Systems Biology Approach for Cancer Vaccine Development and Evaluation
Abstract
:1. Introduction
2. System Vaccinology
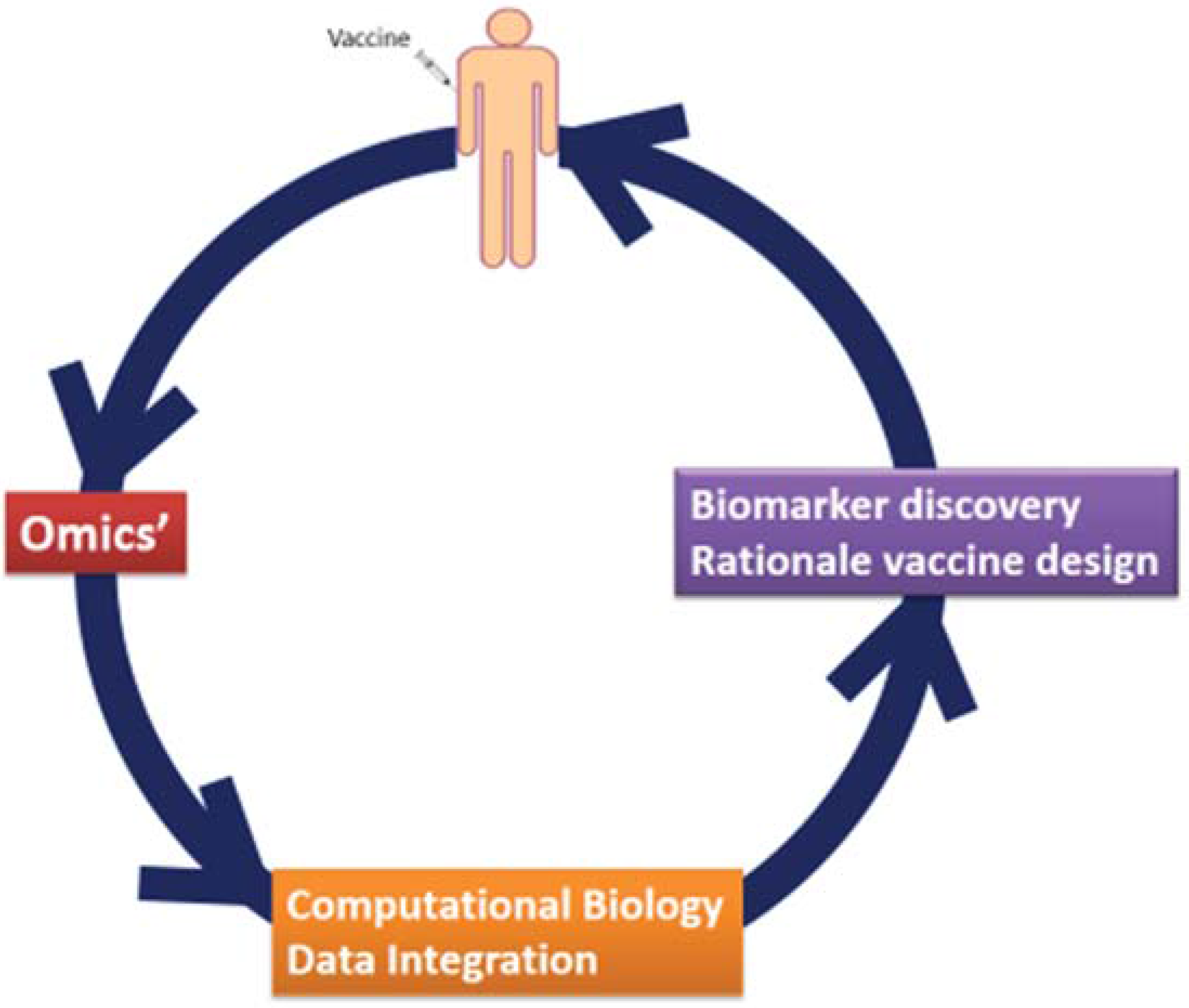
3. System Vaccinology for Infectious Disease Vaccines
4. Biosignatures for Vaccine Safety and Efficacy
5. Systems Vaccinology for Cancer Vaccines Application
5.1. Identification of Tumor Associated Antigens (TAAs)

5.2. Identification of Tumor Associated Antigens (Mutanome)
5.3. Immune Monitoring
6. Conclusions
Acknowledgments
Author Contributions
Conflict of Interests
List of Abbreviations
| Ad5 – Adenovirus 5 |
| CRC – colorectal cancer |
| ELISA – enzyme-linked immunosorbent assay |
| ELISPOT – Enzyme-Linked ImmunoSpot |
| GB – glioblastoma |
| GCSPRI – grating-coupled surface plasmon resonance imaging |
| HepB – Hepatitis B virus vaccine |
| HLA – human leukocyte antigen |
| IFNγ – interferon gamma |
| MHC – major histocompatibility complex |
| MMR-II – measles, mumps, and rubella vaccine |
| MS – Mass-spectrometry |
| PBMC – peripheral blood mononuclear cells |
| RCC – renal cell cancer |
| SNP – single nucleotide polymorphism |
| SPCE – surface plasmon coupled emission |
| TAAs – tumor-associated antigens |
| TCR – T-cell receptor |
| TLR – Toll-like receptor |
| TILs – tumor infiltrating lymphocytes |
References
- Koff, W.C.; Burton, D.R.; Johnson, P.R.; Walker, B.D.; King, C.R.; Nabel, G.J.; Ahmed, R.; Bhan, M.K.; Plotkin, S.A. Accelerating next-generation vaccine development for global disease prevention. Science 2013. [Google Scholar] [CrossRef] [PubMed]
- Nabel, G.J. Designing tomorrow’s vaccines. N. Engl. J. Med. 2013, 368, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Strategic Advisory Group of Experts on immunization. 2014 Assessment report of the Global Vaccine action plan. 2014. Available online: http://www.who.int/immunization/global_vaccine_action_plan/SAGE_DoV_GVAP_Assessment_report_2014_English.pdf?ua=1 (accessed on 1 May 2015).
- Minor, P.D. Live attenuated vaccines: Historical successes and current challenges. Virology 2015, 479 480, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Delany, I.; Rappuoli, R.; de Gregorio, E. Vaccines for the 21st century. EMBO Mol. Med. 2014, 6, 708–720. [Google Scholar] [CrossRef] [PubMed]
- Buonaguro, L.; Tornesello, M.L.; Buonaguro, F.M. Virus-like particles as particulate vaccines. Curr. HIV Res. 2010, 8, 299–399. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; Ricci, E.P.; Mercier, B.C.; Moore, M.J.; Fitzgerald, K.A. Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol. 2014, 14, 361–376. [Google Scholar] [CrossRef] [PubMed]
- Gantier, M.P. New perspectives in MicroRNA regulation of innate immunity. J. Interferon Cytokine Res. 2010, 30, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Kominsky, D.J.; Campbell, E.L.; Colgan, S.P. Metabolic shifts in immunity and inflammation. J. Immunol. 2010, 184, 4062–4068. [Google Scholar] [CrossRef] [PubMed]
