virus VP35 and VP24 Proteins on Global Gene Expression in Human Dendritic Cells. J. Virol. 2015, 89, 7567–7583. [Google Scholar] [CrossRef] [PubMed]
- Wahl-Jensen, V.; Kurz, S.; Feldmann, F.; Buehler, L.K.; Kindrachuk, J.; DeFilippis, V.; da Silva Correia, J.; Früh, K.; Kuhn, J.H.; Burton, D.R.; et al. Ebola virion attachment and entry into human macrophages profoundly effects early cellular gene expression. PLoS Negl. Trop. Dis. 2011, 5, e1359. [Google Scholar] [CrossRef] [PubMed]
- Olejnik, J.; Forero, A.; Deflubé, L.R.; Hume, A.J.; Manhart, W.A.; Nishida, A.; Marzi, A.; Katze, M.G.; Ebihara, H.; Rasmussen, A.L.; et al. Ebolaviruses Associated with Differential Pathogenicity Induce Distinct Host Responses in Human Macrophages. J. Virol. 2017, 91, e00179-17. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.; Davis, K.; Geisbert, T.; Schmaljohn, C.; Huggins, J. A Mouse Model for Evaluation of Prophylaxis and Therapy of Ebola Hemorrhagic Fever. Source J. Infect. Dis. 1998, 178, 651–661. [Google Scholar] [CrossRef]
- Dutta, M.; Robertson, S.J.; Okumura, A.; Scott, D.P.; Chang, J.; Weiss, J.M.; Sturdevant, G.L.; Feldmann, F.; Haddock, E.; Chiramel, A.I.; et al. Systems Approach Reveals MAVS Signaling in Myeloid Cells as Critical for Resistance to Ebola Virus in Murine Models of Infection. Cell Rep. 2017, 18, 816–829. [Google Scholar] [CrossRef] [PubMed]
- Brannan, J.M.; Froude, J.W.; Prugar, L.I.; Bakken, R.R.; Zak, S.E.; Daye, S.P.; Wilhelmsen, C.E.; Dye, J.M. Interferon α/β Receptor–Deficient Mice as a Model for Ebola Virus Disease. J. Infect. Dis. 2015, 212, S282–S294. [Google Scholar] [CrossRef] [PubMed]
- Lüdtke, A.; Oestereich, L.; Ruibal, P.; Wurr, S.; Pallasch, E.; Bockholt, S.; Ip, W.H.; Rieger, T.; Gómez-Medina, S.; Stocking, C.; et al. Ebola Virus Disease in Mice with Transplanted Human Hematopoietic Stem Cells. J. Virol. 2015, 89, 4700–4704. [Google Scholar] [CrossRef] [PubMed]
- Bird, B.H.; Spengler, J.R.; Chakrabarti, A.K.; Khristova, M.L.; Sealy, T.K.; Coleman-McCray, J.D.; Martin, B.E.; Dodd, K.A.; Goldsmith, C.S.; Sanders, J.; et al. Humanized Mouse Model of Ebola Virus Disease Mimics the Immune Responses in Human Disease. J. Infect. Dis. 2016, 213, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, E.; Saijo, M. Animal models for Ebola and Marburg virus infections. Front. Microbiol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Ebihara, H.; Takada, A.; Kobasa, D.; Jones, S.; Neumann, G.; Theriault, S.; Bray, M.; Feldmann, H.; Kawaoka, Y. Molecular Determinants of Ebola Virus Virulence in Mice. PLoS Pathog. 2006, 2, e73. [Google Scholar] [CrossRef] [PubMed]
- Caballero, I.S.; Honko, A.N.; Gire, S.K.; Winnicki, S.M.; Melé, M.; Gerhardinger, C.; Lin, A.E.; Rinn, J.L.; Sabeti, P.C.; Hensley, L.E.; et al. In Vivo Ebola virus infection leads to a strong innate response in circulating immune cells. BMC Genom. 2016. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Lukwiya, M.; Bausch, D.; Mahanty, S.; Sanchez, A.J.; Wagoner, K.D.; Rollin, P.E. Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: Cellular responses, virus load, and nitric oxide levels. J. Virol. 2004, 78, 10370–10377. [Google Scholar] [CrossRef] [PubMed]
