1. Introduction
Influenza A virus in swine (IAV-S) is considered one of the most important infectious disease agents affecting North American swine [
1]. The financial impact of IAV-S to the pork industry comes mainly from reduction in the growth rate of infected pigs [
2]. Additionally, IAV-S is a zoonotic pathogen which can be transmitted between people and pigs with consequences to public health. Farm workers or other persons in contact with livestock may become infected with IAV-S or, conversely, they may transmit human IAV to swine [
3,
4]. It is possible for an IAV-S strain to be adapted to the human host well enough to spread between humans and initiate a pandemic. A version of this occurred in 2009, leading to the H1N1 influenza pandemic. During that time, public misperceptions about the safety of eating pork caused economic losses to the US industry estimated at over $1 billion [
5].
Considering the important consequences of IAV-S, effective control measures are highly desirable. A range of farm management practices can help limit the circulation of IAV-S in herds, including the use of vaccines in sows, nursery pigs, or finishers. Swine veterinarians and producers are likely aware that IAV-S vaccines sometimes have disappointing efficacy in the field [
6]. However, under the right conditions vaccines can reduce or eliminate transmission of IAV-S in herds [
7,
8]. Successful IAV-S vaccination can improve herd health and lower the risk of transmission to other species, including humans. This document was developed to provide veterinarians with a scientifically-based overview of IAV-S biology, immunity to the virus, vaccines currently available, and technologies that show promise for the future. It also describes strategies to optimize the use of available IAV-S vaccines for a given herd, with the aid of modern diagnostics and surveillance programs.
2. Influenza A Virus Structure and Function
Influenza A viruses (family Orthomyxoviridae) are highly variable, enveloped viruses with negative-sense, single-stranded, segmented RNA genomes. Most influenza A viruses circulate among birds, particularly species that live in aquatic environments [
9,
10]. Some strains are adapted for efficient replication and sustained transmission in particular mammalian species, including humans and swine [
9,
11,
12,
13,
14]. Once a virus has become adapted to the swine host, it is considered to be IAV-S, regardless of its ancestral origins.
2.1. Roles and Functions of Influenza A Virus Proteins
The influenza A virus genome consists of eight segments, which encode for at least 12 proteins (
Table 1) [
15,
16]. Three of these proteins—the viral hemagglutinin (HA), neuraminidase (NA), and matrix 2 (M2) proteins—are incorporated into the envelope of the virus (
Figure 1). The HA and NA are glycoproteins with stem and head structures that protrude from the surface of the virus. The HA is the most abundant of the envelope proteins, up to 80% of the total [
15].
Table 1.
Influenza virus proteins.
Table 1.
Influenza virus proteins.
| Abbreviation | Protein | Location | Significance in IAV-S Vaccines |
|---|
| HA | Hemagglutinin | Envelope | Subtype of HA in inactivated virus vaccine must match challenge virus. Vaccines with more closely related HA are more likely to be protective. |
| NA | Neuraminidase | Envelope | NA content in vaccines is not standardized, but studies indicate a role in protection [17,18,19,20]. |
| M2 | Matrix 2 | Envelope | Experimental vaccines were directed against highly conserved M2 [21,22]. |
| M1 | Matrix 1 | Internal | |
| NP | Nucleoprotein | Internal | |
| NS1 | Nonstructural protein 1 | Nonstructural | Gene deletion in NS1confers attenuation in a candidate vaccine [23,24]. |
| NS2/ NEP | Nonstructural protein 2/Nuclear export protein | Nonstructural or internal | |
| PA | Polymerase acidic protein | Internal | |
| PB1 | Polymerase basic protein 1 | Internal | Mutations in PB1 and PB2 confer attenuation in a candidate vaccine [25]. |
| PB1-F2 | | Internal | |
| PB2 | Polymerase basic protein 2 | Internal | Mutations in PB1 and PB2 confer attenuation in a candidate vaccine [25]. |
2.1.1. Influenza A Virus Hemagglutinin
The HA protein is responsible for binding influenza virions to host cells [
14,
15]. While binding is complex and is still incompletely understood [
26,
27], the HA interacts initially with sialic acids that are linked to host cell proteins (
Figure 1) [
15,
26,
27]. Influenza A viruses adapted to either people or swine tend to preferentially bind to sialic acid receptors that have α(2,6) linkages with the carbohydrate moiety [
26,
27,
28]. In contrast, most avian influenza viruses have a binding preference for α(2,3) linked sialic acids. The binding preference of each influenza virus tends to match the predominant receptors in the host tissues where it normally replicates. Most of the sialic acid receptors in the nasal epithelium, trachea and bronchi of the human respiratory tract are of the α(2,6) linked form [
26]. The absence of α(2,3) sialic acid linkages in the upper respiratory tract is thought to limit the binding and person-to-person transmission of avian influenza viruses. The types of influenza virus receptors in the pig are still incompletely understood. One widely quoted study reported that both α(2,3) and α(2,6) linked sialic acids occur in the upper respiratory tract of swine [
29]. Some newer studies suggest that the pattern may instead resemble that of humans [
28,
30,
31]. Once the influenza virion has bound to the cell surface, it enters the host cell in an endosome [
15]. The acidic pH in the endosome triggers a conformational change in the HA, which results in the fusion of the viral and endosomal membranes.