- Bensinger, S.J.; Tontonoz, P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 2008, 454, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Hagan, T.; Nakaya, H.I.; Subramaniam, S.; Pulendran, B. Systems vaccinology: Enabling rational vaccine design with systems biological approaches. Vaccine 2015. [Google Scholar] [CrossRef] [PubMed]
- Buonaguro, L.; Pulendran, B. Immunogenomics and systems biology of vaccines. Immunol. Rev. 2011, 239, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Petrizzo, A.; Tagliamonte, M.; Tornesello, M.L.; Buonaguro, F.M.; Buonaguro, L. Prediction of individual immune responsiveness to a candidate vaccine by a systems vaccinology approach. J. Transl. Med. 2014. [Google Scholar] [CrossRef] [PubMed]
- Oberg, A.L.; McKinney, B.A.; Schaid, D.J.; Pankratz, V.S.; Kennedy, R.B.; Poland, G.A. Lessons learned in the analysis of high-dimensional data in vaccinomics. Vaccine 2015. [Google Scholar] [CrossRef] [PubMed]
- Poland, G.A.; Ovsyannikova, I.G.; Kennedy, R.B.; Lambert, N.D.; Kirkland, J.L. A systems biology approach to the effect of aging, immunosenescence and vaccine response. Curr. Opin. Immunol. 2014, 29, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Querec, T.D.; Akondy, R.S.; Lee, E.K.; Cao, W.; Nakaya, H.I.; Teuwen, D.; Pirani, A.; Gernert, K.; Deng, J.; Marzolf, B.; et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009, 10, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, D.; Therrien, R.; Kettaf, N.; Angermann, B.R.; Boucher, G.; Filali-Mouhim, A.; Moser, J.M.; Mehta, R.S.; Drake, D.R., III; Castro, E.; et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 2008, 205, 3119–3131. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, H.I.; Wrammert, J.; Lee, E.K.; Racioppi, L.; Marie-Kunze, S.; Haining, W.N.; Means, A.R.; Kasturi, S.P.; Khan, N.; Li, G.M.; et al. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011, 12, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Bucasas, K.L.; Franco, L.M.; Shaw, C.A.; Bray, M.S.; Wells, J.M.; Nino, D.; Arden, N.; Quarles, J.M.; Couch, R.B.; Belmont, J.W. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J. Infect. Dis. 2011, 203, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Vahey, M.T.; Wang, Z.; Kester, K.E.; Cummings, J.; Heppner, D.G., Jr.; Nau, M.E.; Ofori-Anyinam, O.; Cohen, J.; Coche, T.; Ballou, W.R.; et al. Expression of genes associated with immunoproteasome processing of major histocompatibility complex peptides is indicative of protection with adjuvanted RTS, S malaria vaccine. J. Infect. Dis. 2010, 201, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Andersen-Nissen, E.; Heit, A.; McElrath, M.J. Profiling immunity to HIV vaccines with systems biology. Curr. Opin. HIV AIDS 2012, 7, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Monaco, A.; Marincola, F.M.; Sabatino, M.; Pos, Z.; Tornesello, M.L.; Stroncek, D.F.; Wang, E.; Lewis, G.K.; Buonaguro, F.M.; Buonaguro, L. Molecular immune signatures of HIV-1 vaccines in human PBMCs. FEBS Lett. 2009, 583, 3004–3008. [Google Scholar] [CrossRef] [PubMed]