- Fisher-Hoch, S.P.; Lloyd, G.; Platt, G.S.; Simpson, D.I.H.; Neild, G.H.; Barrett, A.J. Haematological and biochemical monitoring of ebola infection in rhesus monkeys: implications for patient management. Lancet 1983, 322, 1055–1058. [Google Scholar] [CrossRef]
- Rasmussen, A.L.; Okumura, A.; Ferris, M.T.; Green, R.; Feldmann, F.; Kelly, S.M.; Scott, D.P.; Safronetz, D.; Haddock, E.; LaCasse, R.; et al. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 2014. [Google Scholar] [CrossRef] [PubMed]
- Churchill, G.A.; Airey, D.C.; Allayee, H.; Angel, J.M.; Attie, A.D.; Beatty, J.; Beavis, W.D.; Belknap, J.K.; Bennett, B.; Berrettini, W.; et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004, 36, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Cilloniz, C.; Ebihara, H.; Ni, C.; Neumann, G.; Korth, M.J.; Kelly, S.M.; Kawaoka, Y.; Feldmann, H.; Katze, M.G. Functional genomics reveals the induction of inflammatory response and metalloproteinase gene expression during lethal Ebola virus infection. J. Virol. 2011, 85, 9060–9068. [Google Scholar] [CrossRef] [PubMed]
- Panchal, R.G.; Bradfute, S.B.; Peyser, B.D.; Warfield, K.L.; Ruthel, G.; Lane, D.; Kenny, T.A.; Anderson, A.O.; Raschke, W.C.; Bavari, S. Reduced Levels of Protein Tyrosine Phosphatase CD45 Protect Mice from the Lethal Effects of Ebola Virus Infection. Cell Host Microbe 2009, 6, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Bradfute, S.B.; Warfield, K.L.; Bray, M. Mouse models for filovirus infections. Viruses 2012, 4, 1477–1508. [Google Scholar] [CrossRef] [PubMed]
- Warfield, K.L.; Perkins, J.G.; Swenson, D.L.; Deal, E.M.; Bosio, C.M.; Aman, M.J.; Yokoyama, W.M.; Young, H.A.; Bavari, S. Role of natural killer cells in innate protection against lethal ebola virus infection. J. Exp. Med. 2004, 200, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Cimini, E.; Viola, D.; Cabeza-Cabrerizo, M.; Romanelli, A.; Tumino, N.; Sacchi, A.; Bordoni, V.; Casetti, R.; Turchi, F.; Martini, F.; et al. Different features of Vδ2 T and NK cells in fatal and non-fatal human Ebola infections. PLoS Negl. Trop. Dis. 2017, 11, e0005645. [Google Scholar] [CrossRef] [PubMed]
- Larsen, T.; Stevens, E.L.; Davis, K.J.; Geisbert, J.B.; Daddario-DiCaprio, K.M.; Jahrling, P.B.; Hensley, L.E.; Geisbert, T.W. Pathologic Findings Associated with Delayed Death in Nonhuman Primates Experimentally Infected with Zaire Ebola Virus. J. Infect. Dis. 2007, 196, S323–S328. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Blancett, C.D.; Koistinen, K.A.; Schellhase, C.W.; Bearss, J.J.; Radoshitzky, S.R.; Honnold, S.P.; Chance, T.B.; Warren, T.K.; Froude, J.W.; et al. Identification and pathological characterization of persistent asymptomatic Ebola virus infection in rhesus monkeys. Nat. Microbiol. 2017, 2, 17113. [Google Scholar] [CrossRef] [PubMed]