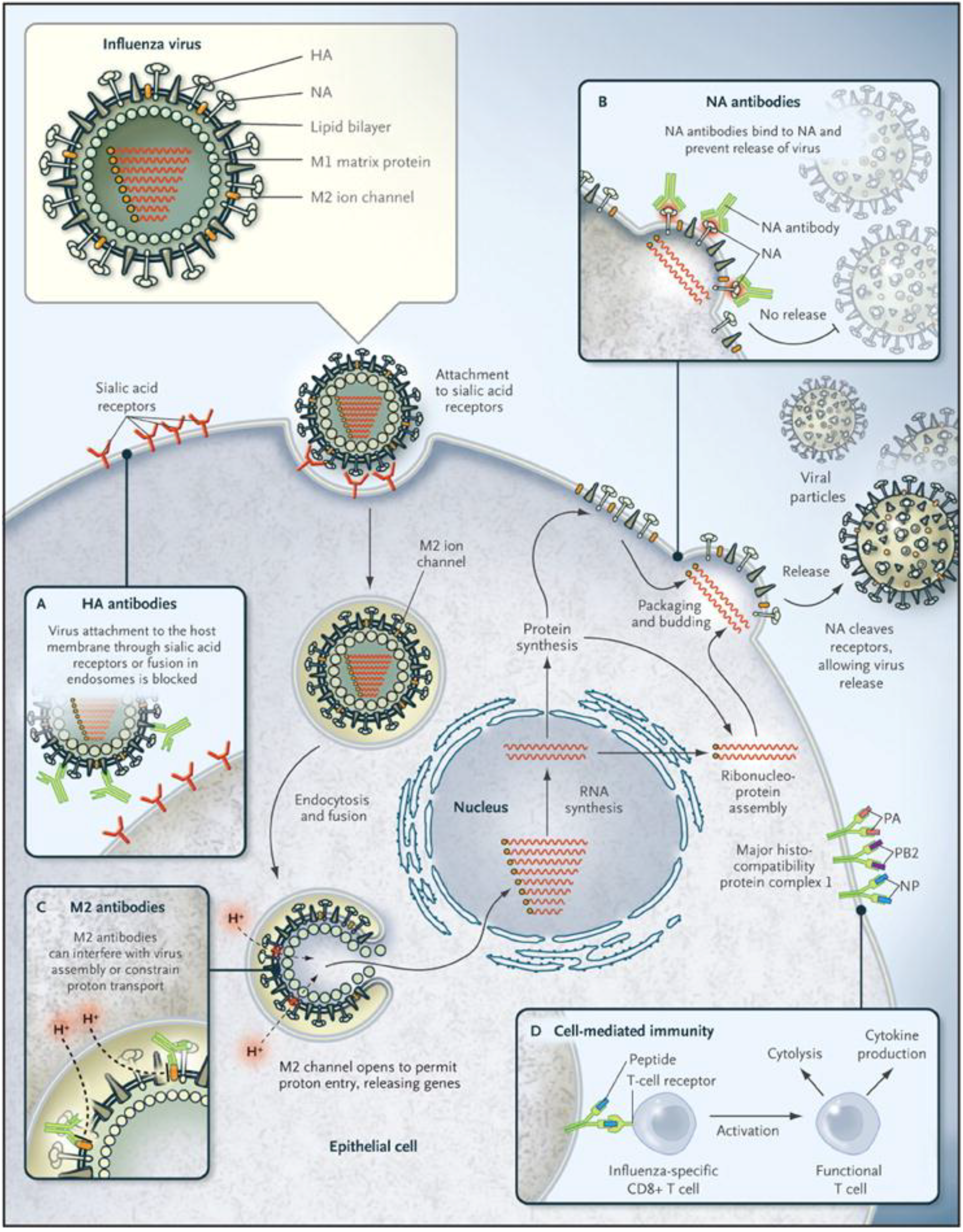
Figure 1.
Influenza virus infection cycle. Basic structural features of an influenza virus are diagrammed in the top left corner. Infection begins with the binding of hemagglutinin (HA) proteins to receptor molecules on the cell surface. The cycle is completed when new particles, each containing eight RNA segments, bud off from the cell membrane. Neuraminidase (NA) protein cleaves the bonds between HA and sialic acid molecules, allowing new virus to disperse. Boxes labeled A–D indicate points in the cycle that may be inhibited by antibodies or T cells. (Figure used with permission from the New England Journal of Medicine, Linda C. Lambert and Anthony S. Fauci, Influenza Vaccines for the Future, Vol. 363:2039. © 2010 Massachusetts Medical Society).
Figure 1.
Influenza virus infection cycle. Basic structural features of an influenza virus are diagrammed in the top left corner. Infection begins with the binding of hemagglutinin (HA) proteins to receptor molecules on the cell surface. The cycle is completed when new particles, each containing eight RNA segments, bud off from the cell membrane. Neuraminidase (NA) protein cleaves the bonds between HA and sialic acid molecules, allowing new virus to disperse. Boxes labeled A–D indicate points in the cycle that may be inhibited by antibodies or T cells. (Figure used with permission from the New England Journal of Medicine, Linda C. Lambert and Anthony S. Fauci, Influenza Vaccines for the Future, Vol. 363:2039. © 2010 Massachusetts Medical Society).
2.1.2. Other Influenza Surface Proteins: Neuraminidase and Matrix 2
The function of the influenza A virus NA protein is to cleave sialic acids on host cells, allowing newly made virions to be released efficiently from the infected cell (
Figure 1) [
14,
15]. This protein is required for influenza A viruses to spread efficiently [
14], and antibodies to it are likely to impede the spread of virus to uninfected cells. The enzymatic activity of the NA might also facilitate the infection of cells, by helping the virion penetrate the blanket of mucus in the respiratory tract or the glycocalyx surrounding the cell [
32].
The transmembrane matrix 2 (M2) protein is a proton-selective ion channel, and is required for efficient uncoating of influenza A viruses [
14]. M2 acidifies the viral core after the virus enters the host cell, allowing the viral ribonucleoprotein complexes (vRNPs) containing the gene segments to be released into the cytoplasm (
Figure 1) [
15].
2.1.3. Internal Proteins of Influenza A Viruses
The internal proteins of influenza A viruses have structural roles, are involved in nucleic acid replication, alter antiviral responses in the host cell, and influence virulence [
14,
15,
16,
33]. Matrix 1 (M1) is an abundant protein that forms the internal matrix of the virion [
15,
33]. During replication, it is thought to be important in virus assembly and budding [
14]. The viral ribonucleoprotein complexes, contained within the matrix, consist of viral RNA wrapped around the nucleoprotein (NP) [
15,
33]. The vRNPs associate with the three polymerase proteins, polymerase acidic protein (PA), polymerase basic protein 1 (PB1) and polymerase basic protein 2 (PB2) [
33]. Once the virus uncoats, the vRNPs are transported to the nucleus of the host cell, where the viral RNA-dependent RNA polymerase carries out replication of the genome. Nuclear export protein has an important role in exporting newly made viral RNA from the nucleus [
14,
33]. The nonstructural protein NS1 is a multifunctional protein, which has been implicated in the inhibition of interferon-mediated host defenses [
33,
34,
35]. An additional protein, PB1-F2, can be found in either a truncated or full length form in different influenza A virus strains [
16]. The full length form of PB1-F2 does not seem to be necessary for virus function, and truncation is common in swine H1N1 viruses [
16]. This protein appears to modulate immune responses and may increase pathogenicity in some virus/host combinations [
16,
36].