- Buonaguro, L.; Monaco, A.; Arico, E.; Wang, E.; Tornesello, M.L.; Lewis, G.K.; Marincola, F.M.; Buonaguro, F.M. Gene expression profile of peripheral blood mononuclear cells in response to HIV-VLPs stimulation. BMC. Bionformatics 2008. [Google Scholar] [CrossRef] [PubMed]
- Obermoser, G.; Presnell, S.; Domico, K.; Xu, H.; Wang, Y.; Anguiano, E.; Thompson-Snipes, L.; Ranganathan, R.; Zeitner, B.; Bjork, A.; et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity 2013, 38, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.M.; Bett, A.J.; Cristescu, R.; Loboda, A.; Ter, M.J. Transcriptional profiling of vaccine-induced immune responses in humans and non-human primates. Microb. Biotechnol. 2012, 5, 177–187. [Google Scholar] [CrossRef] [PubMed]
- O'Connor, D.; Pollard, A.J. Characterizing vaccine responses using host genomic and transcriptomic analysis. Clin. Infect. Dis. 2013, 57, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, N.; Ovsyannikova, I.G.; Vierkant, R.A.; Ryan, J.E.; Pankratz, V.S.; Jacobson, R.M.; Poland, G.A. Associations between SNPs in toll-like receptors and related intracellular signaling molecules and immune responses to measles vaccine: Preliminary results. Vaccine 2008, 26, 1731–1736. [Google Scholar] [CrossRef] [PubMed]
- Ovsyannikova, I.G.; Dhiman, N.; Haralambieva, I.H.; Vierkant, R.A.; O'Byrne, M.M.; Jacobson, R.M.; Poland, G.A. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Hum. Genet. 2010, 127, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Banus, S.; Bottema, R.W.; Siezen, C.L.; Vandebriel, R.J.; Reimerink, J.; Mommers, M.; Koppelman, G.H.; Hoebee, B.; Thijs, C.; Postma, D.S.; et al. Toll-like receptor 4 polymorphism associated with the response to whole-cell pertussis vaccination in children from the KOALA study. Clin. Vaccine Immunol. 2007, 14, 1377–1380. [Google Scholar] [CrossRef] [PubMed]
- Kimman, T.G.; Banus, S.; Reijmerink, N.; Reimerink, J.; Stelma, F.F.; Koppelman, G.H.; Thijs, C.; Postma, D.S.; Kerkhof, M. Association of interacting genes in the toll-like receptor signaling pathway and the antibody response to pertussis vaccination. PLoS One 2008, 3, e3665. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Pastorino, R.; Di, G.P.; Ianuale, C.; Amore, R.; Ricciardi, W.; Boccia, S. The link between genetic variation and variability in vaccine responses: systematic review and meta-analyses. Vaccine 2014, 32, 1661–1669. [Google Scholar] [CrossRef] [PubMed]
- Gray, G.E.; Allen, M.; Moodie, Z.; Churchyard, G.; Bekker, L.G.; Nchabeleng, M.; Mlisana, K.; Metch, B.; de Bruyn, G.; Latka, M.H.; et al. Safety and efficacy of the HVTN 503/Phambili study of a clade-B-based HIV-1 vaccine in South Africa: A double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet Infect. Dis. 2011, 11, 507–515. [Google Scholar] [CrossRef]