- Shantha, J.G.; Yeh, S.; Nguyen, Q.D. Ebola virus disease and the eye. Curr. Opin. Ophthalmol. 2016, 27, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Billioux, B.J.; Smith, B.; Nath, A. Neurological Complications of Ebola Virus Infection. Neurotherapeutics 2016, 13, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Khiabanian, H.; Kugelman, J.; Elliott, O.; Nagle, E.; Yu, G.-Y.; Warren, T.; Palacios, G.; Rabadan, R. Transcriptome reconstruction and annotation of cynomolgus and African green monkey. BMC Genom. 2014, 15, 846. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Thierry-Mieg, J.; Thierry-Mieg, D.; Nishida, A.; Pipes, L.; Bozinoski, M.; Thomas, M.J.; Kelly, S.; Weiss, J.M.; Raveendran, M.; et al. Tissue-specific transcriptome sequencing analysis expands the non-human primate reference transcriptome resource (NHPRTR). Nucleic Acids Res. 2015, 43, D737–D742. [Google Scholar] [CrossRef] [PubMed]
- Zimin, A.V.; Cornish, A.S.; Maudhoo, M.D.; Gibbs, R.M.; Zhang, X.; Pandey, S.; Meehan, D.T.; Wipfler, K.; Bosinger, S.E.; Johnson, Z.P.; et al. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol. Direct. 2014, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Rubins, K.H.; Hensley, L.E.; Wahl-Jensen, V.; Daddario DiCaprio, K.M.; Young, H.A.; Reed, D.S.; Jahrling, P.B.; Brown, P.O.; Relman, D.A.; Geisbert, T.W. The temporal program of peripheral blood gene expression in the response of nonhuman primates to Ebola hemorrhagic fever. Genome Biol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Yen, J.Y.; Garamszegi, S.; Geisbert, J.B.; Rubins, K.H.; Geisbert, T.W.; Honko, A.; Xia, Y.; Connor, J.H.; Hensley, L.E. Therapeutics of Ebola hemorrhagic fever: Whole-genome transcriptional analysis of successful disease mitigation. J. Infect. Dis. 2011. [Google Scholar] [CrossRef] [PubMed]
- Menicucci, A.R.; Sureshchandra, S.; Marzi, A.; Feldmann, H.; Messaoudi, I. Transcriptomic analysis reveals a previously unknown role for CD8(+) T-cells in rVSV-EBOV mediated protection. Sci. Rep. 2017, 7, 919. [Google Scholar] [CrossRef] [PubMed]
- Zaas, A.K.; Chen, M.; Varkey, J.; Veldman, T.; Hero, A.O.; Lucas, J.; Huang, Y.; Turner, R.; Gilbert, A.; Lambkin-Williams, R.; et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 2009. [Google Scholar] [CrossRef] [PubMed]
- Woods, C.W.; McClain, M.T.; Chen, M.; Zaas, A.K.; Nicholson, B.P.; Varkey, J.; Veldman, T.; Kingsmore, S.F.; Huang, Y.; Lambkin-Williams, R.; et al. Host Transcriptional Signature for Presymptomatic Detection of Infection in Humans Exposed to Influenza H1N1 or H3N2. PLoS ONE 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobasa, D.; Jones, S.M.; Shinya, K.; Kash, J.C.; Copps, J.; Ebihara, H.; Hatta, Y.; Hyun, K.J.; Halfmann, P.; Hatta, M.; et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 2007, 445, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Caballero, I.S.; Yen, J.Y.; Hensley, L.E.; Honko, A.N.; Goff, A.J.; Connor, J.H. Lassa and Marburg viruses elicit distinct host transcriptional responses early after infection. BMC Genom. 2014, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Connor, J.H.; Yen, J.; Caballero, I.S.; Garamszegi, S.; Malhotra, S.; Lin, K.; Hensley, L.; Goff, A.J. Transcriptional Profiling of the Immune Response to Marburg Virus Infection. J. Virol. 2015, 89, 9865–9874. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, S.; Yen, J.Y.; Honko, A.N.; Garamszegi, S.; Caballero, I.S.; Johnson, J.C.; Mucker, E.M.; Trefry, J.C.; Hensley, L.E.; Connor, J.H. Transcriptional profiling of the circulating immune response to lassa virus in an aerosol model of exposure. PLoS Negl. Trop. Dis. 2013, 7, e2171. [Google Scholar] [CrossRef] [PubMed]