2.2. Influenza A Virus Subtypes
The HA and NA proteins are very diverse, and this diversity is used to classify influenza viruses into subtypes. Currently, at least 18 hemagglutinin antigens (H1 to H18) and nine neuraminidase antigens (N1 to N9) have been recognized [
10,
37]. Most HA types (H1 through H16) occur in avian influenza viruses, but H17 and H18 were discovered and found exclusively in bats to date [
37]. In contrast, only a limited number of subtypes have adapted to circulate in any mammalian species. Currently, most of the influenza A viruses in North American swine populations belong to the subtypes H1N1, H1N2 and H3N2, although other variants (e.g., H3N1 and H2N3) have occasionally been detected (
Section 2.4) [
38,
39].
2.3. Generation of Diversity in Influenza A Viruses
Diversity in influenza A viruses can be produced by mechanisms that fall under two categories, “antigenic shift” and “antigenic drift”. The least complicated form of antigenic shift occurs if pigs (or another host population, such as humans) are infected with an influenza A virus that transmits “whole” from another species, such as a bird or a human [
13,
40] (
Figure 2A). This type of event generally requires a period of adaptation, during which the virus gains efficiency at replicating in the new host. Another form of antigenic shift is a consequence of the unique influenza virus genome, which has eight gene segments. This makes possible the exchange of gene segments between two influenza viruses that infect the same individual animal (called “gene reassortment”). Reassortment generates novel viruses that express proteins in new combinations (
Figure 2B), including variants that contain either a new HA, a new NA, or both. Antigenic shift variants, whether resulting from whole-virus interspecies transmission or gene reassortment, can completely evade the pre-existing immunity in the host population.
Figure 2.
Antigenic shift. There are two ways that an influenza virus with new antigenic properties may enter the pig population. (A) Virus that was previously adapted to another animal host, such as avian species, enters pigs and adapts to circulate efficiently in swine. The diagram portrays the inter-species transmission of an avian H1N1 virus, which became established in European swine populations; (B) Virus previously adapted to another host, such as birds or humans, co-infects a pig along with a common swine-adapted strain. This can lead to gene reassortment, producing a new “reassortant” virus that contains an HA and/or NA antigenically different from those that previously circulated in swine. The diagram portrays reassortment between human seasonal H1N1 and swine H3N2 viruses. In both (A) and (B), the swine population lacks antibodies to important surface proteins of the new virus.
Figure 2.
Antigenic shift. There are two ways that an influenza virus with new antigenic properties may enter the pig population. (A) Virus that was previously adapted to another animal host, such as avian species, enters pigs and adapts to circulate efficiently in swine. The diagram portrays the inter-species transmission of an avian H1N1 virus, which became established in European swine populations; (B) Virus previously adapted to another host, such as birds or humans, co-infects a pig along with a common swine-adapted strain. This can lead to gene reassortment, producing a new “reassortant” virus that contains an HA and/or NA antigenically different from those that previously circulated in swine. The diagram portrays reassortment between human seasonal H1N1 and swine H3N2 viruses. In both (A) and (B), the swine population lacks antibodies to important surface proteins of the new virus.
More gradual changes in the HA and NA, or “antigenic drift”, result from accumulated mutations in the HA or NA proteins of the virus (
Figure 3). The influenza A virus polymerase is error-prone, facilitating such mutations. Antigenic drift also allows the virus to evade pre-existing immune responses, although this occurs more slowly than with antigenic shifts.
The internal proteins of influenza A viruses tend to accumulate fewer mutations than the HA and NA. Nevertheless, there is some variability, which is thought to contribute to the adaptation of the virus to a host species [
26]. One particular combination of internal viral genes, called the TRIG cassette (
Section 2.4.1), has become widespread in North American influenza viruses of swine, and seems to promote their acquisition of new HA and NA proteins [
39].
Figure 3.
Antigenic drift. Over time, random mutations in HA and NA genes of an influenza A virus in swine (IAV-S) strain may cause significant changes in antigenic properties. A swine herd with population immunity to IAV-S has neutralizing antibodies specific to a strain that was previously encountered through infection or vaccination. However, if antigenic drift produces a new variant strain that pre-existing antibodies in the herd are unable to neutralize, the pigs become susceptible to reinfection.
Figure 3.
Antigenic drift. Over time, random mutations in HA and NA genes of an influenza A virus in swine (IAV-S) strain may cause significant changes in antigenic properties. A swine herd with population immunity to IAV-S has neutralizing antibodies specific to a strain that was previously encountered through infection or vaccination. However, if antigenic drift produces a new variant strain that pre-existing antibodies in the herd are unable to neutralize, the pigs become susceptible to reinfection.
2.4. Subtypes of Influenza A Virus in Swine
2.4.1. North America
The populations of IAV-S found in North America are currently very diverse. They include H3N2 viruses, H1N1 and H1N2 viruses (
Table 2), and occasionally other subtypes with a limited distribution.
Table 2.
Major IAV-S lineages that have been found among pigs in North America.
Table 2.
Major IAV-S lineages that have been found among pigs in North America.
| Virus | Origin of HA | Origin of NA | Internal Genes (Typical) |
|---|
| Classical H1N1 | swine adapted (1918) | swine adapted (1918) | swine adapted (1918) |
| Reassortant H1N1 (rH1N1) | classical H1N1 | classical H1N1 | TRIG |
| Human-like H1N1 | human IAV | human IAV | TRIG |
| 2009 Pandemic H1N1 | classical H1N1 | Eurasian IAV-S | TRIG lineage; M gene from Eurasian virus |
| H1N2 | classical H1N1 | human IAV | TRIG |
| Human-like H1N2 | human IAV | human IAV | TRIG |
| H3N2 | human IAV | human IAV | TRIG |
2.4.1.1. Classical H1N1 Virus
The first influenza virus known to have infected pigs is called the “classical” H1N1. It entered swine populations in 1918, concurrently with the highly virulent H1N1 Spanish flu epidemic in people [
9,
11,
12,
39]. During the 1918 human pandemic, the H1N1 human influenza virus was transmitted between people and pigs; outbreaks in farm families were often followed immediately by outbreaks in their swine herds, and outbreaks in pigs were sometimes followed by illness among humans on the farm [
3]. While some authors suggest that the classical H1N1 virus might have entered swine populations from humans, the origins of this virus and direction of transfer are still unclear. The virus circulated in both humans and pigs after this time, but diverged significantly in the two host populations [
41,
42]. H1N1 viruses in people continued to change through antigenic drift. In contrast, classical H1N1 virus remained antigenically stable while it circulated in swine populations in North America [
39].