- Tameris, M.D.; Hatherill, M.; Landry, B.S.; Scriba, T.J.; Snowden, M.A.; Lockhart, S.; Shea, J.E.; McClain, J.B.; Hussey, G.D.; Hanekom, W.A.; et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet 2013, 381, 1021–1028. [Google Scholar] [CrossRef]
- Sabchareon, A.; Wallace, D.; Sirivichayakul, C.; Limkittikul, K.; Chanthavanich, P.; Suvannadabba, S.; Jiwariyavej, V.; Dulyachai, W.; Pengsaa, K.; Wartel, T.A.; et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: A randomised, controlled phase 2b trial. Lancet 2012, 380, 1559–1567. [Google Scholar] [CrossRef]
- Zak, D.E.; Andersen-Nissen, E.; Peterson, E.R.; Sato, A.; Hamilton, M.K.; Borgerding, J.; Krishnamurty, A.T.; Chang, J.T.; Adams, D.J.; Hensley, T.R.; et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8+ T-cell responses but is attenuated by preexisting Ad5 immunity. Proc. Natl. Acad. Sci. USA 2012, 109, E3503–E3512. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, E. The makings of a tumor rejection antigen. Immunity 1999, 11, 263–270. [Google Scholar] [CrossRef]
- Buonaguro, L.; Petrizzo, A.; Tornesello, M.L.; Buonaguro, F.M. Translating tumor antigens into cancer vaccines. Clin. Vaccine Immunol. 2011, 18, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. Inducing autoimmune disease to treat cancer. Proc. Natl. Acad. Sci. USA 1999, 96, 5340–5342. [Google Scholar] [CrossRef] [PubMed]
- Nambiar, P.R.; Gupta, R.R.; Misra, V. An "Omics" based survey of human colon cancer. Mutat. Res. 2010, 693, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Habermann, J.K.; Paulsen, U.; Roblick, U.J.; Upender, M.B.; McShane, L.M.; Korn, E.L.; Wangsa, D.; Kruger, S.; Duchrow, M.; Bruch, H.P.; et al. Stage-specific alterations of the genome, transcriptome, and proteome during colorectal carcinogenesis. Genes Chromosomes Cancer 2007, 46, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Habermann, J.K.; Bundgen, N.K.; Gemoll, T.; Hautaniemi, S.; Lundgren, C.; Wangsa, D.; Doering, J.; Bruch, H.P.; Nordstroem, B.; Roblick, U.J.; et al. Genomic instability influences the transcriptome and proteome in endometrial cancer subtypes. Mol. Cancer 2011. [Google Scholar] [CrossRef] [PubMed]
- Rammensee, H.; Bachmann, J.; Emmerich, N.P.; Bachor, O.A.; Stevanovic, S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 1999, 50, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Patronov, A.; Doytchinova, I. T-cell epitope vaccine design by immunoinformatics. Open. Biol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Singh-Jasuja, H.; Emmerich, N.P.; Rammensee, H.G. The Tubingen approach: Identification, selection, and validation of tumor-associated HLA peptides for cancer therapy. Cancer Immunol. Immunother. 