- Garamszegi, S.; Yen, J.Y.; Honko, A.N.; Geisbert, J.B.; Rubins, K.H.; Geisbert, T.W.; Xia, Y.; Hensley, L.E.; Connor, J.H. Transcriptional correlates of disease outcome in anticoagulant-treated non-human primates infected with ebolavirus. PLoS Negl. Trop. Dis. 2014, 8, e3061. [Google Scholar] [CrossRef] [PubMed]
- Rhein, B.A.; Powers, L.S.; Rogers, K.; Anantpadma, M.; Singh, B.K.; Sakurai, Y.; Bair, T.; Miller-Hunt, C.; Sinn, P.; Davey, R.A.; et al. Interferon-γ Inhibits Ebola Virus Infection. PLoS Pathog. 2015, 11, e1005263. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Greer, P.; Mahanty, S.; Shieh, W.-J.; Zaki, S.R.; Ahmed, R.; Rollin, P.E. CD8-Mediated Protection against Ebola Virus Infection Is Perforin Dependent. J. Immunol. 2005, 174, 4198–4202. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.M.; Feldmann, H.; Stroher, U.; Geisbert, J.B.; Fernando, L.; Grolla, A.; Klenk, H.D.; Sullivan, N.J.; Volchkov, V.E.; Fritz, E.A.; et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 2005, 11. [Google Scholar] [CrossRef] [PubMed]
- Geisbert, T.W.; Feldmann, H. Recombinant vesicular stomatitis virus-based vaccines against Ebola and Marburg virus infections. J. Infect. Dis. 2011, 204, S1075–S1081. [Google Scholar] [CrossRef] [PubMed]
- Marzi, A.; Feldmann, H.; Geisbert, T.W.; Falzarano, D. Vesicular Stomatitis Virus-Based Vaccines for Prophylaxis and Treatment of Filovirus Infections. J. Bioterror. Biodef. 2011. [Google Scholar] [CrossRef] [PubMed]
- Henao-Restrepo, A.M.; Camacho, A.; Longini, I.M.; Watson, C.H.; Edmunds, W.J.; Egger, M.; Carroll, M.W.; Dean, N.E.; Diatta, I.; Doumbia, M.; et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!). Lancet 2017, 389, 505–518. [Google Scholar] [CrossRef]
- Barrenas, F.; Green, R.R.; Thomas, M.J.; Law, G.L.; Proll, S.C.; Engelmann, F.; Messaoudi, I.; Marzi, A.; Feldmann, H.; Katze, M.G. Next generation sequencing reveals a controlled immune response to Zaire Ebola virus challenge in cynomolgus macaques immunized with VSVΔG/EBOVgp. Clin. Vaccine Immunol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Speranza, E.; Muñoz-Fontela, C.; Haldenby, S.; Rickett, N.Y.; Garcia-Dorival, I.; Fang, Y.; Hall, Y.; Zekeng, E.-G.; Lüdtke, A.; et al. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol. 2017, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Kash, J.C.; Walters, K.-A.; Kindrachuk, J.; Baxter, D.; Scherler, K.; Janosko, K.B.; Adams, R.D.; Herbert, A.S.; James, R.M.; Stonier, S.W.; et al. Longitudinal peripheral blood transcriptional analysis of a patient with severe Ebola virus disease. Sci. Transl. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ellebedy, A.H.; Jackson, K.J.L.; Kissick, H.T.; Nakaya, H.I.; Davis, C.W.; Roskin, K.M.; McElroy, A.K.; Oshansky, C.M.; Elbein, R.; Thomas, S.; et al. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol. 2016, 17, 1226–1234. [Google Scholar] [CrossRef] [PubMed]