2.4.1.2. Triple Reassortant H3N2 Viruses and the Emergence of the TRIG Cassette
The classical H1N1 virus was the major IAV-S in North American pigs for 70 years [
39]. Although some human H3 viruses were also found at low levels in pigs, no virus became established as a stable lineage during this time [
39]. Triple reassortant H3N2 viruses were first detected in U.S. pigs in the late 1990s. They appeared first mainly in the Midwest [
12,
43,
44,
45], and they have been detected in Canada since 2005 [
46,
47,
48]. These viruses contain the HA and NA proteins from human influenza viruses, and internal proteins from the classical IAV-S (NS, NP, M), avian influenza viruses (PB2, PA) and human influenza viruses (PB1) [
39,
44]. This particular combination of internal genes is known as the triple reassortant internal gene (TRIG) cassette.
The TRIG cassette seems to be particularly efficient in generating viruses with new HA and NA gene segments, and viruses carrying this cassette have reassorted with additional human influenza viruses, as well as with other influenza viruses in swine [
39,
49,
50]. Many new viruses containing the TRIG cassette have been detected in North America since the triple reassortant H3N2 viruses emerged, and certain H3N2, H1N1 and H1N2 viruses have become endemic in swine populations. The H3 viruses arose from at least three separate introductions of H3 human influenza viruses into pigs [
39]. HA of the H3N2 viruses in pigs can be divided into 4 distinct genetic clusters (I, II, III and IV) [
39,
51]; most of those circulating in recent years are in cluster IV.
2.4.1.3. H1N1 and H1N2 Viruses with the TRIG Cassette
Reassortant H1N1 viruses (rH1N1), which contain the same neuraminidase and hemagglutinin as the classical H1N1 virus, but with the TRIG cassette, became common in North American pigs after 1998 [
39,
49,
50,
52]. H1N2 viruses with the TRIG cassette contain H1 from the classical H1N1 virus and human-origin N2 from the triple reassortant H3N2 viruses [
39]. Since 2005, TRIG-containing H1N1 and H1N2 viruses with human-like H1, distinct from classical H1, have also become established in North American swine populations [
39,
50]. The N1 or N2 of these viruses is also of human lineage. Such “human-like” IAV-S viruses are often isolated from pigs with respiratory signs [
39]. There are now four phylogenetic clusters of H1 viruses—alpha, beta, gamma and delta—endemic among pigs in the U.S., with variable cross-reactivity and cross protection between the clusters [
50]. The δ cluster has been divided into δ1 and δ2, from what appears to be independent introductions of human seasonal H1 influenza viruses into pigs [
53].
The N2 genes of current swine H1N2 and H3N2 viruses fall into two distinct lineages [
54]. One of these (N2-1998) was introduced from human seasonal H3N2 during the original triple-reassortment event of the late 1990s. The other (N2-2002) was acquired from a human seasonal H3N2 virus that circulated around 2001–2002.
2.4.1.4. 2009 Pandemic H1N1 and Its Reassortants
Many herds were also infected with the 2009 pandemic H1N1 virus circulating among people. This virus entered human populations in 2009, and spread throughout the world. It originated from North American and Eurasian IAV-S strains that underwent gene reassortment [
55,
56]. The HA of this virus appears to come from classical H1N1 IAV-S, and the NA from an avian-like H1N1 virus that circulates among pigs in Eurasia. The pandemic H1N1 virus has been introduced repeatedly into swine herds throughout the world [
13,
40,
57,
58,
59]. This virus has undergone reassortment with IAV-S in the US, Canada, and other locations around the world [
13,
40,
54,
58,
59,
60,
61,
62]. The M gene and other internal genes from the pandemic virus have been found in many H1 and H3 IAV-S strains [
54].
2.4.1.5. Other Subtypes
Other variants of the dominant subtypes are also reported periodically among pigs (e.g., [
43]), and additional subtypes have been found occasionally. H2N3 viruses were isolated from pigs on two farms in the central U.S. in 2006 [
63]. The H2 and N3 in these viruses were from avian influenza viruses in waterfowl (possibly different bird species), and the PA was also of avian lineage, but they contained the other genes from the TRIG cassette [
63,
64]. This virus did not seem to affect other herds [
64]. However, its H2 bound well to mammalian receptors, and it might be able to replicate readily in mammals [
39]. H3N1 viruses have also been found [
65,
66], but were composed of swine-lineage H3 and N1 and thus, less of a risk to immune pig populations than truly novel subtypes such as the H2N3.
2.4.2. Influenza A Viruses in Swine on Other Continents
With the globalization of world economies, there is a risk of introducing influenza viruses from other regions into the U.S. H3N2, H1N1 and H1N2 viruses circulate among pigs in Europe and Asia, and other subtypes such as H3N1 have been found [
11,
12,
13,
40,
67,
68]. Despite the superficial similarity in subtypes, many of these viruses are different from the viruses found in North America. For example, an H1N1 virus, which has all gene segments of avian origin, has circulated among swine in Europe since the 1980s [
40]. H3N2 viruses in Europe have HA and NA of human influenza A virus origin, but do not contain the TRIG cassette [
11,
40]. Pandemic H1N1 has infected swine herds throughout the world, and in some cases, this virus has undergone reassortment with local IAV-S strains [
13,
40].There is currently little information about IAV-S subtypes in Mexico or South America where H3N2 and H1N1 viruses are known to circulate, but genetic characterization has rarely been reported [
69]. The prevalence of IAV-S subtypes and lineages in African countries is also not well characterized [
70].
3. Mechanisms of Immunity to Influenza A Viruses
Immunity to viruses is a complex process involving both nonspecific (innate) protective responses, and specific (adaptive) humoral and cell-mediated immunity directed against the specific virus. The various cells and soluble mediators of the immune system interact in many ways to eliminate the pathogen. The lungs present a particular challenge for immune responses, as viruses must be eliminated, but severe inflammation must also be avoided to minimize tissue damage.