2004, 53, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Dutoit, V.; Herold-Mende, C.; Hilf, N.; Schoor, O.; Beckhove, P.; Bucher, J.; Dorsch, K.; Flohr, S.; Fritsche, J.; Lewandrowski, P.; et al. Exploiting the glioblastoma peptidome to discover novel tumour-associated antigens for immunotherapy. Brain 2012, 135, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Rammensee, H.G.; Singh-Jasuja, H. HLA ligandome tumor antigen discovery for personalized vaccine approach. Expert Rev. Vaccines 2013, 12, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Castle, J.C.; Kreiter, S.; Diekmann, J.; Lower, M.; van de, R.N.; de, G.J.; Selmi, A.; Diken, M.; Boegel, S.; Paret, C.; et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012, 72, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Overwijk, W.W.; Wang, E.; Marincola, F.M.; Rammensee, H.G.; Restifo, N.P. Mining the mutanome: developing highly personalized Immunotherapies based on mutational analysis of tumors. J. Immunother. Cancer 2013. [Google Scholar] [CrossRef] [PubMed]
- Heemskerk, B.; Kvistborg, P.; Schumacher, T.N. The cancer antigenome. EMBO J. 2013, 32, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Gubin, M.M.; Zhang, X.; Schuster, H.; Caron, E.; Ward, J.P.; Noguchi, T.; Ivanova, Y.; Hundal, J.; Arthur, C.D.; Krebber, W.J.; et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014, 515, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Van, R.N.; van Buuren, M.M.; Philips, D.; Velds, A.; Toebes, M.; Heemskerk, B.; van Dijk, L.J.; Behjati, S.; Hilkmann, H.; El, A.D.; et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 2013, 31, e439–e442. [Google Scholar]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Li, M.; Zhang, X.; Jones, S.; Leary, R.J.; Lin, J.C.; Boca, S.M.; Carter, H.; Samayoa, J.; Bettegowda, C.; et al. The genetic landscape of the childhood cancer medulloblastoma. Science 2011, 331, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Jiang, Z.; Liu, J.; Haverty, P.M.; Guan, Y.; Stinson, J.; Yue, P.; Zhang, Y.; Pant, K.P.; Bhatt, D.; et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 2010, 465, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Timmermann, B.; Kerick, M.; Roehr, C.; Fischer, A.; Isau, M.; Boerno, S.T.; Wunderlich, A.; Barmeyer, C.; Seemann, P.; Koenig, J.; et al. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS ONE 2010, 5, e15661. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Walia, V.; Lin, J.C.; Teer, J.K.; Prickett, T.D.; Gartner, J.; Davis, S.; Stemke-Hale, K.; Davies, M.A.; Gershenwald, J.E.; et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 2011, 43, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Disis, M.L. Immunologic biomarkers as correlates of clinical response to cancer immunotherapy. Cancer Immunol. Immunother. 2011, 60, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Keilholz, U.; Martus, P.; Scheibenbogen, C. Immune monitoring of T-cell responses in cancer vaccine development. Clin. Cancer Res. 2006, 12, 2346s–2352s. [Google Scholar] [CrossRef] [PubMed]
- Gratama, J.W.; Kern, F.; Manca, F.; Roederer, M. Measuring antigen-specific immune responses, 2008 update. Cytometry A 2008, 73, 971–974. [Google Scholar] [CrossRef] [PubMed]