- Duy, J.; Koehler, J.W.; Honko, A.N.; Schoepp, R.J.; Wauquier, N.; Gonzalez, J.-P.; Pitt, M.L.; Mucker, E.M.; Johnson, J.C.; O’Hearn, A.; et al. Circulating microRNA profiles of Ebola virus infection. Sci. Rep. 2016, 6, 24496. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Hensley, L.E.; Geisbert, T.W.; Johnson, J.; Stossel, A.; Honko, A.; Yen, J.Y.; Geisbert, J.; Paragas, J.; Fritz, E.; et al. Interferon-β Therapy Prolongs Survival in Rhesus Macaque Models of Ebola and Marburg Hemorrhagic Fever. J. Infect. Dis. 2013, 208, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Konde, M.K.; Baker, D.P.; Traore, F.A.; Sow, M.S.; Camara, A.; Barry, A.A.; Mara, D.; Barry, A.; Cone, M.; Kaba, I.; et al. Interferon β-1a for the treatment of Ebola virus disease: A historically controlled, single-arm proof-of-concept trial. PLoS ONE 2017, 12, e0169255. [Google Scholar] [CrossRef] [PubMed]
- Broadhurst, M.J.; Brooks, T.J.G.; Pollock, N.R. Diagnosis of Ebola Virus Disease: Past, Present, and Future. Clin. Microbiol. Rev. 2016, 29, 773–793. [Google Scholar] [CrossRef] [PubMed]
- Tsalik, E.L.; Henao, R.; Nichols, M.; Burke, T.; Ko, E.R.; McClain, M.T.; Hudson, L.L.; Mazur, A.; Freeman, D.H.; Veldman, T.; et al. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci. Transl. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Sampson, D.L.; Fox, B.A.; Yager, T.D.; Bhide, S.; Cermelli, S.; McHugh, L.C.; Seldon, T.A.; Brandon, R.A.; Sullivan, E.; Zimmerman, J.J.; et al. A Four-Biomarker Blood Signature Discriminates Systemic Inflammation Due to Viral Infection Versus Other Etiologies. Sci. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Navalkar, K.; Kim, T.-K.; Gelinas, R. Pre-Symptomatic Diagnosis of Ebola Virus Infection. J. Emerg. Virol. Dis. 2013, 3. [Google Scholar] [CrossRef]
- Scott, J.T.; Sesay, F.R.; Massaquoi, T.A.; Idriss, B.R.; Sahr, F.; Semple, M.G. Post-Ebola Syndrome, Sierra Leone. Emerg. Infect. Dis. 2016, 22, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Varkey, J.B.; Shantha, J.G.; Crozier, I.; Kraft, C.S.; Lyon, G.M.; Mehta, A.K.; Kumar, G.; Smith, J.R.; Kainulainen, M.H.; Whitmer, S.; et al. Persistence of Ebola Virus in Ocular Fluid during Convalescence. N. Engl. J. Med. 2015, 372, 2423–2427. [Google Scholar] [CrossRef] [PubMed]

| Gene Group | Immortalized Cells | Primary Cells | Mouse | NHP | Human |
|---|---|---|---|---|---|
| ISGs | Suppressed in most cell lines | Upregulated in Macrophages | Upregulated in spleens | Early Response Upregulated | Upregulated |
| Pro-inflammatory Cytokines | Mostly suppressed | Upregulated in Macrophages | Upregulated | Mid Response Upregulated | Upregulated |
| Coagulation | Suppressed | Upregulated | |||
| Neutrophil associated Transcripts | N/A | N/A | Upregulated in spleens | Upregulated in PBMCs | No Significant Change |
| Natural Killer Cell Transcripts | N/A | N/A | Upregulated in Surviving Animals | No Significant Change | Upregulated in Survivors |
| Acute Phase Response | Suppressed | N/A | N/A | Upregulated | Upregulated |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Speranza, E.; Connor, J.H. Host Transcriptional Response to Ebola Virus Infection. Vaccines 2017, 5, 30. https://doi.org/10.3390/vaccines5030030
Speranza E, Connor JH. Host Transcriptional Response to Ebola Virus Infection. Vaccines. 2017; 5(3):30. https://doi.org/10.3390/vaccines5030030
Chicago/Turabian StyleSperanza, Emily, and John H Connor. 2017. "Host Transcriptional Response to Ebola Virus Infection" Vaccines 5, no. 3: 30. https://doi.org/10.3390/vaccines5030030