3.1. Innate Responses to Influenza A Viruses
Innate responses include chemical, physical and cellular responses that are immediately protective against a broad range of invading microorganisms. These reactions are often triggered by the “recognition” of certain motifs that are found in microorganisms, but are not normally present in the host. For example, the viral RNA of influenza A viruses is recognized by pattern recognition molecules in infected cells, and triggers the production of proteins called type I interferons (interferon α and interferon β) [
39,
71]. Type I interferons, in turn, have a number of antiviral properties that inhibit influenza virus replication [
39]. Conversely, the influenza A virus protein NS1 can inhibit interferon-mediated host defenses [
24,
25,
26,
72]. Other components of innate responses, including macrophages [
73] and natural killer (NK) cells [
74] also play roles during influenza A virus infections. Innate immune responses contribute to the activation of adaptive immunity [
72].
3.2. Adaptive Immune Responses to Influenza A Viruses
Adaptive immune responses are traditionally divided into humoral immunity, which includes those mechanisms that are dependent on antibodies, and cell-mediated immunity (CMI), in which immune cells (cytotoxic T cells) directly recognize and destroy infected host cells. Antibody responses can be induced by any protein that enters the body, including those of killed viruses (e.g., in inactivated influenza vaccines for swine). Cytotoxic T cell responses are induced optimally by live organisms (such as IAV) replicating inside cells.
The critical immune cell types for both humoral immunity and CMI are various types of lymphocytes (B or T lymphocytes). Each lymphocyte responds very specifically to the immunogenic portions of that virus, called antigens. Most antigens are proteins. In reality, each lymphocyte actually responds to a tiny piece of the viral protein, called an epitope. Epitopes may be shared between strains of influenza viruses, or may be unique to a strain. Depending on the epitopes that are recognized, immune responses may be more or less broadly protective against different viral strains. Some subpopulations of responding lymphocytes also differentiate into memory cells, which respond more strongly when they encounter the same virus a second time.
3.2.1. Adaptive Immunity: Humoral Response
3.2.1.1. The Generation of Humoral Immune Responses to Influenza A Viruses
Antibodies are important in preventing influenza virus infections and in reducing the severity of disease [
39]. Antibodies are made by B cells (also called B lymphocytes). B cells can make different isotypes of antibodies. They initially make IgM, then switch to producing other isotypes such as IgG or IgA. Antibody isotypes differ in their ability to carry out the various humoral immune responses that defend the body from pathogens. Helper T cells are critical for the production of high quality IgG and IgA antibodies that have high affinity to the antigen.
Antibodies to the HA of IAV-S can be found in the respiratory tract of pigs as soon as 4–5 days after infection with IAV-S [
75,
76], and serum titers appear in approximately 7 days [
77,
78,
79], although peak levels occur later (e.g., 2 weeks [
75]). Both local IgA and IgG are found in the respiratory tract of influenza virus-infected pigs [
39,
75,
76,
78,
80,
81]. Protective antibody responses in the lungs are often, but not always, correlated with the serum antibody titers in the blood [
39,
82,
83]. Even though antibody titers measured in the blood fall gradually over time, influenza-specific B cells might be maintained throughout a pig’s life. In laboratory animals such as mice, memory B cells specific for influenza A viruses can be found for many months in the respiratory tract and other tissues [
71]. Humans can develop long-term antibody responses to some antigens, possibly spanning many decades [
71].
3.2.1.2. Antibody Responses to the HA Protein
The influenza A virus HA usually induces the strongest antibody responses after infection [
84], probably because this protein is present in large amounts on the surface of the virus. Antibody titers to the HA are, however, affected by the dose of virus [
71], and possibly its subtype. In some studies, pigs infected with H3N2 viruses had higher serum antibody titers than pigs infected with H1N1 or H1N2 viruses [
33,
79,
85].
Some antibodies to the HA, called neutralizing antibodies, can block this protein from attaching to its sialic acid receptor, and thus prevent the virus from infecting cells. Well-matched antibodies to the HA can be sufficient to prevent an influenza virus infection, and also contribute to clearing the virus from the lungs [
71,
86]. Whether the antibody response to the HA can protect the animal from a different viral strain depends on the similarity between the HA proteins of the two viruses [
72]. This is influenced not only by the overall genetic similarity between these proteins, but on how well the individual epitopes match; changes in some parts of the HA can influence protection more than others [
87]. Neutralizing antibody responses are thought to be the most important immune responses for protecting pigs against closely related IAV-S of the same subtype [
71,
80,
83].Protective immunity mediated by neutralizing antibodies can be predicted by the magnitude of serum antibody activity against the particular IAV-S strain, e.g., by the serum hemagglutination inhibition (HI) titer [
39] or by serum neutralization (SN) tests [
88,
89].
3.2.1.3. Antibody Responses to the NA Protein
Animals exposed to influenza viruses normally make fewer antibodies to the NA than the HA [
84]. Humoral responses to the NA have not been studied as extensively, but they may also be important in protection [
28,
29,
30,
31,
72]. Although they do not prevent infections, antibodies to NA seem to contribute to reduced virus shedding and/or less severe illness [
28,
29,
30,
31,
80,
86]. Antibodies to the NA are thought to reduce the ability of the virus to spread from cell to cell, by preventing this enzyme from cleaving sialic acids [
72,
86]. They might also prevent the NA from clearing a path through the respiratory tract mucus layer and cell glycocalyx [
86]. Similarly to the HA, mutations in the NA can reduce the protective effects of pre-existing antibodies [
72], although the NA has a somewhat slower mutation rate [
90,
91,
92].
3.2.1.4. Other Roles for Antibody
Antibodies might also contribute to immunity against influenza viruses by alternative mechanisms. In some cases, antibodies can promote the ingestion of viruses by phagocytes (cells, such as macrophages, which engulf and destroy many pathogens). These cells have receptors that can recognize part of the antibody molecule, essentially tethering the virus to the cell via the antibody. The virus is then ingested and destroyed. Similarly, some immune cells, such as macrophages, can recognize antibodies attached to viral antigens on infected cells, and destroy the infected cell. This mechanism is known as antibody-dependent cell cytotoxicity (ADCC). Antibodies to both the viral HA and NA can mediate ADCC, and may contribute to clearance of influenza viruses [
72].