- Moodie, Z.; Price, L.; Gouttefangeas, C.; Mander, A.; Janetzki, S.; Lower, M.; Welters, M.J.; Ottensmeier, C.; van der Burg, S.H.; Britten, C.M. Response definition criteria for ELISPOT assays revisited. Cancer Immunol. Immunother. 2010, 59, 1489–1501. [Google Scholar] [CrossRef] [PubMed]
- Maecker, H.T.; McCoy, J.P., Jr.; Amos, M.; Elliott, J.; Gaigalas, A.; Wang, L.; Aranda, R.; Banchereau, J.; Boshoff, C.; Braun, J.; et al. A model for harmonizing flow cytometry in clinical trials. Nat. Immunol. 2010, 11, 975–978. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Wagner, K.; Wolchok, J.D.; Allison, J.P. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nat. Rev. Cancer 2011, 11, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Freeman, J.D.; Warren, R.L.; Webb, J.R.; Nelson, B.H.; Holt, R.A. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009, 19, 1817–1824. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.L.; Freeman, J.D.; Zeng, T.; Choe, G.; Munro, S.; Moore, R.; Webb, J.R.; Holt, R.A. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome. Res. 2011, 21, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Klinger, M.; Kong, K.; Moorhead, M.; Weng, L.; Zheng, J.; Faham, M. Combining next-generation sequencing and immune assays: A novel method for identification of antigen-specific T cells. PLoS ONE 2013, 8, e74231. [Google Scholar] [CrossRef] [PubMed]
- Soen, Y.; Chen, D.S.; Kraft, D.L.; Davis, M.M.; Brown, P.O. Detection and characterization of cellular immune responses using peptide-MHC microarrays. PLoS Biol. 2003, 1, e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, C.; Oelke, M.; Paulaitis, M.E.; Schneck, J.P. Novel cellular microarray assay for profiling T-cell peptide antigen specificities. J. Proteome. Res. 2010, 9, 5629–5637. [Google Scholar] [CrossRef] [PubMed]
- Rice, J.M.; Stern, L.J.; Guignon, E.F.; Lawrence, D.A.; Lynes, M.A. Antigen-specific T cell phenotyping microarrays using grating coupled surface plasmon resonance imaging and surface plasmon coupled emission. Biosens. Bioelectron. 2012, 31, 264–269. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Circelli, L.; Petrizzo, A.; Tagliamonte, M.; Tornesello, M.L.; Buonaguro, F.M.; Buonaguro, L. Systems Biology Approach for Cancer Vaccine Development and Evaluation. Vaccines 2015, 3, 544-555. https://doi.org/10.3390/vaccines3030544
Circelli L, Petrizzo A, Tagliamonte M, Tornesello ML, Buonaguro FM, Buonaguro L. Systems Biology Approach for Cancer Vaccine Development and Evaluation. Vaccines. 2015; 3(3):544-555. https://doi.org/10.3390/vaccines3030544
Chicago/Turabian StyleCircelli, Luisa, Annacarmen Petrizzo, Maria Tagliamonte, Maria Lina Tornesello, Franco M. Buonaguro, and Luigi Buonaguro. 2015. "Systems Biology Approach for Cancer Vaccine Development and Evaluation" Vaccines 3, no. 3: 544-555. https://doi.org/10.3390/vaccines3030544