3.2.3. Immunity from Vaccination
Although other vaccine types are in development, most IAV-S vaccines licensed in the U.S. contain inactivated whole viruses of the H1 and H3 subtypes [
38]. Factors that may influence vaccine efficacy include homology between the vaccine and challenge strains, the immunogenicity of vaccine components, the quantity of antigen included, and the adjuvant used. Matching of the viral HA is a critical component in vaccination because the goal of inactivated vaccines is mainly to produce neutralizing antibodies to this protein [
80]. Responses to the NA may also be protective [
28,
29,
30,
31]. While there have been reports of CMI responses to inactivated influenza vaccines in swine [
106], CMI and mucosal immune responses to these vaccines are generally thought to be very limited [
94,
107,
108]. A poor CMI response to an inactivated vaccine limits its ability to cross-protect against strains with antigenically divergent HA and NA proteins. A recent study tested an intranasal IAV-S vaccine containing inactivated virus and a synthetic adjuvant called poly (I:C) [
109]. Compared with a conventional inactivated vaccine, the intranasal poly (I:C) adjuvanted vaccine induced higher cross-reactive serum HI antibody titers and somewhat greater protection against heterologous challenge.
4. Maternal Antibodies to Influenza A Viruses in Swine
Newborn piglets have no maternal antibodies at birth, as these proteins cannot cross the placenta in swine [
1]. Although neonatal piglets are able to mount immune responses, their immune system appears to be underdeveloped at birth, and less able to respond to pathogens and vaccines for the first few weeks [
110]. Maternal IgA, IgG and IgM antibodies to IAV-S and other pathogens are transferred to piglets in colostrum from the dam, usually during the first 36 h of life [
110]. Lymphocytes and other cells are also present in the colostrum, and can cross the intestinal lining in the neonate during this “open gut” period [
110]. In addition, immunomodulatory factors received in colostrum might aid the development of the newborn’s own immune system [
110]. Although the gut closes to the absorption of antibodies and cells after approximately 36 hours, piglets continue to receive passive mucosal IgA antibodies in their gastrointestinal tract from the milk until weaning [
1].
A common strategy to control IAV-S is to vaccinate sows, which then transfer this protection to their piglets in colostrum. As the maternal antibodies decline, however, the piglets become susceptible to infection. In the field, where most sows have been infected with IAV-S, vaccination often results in significantly higher and relatively uniform HI titers (often ≥160) compared to unvaccinated sows [
79]. In the offspring of these sows with high titers, maternal antibodies have been found to persist until 14–16 weeks, while they often disappear around 6 weeks in piglets born to exposed but unvaccinated dams ([
111] cited in [
79]).
4.1. Maternal Antibody Inhibition of Active Immune Responses in Piglets
Although they may protect the piglet, maternal antibodies can interfere with the development of active immunity after infection or vaccination [
110]. In several studies, piglets with maternal antibodies had no rise in HI titers after they were exposed to IAV-S [
112,
113,
114,
115,
116]. Other groups reported that such piglets did respond to infection or vaccination, though more weakly. In one study, 7-week-old piglets with low levels of maternal antibodies developed lower HI titers, virus-specific immunoglobulins, and T cell proliferative responses compared to piglets without maternal antibodies, when they were infected with the same H1N1 virus that had been used to vaccinate their dams [
117]. Nevertheless, these pigs did mount an immune response that resulted in less severe clinical signs, compared to naive pigs, when they were re-exposed to the virus at 15 weeks of age. In another study, piglets vaccinated with a bivalent inactivated vaccine at 1 and 4 weeks, 1 and 8 weeks, 4 and 8 weeks, 8 and 10 weeks, or 8 and 12 weeks of age had detectable HI titers after the second dose [
118]. The titers were lower in piglets with passive maternal antibodies. Other studies have shown that it is possible for piglets to develop cell-mediated immune responses (CMI) to antigens despite the presence of maternal antibodies [
110]. Results of another study suggest that inactivated IAV-S can induce greater HI antibody titers and cross-protection in maternal antibody-positive piglets if it is formulated with poly (I:C) adjuvant and administered intranasally [
109].
4.2. Maternal Antibody-Mediated Protection from Disease
Maternal antibodies sometimes provide pigs with partial or complete protection from disease after inoculation with IAV-S [
112,
113,
116,
117], although protection may vary depending on the antibody titer [
112]. One of these studies examined overall growth rates, as well as clinical signs, after two exposures to the same influenza virus [
117]. In this study, 7-week-old piglets without maternal antibodies grew more slowly immediately after they were inoculated with IAV-S, compared to piglets with maternal antibodies; however, the animals in the two groups were similar in size 3 weeks later. When both groups of piglets were exposed to this virus again at 15 weeks of age, the piglets without maternal antibodies seemed to have higher growth rates overall. The explanation appeared to be that these animals were less affected by the second exposure to the virus. When piglets’ IAV-S specific maternal antibodies are poorly matched to a strain that subsequently infects them, there may be a risk of vaccine-associated enhanced respiratory disease (VAERD), a phenomenon described in
Section 9.
4.3. Maternal Antibody Protection from Infection and Transmission of Virus
Piglets protected from clinical signs by maternal antibodies may still become infected and shed virus [
114,
116,
117,
119]. In some experiments, piglets with low levels of maternal antibodies shed IAV-S at least as long [
112] or longer [
117] than piglets without maternal antibodies. In contrast, high maternal antibody titers inhibited virus shedding in one of these two studies [
112]. Neonatal piglets that become infected might act as reservoirs for influenza viruses under some field conditions even when viruses are not detected in the breeding sows [
120,
121]. An H1N1 virus could be detected for up to 70 days in one group of growing pigs housed in a finishing barn, when no additional animals were added to the group [
120]. Piglets with waning maternal antibodies could be infected with IAV-S from older pigs or other sources of the virus, and these viruses may be transmitted relatively slowly among the growing pigs. A small number of animals infected at weaning might act as the source of virus for the rest of the cohort, and could also transport the virus to distant production units. Vaccination of sows might be able to decrease virus transmission in some cases. In one study, high and uniform titers of maternal antibodies to an H1N1 virus reduced the transmission of the same virus, although it did not prevent the transmission of an H1N1 virus that belonged to a different phylogenetic cluster [
122]. Supportive evidence is also provided by a report from the field, in a herd where mass vaccination of sows was able to boost maternal antibody levels sufficiently to stop an outbreak of respiratory disease among suckling and nursery piglets [
7]. Based on these studies, sow vaccination often provides piglets a measure of protection, but stopping virus transmission in the herd requires a close antigenic match between the sow vaccine and the local circulating strain(s).
5. Influenza Surveillance in Swine
New subtypes or strains of influenza resulting from antigenic shift or drift may produce an increased threat to animal and human health. These emerging influenza variants can be identified by monitoring changes in circulating influenza virus strains in swine. The integrated and coordinated USDA Swine Influenza Virus Surveillance System strategy was developed due to concerns about antigenic drift and gene reassortment of IAV-S strains, as well as sporadic reports of human infections with IAV-S. Program details are available at USDA’s Swine Influenza Surveillance website [
123]. Documents included at this website are as follows:
Influenza Surveillance in Swine Procedures Manual [
124]:
Appendix A: National Surveillance Plan for Swine Influenza Virus in Pigs [
125]
Appendix B: AVIC and State Veterinarian Directory
Appendix C: Testing Guidelines, Forms, Submission Instructions
Appendix D: Designated NAHLN SIV Testing Laboratories
Appendix E: Notification Plan
Appendix F: SIV Specimen Submission Form for Regulatory Veterinarians
Quick Reference Guide on USDA Swine Influenza Virus (SIV) Surveillance
Influenza Virus Surveillance in Swine - Program Overview for Veterinarians (color brochure)
Producer Guide to Influenza Virus Surveillance in Pigs (color brochure)
5.1. Program Objectives
The surveillance program was designed by the USDA Animal and Plant Health Inspection Service (APHIS), the USDA Animal Research Service (ARS), industry stakeholders and public health officials with the intent to foster robust scientific research that would address veterinary and public health concerns [
123,
125]. Implementation of the program formally began in 2010. The objectives of this surveillance program are to:
Monitor genetic evolution of endemic influenza in swine to better understand endemic and emerging influenza virus ecology;
Make available influenza isolates for research and to establish an objective database for genetic analysis of these isolates and related information; and
Select proper isolates for the development of relevant diagnostic reagents, updated diagnostic assays, and vaccine seed stock products.
In designing the program to meet these objectives, special care was taken to give veterinarians and producers flexibility to participate without concern that IAV-S detection would bring scrutiny to their operations. Sharing of data and viral isolates is encouraged so that public health authorities, researchers and manufacturers will be equipped with information and materials that are as contemporary as possible. To meet both of these goals, veterinarians and producers are given the option to submit specimens anonymously.
5.2. Targeted Swine Populations
In order to optimize resources and provide a level of uniformity in sampling, the surveillance program is focused toward sampling from swine that meet specific criteria. The following swine populations are targeted for the IAV-S Surveillance Program [
123]:
Case-compatible swine accessions submitted to veterinary diagnostic laboratories. This stream includes samples submitted by producers, veterinarians or other personnel who observe pigs exhibiting influenza-like illness (ILI) on farms. Samples collected for routine diagnostic testing may also be tested at the laboratory for influenza as part of the surveillance program.
Swine exhibiting ILI at first points of concentration or commingling events such as auctions, markets, fairs or other swine exhibition events. Animal health officials or licensed veterinarians may observe pigs with ILI at events with an increased potential for influenza transmission and/or elevated human exposure. The veterinarian in charge or animal health official may segregate, examine and collect appropriate samples from pigs exhibiting ILI. These samples would be submitted to a veterinary diagnostic lab and tested for IAV-S as part of the surveillance program.
Swine populations epidemiologically linked to a confirmed isolation of IAV-Sin a human. When a person tests positive for influenza that may be linked to swine exposure, that individual’s identity is known to public health officials and possible sources for infection will be known to them. The U.S. influenza surveillance system for humans collects influenza activity information submitted voluntarily by public health partners and health-care providers. The surveillance information is divided into five categories including virological surveillance, outpatient illness surveillance, mortality surveillance, hospitalization surveillance and summary of the geographic spread of influenza [
126]. Occasionally, an epidemiological link may be suspected when human influenza cases have a recent history of exposure to pigs. Samples are collected in cooperation with the owner/producer and herd veterinarian from swine that are epidemiologically linked with a human influenza A case. The number of samples collected will be determined on a case-by-case basis.
5.3. Sample Collection
Samples may be collected and submitted from producers, veterinarians, animal health officials or other personnel [
123]. Animals to be sampled should be febrile if possible (some IAV-S strains induce a minimal febrile response) with serous nasal discharge and cough. Samples may include nasal swabs, oral fluids, and lung tissue. Up to ten samples from animals which fit the case definition may be sampled per diagnostic case under the IAV-S Surveillance Program. Lung tissue collected from mortalities or euthanized animals may be submitted in plastic bags or screw-top plastic tubes. When collecting samples using nasal swabs, polyester or flocked swabs are inserted deep into the nasal cavity while the pig is restrained. The swab is rotated to collect surface epithelium as well as mucosal secretions from both nostrils. Blood on the swabs may interfere with testing, so caution should be taken in rotating too vigorously or inserting the swab too deeply. Swabs should be inserted into virus transport media. Once the samples are collected, they should be chilled until shipped or shipped as soon as possible with ice packs.
5.4. Testing, Reporting and Response
Testing for the IAV-S Surveillance Program is performed through the National Animal Health Laboratory Network (NAHLN) which is composed of state or university laboratories [
123,
124]. The network is organized and supported so that it has the capacity to respond to animal-disease outbreaks nationwide and is the model for effective diagnostic networks, able to respond and communicate diagnostic outcomes to decision makers quickly. NAHLN laboratories perform routine diagnostic tests for endemic animal diseases (like IAV-S) as well as targeted surveillance and response testing for foreign animal diseases in the event of an outbreak. NAHLN laboratories that conduct the testing for the IAV-S Surveillance Program are listed online [
123]. Samples are tested from the following swine populations.
5.4.1. Case-Compatible Swine Accessions
As with any routine diagnostic work-up, NAHLN diagnostic labs conduct tests requested by the veterinarian and report test results to the submitting veterinarian as per the usual process. In addition to the routine testing, NAHLN labs may conduct additional testing of samples for the IAV-S Surveillance Program [
123,
124].
IAV-S Surveillance Program test results are reported as anonymous unless the producer is willing to participate in the traceable surveillance option [
123,
124]. Producers participating in the traceable surveillance option receive surveillance test results. No additional charges to the submitter result from testing conducted in accordance with either the anonymous or traceable surveillance streams of the IAV-S Surveillance Program.
Data are collected by NAHLN, or through direct messaging between laboratory information management systems. Data are then monitored and analyzed by the USDA, in collaboration with industry stakeholders and the office of the State Animal Health Official (SAHO), to identify sequences and data that may initiate further research or targeted surveillance. Data confidentiality and security are priorities. When submitted through traceable surveillance, the response to unusual test results is determined on a case-by-case basis.
5.4.2. Targeted Surveillance of Sick Pigs at First Points of Concentration or Commingling Events
If a SAHO determines samples should be collected, the attending veterinarian and/or regulatory officials collect and submit samples from these animals to a NAHLN lab for IAV-S testing. As with the case-compatible swine accessions, unless written permission is provided by the owners of pigs sampled for traceable surveillance, test results will be entered through the anonymous surveillance stream. The SAHO determines the control measures, if any, which will be utilized at the event prior to or after receiving test results.
5.4.3. Surveillance of Swine Populations Epidemiologically Linked to a Human Case of IAV-S
With producer permission and under the direction of the SAHO, personnel collect samples from swine exhibiting ILI. Samples are submitted to the National Veterinary Services Laboratories (NVSL) through traceable surveillance. Under the herd’s licensed veterinarian’s supervision, movement of the animals occurs when clinical signs have resolved to satisfactory levels.
5.5. Benefits of the IAV-S Surveillance Program
Perceived and real risks to the health of pigs and people from influenza infection fuels the need for the information gained from the surveillance efforts. Testing field samples submitted under the IAV-S Surveillance Program has several immediate and long term benefits to both producers and the public. In the short-term, the information is used to make decisions addressing disease control and prevention measures, human health concerns and trade issues. In addition, researchers and the animal health industry can use the information to develop and update relevant diagnostic reagents, targeted influenza diagnostic assays, effective vaccines and response plans.
During FY2013 (October 2012-September 2013), 21725 pigs were tested through diagnostic laboratory submissions from 4991 accessions. Of the 1579 Influenza A positive accessions, the accessions with each subtype tested as follows: 519 H1N1, 380 H1N2 and 286 H3N2. Fifty-four accessions contained mixed subtypes [
127]. An increase in swine diagnostic lab submissions through the IAV-S Surveillance Program will continue to provide valuable information to the industry, researchers and public health officials. Sequenced virus information from samples submitted through the IAV-S Surveillance Program is available to researchers via the public database GenBank
® (
http://www.ncbi.nlm.nih.gov/genbank/). IAV-S isolates in the NVSL repository are available to researchers and to firms developing vaccines and diagnostic kits.
Over the long-term, the information may be used to help understand influenza infection in pigs, to gain a better understanding of epidemiological factors affecting the mutation and spread of IAV-S in the swine population, and to provide information for broader initiatives such as Veterinary Services 2015 Project, Surveillance for Action, a stream-based comprehensive and integrated animal disease surveillance system.
While the IAV-S Surveillance Program was initiated in 2010 in the U.S. with a focus on monitoring the pandemic H1N1 2009 (H1N1pdm09), IAV-S surveillance has also been occurring in other areas of the world. Internationally from 2009–2011, introductions of the H1N1pdm09 influenza virus from humans into swine were observed in 12 countries and regions. Due to the high level of surveillance and transparency, a majority of reported introductions occurred in the U.S. and Canada (19 and 8, respectively) [
128]. Transmission of seasonal influenza viruses from humans to swine is also being monitored in the U.S. and globally. From 1990 to 2011, at least 23 separate introductions were documented, with six first identified in the U.S. and three in Canada [
128].
Monitoring the transmission of influenza viruses from humans to pigs aids in understanding the contribution of human-origin influenza viruses to the genetic diversity of IAV-S. There have been several instances in the past 15 years when gene reassortment between human seasonal and swine viruses produced new IAV-S lineages that became established in the swine population [
54]. When virus transmission increases between humans and pigs, measures to prevent transmission need to be followed. Increased biosecurity measures and utilization of influenza vaccination for swine workers and pigs may be necessary to lower the risk of reassortment between human and swine viruses.
A surveillance program is most useful (e.g., for selection of well-matched vaccine strains) if participation is high and sampling bias is minimal. The IAV-S Surveillance Program currently focuses on testing specimens submitted from swine herds affected by respiratory disease. If some regions are under-reported or over-reported it may skew the estimation of strain frequencies, which could affect the selection of widely representative vaccine strains. Complementary data could be gained through a sentinel surveillance component, which involves sampling representative herds at regular intervals, independently of disease incidence. Sentinel surveillance could therefore help to identify a full cross-section of circulating IAV-S strains.
7. Best Use of Current Commercial Vaccines for IAV-S Control
7.1. Inactivated Influenza Virus Vaccines and Antigenic Matching
All of the licensed, conventional IAV-S vaccines in the United States (
Table 3) contain inactivated (killed) virus. Each strain of virus is cultivated in eggs or cell culture and inactivated with chemical agents, such as formaldehyde or binary ethylenimine [
134]. Inactivated viruses of multiple strains are blended and formulated with an adjuvant. Regulatory approval of an inactivated virus vaccine requires successful demonstration of safety and efficacy [
134]. Efficacy is determined by experimental challenge of seronegative animals with the strain of virus identical to each vaccine seed virus, and sometimes with a heterologous strain. Since IAV-S strains are antigenically diverse and evolving, and many young pigs have maternal antibodies that can interfere with vaccines, the field efficacy of an approved product cannot be taken for granted. There are commercial vaccines that combine IAV-S antigens with antigens for other agents, such as
Erysipelothrix rhusiopathiae,
Mycoplasma hyopneumoniae,
Leptospira species, or porcine parvovirus. Manufacturers must demonstrate that antigens in a combination product do not interfere with each other in terms of efficacy [
146].
Conventional inactivated vaccines are effective in inducing protective immunity against antigenically identical or very similar strains [
82,
147]. In this ideal situation, the vaccine elicits systemic antibodies that can neutralize the virus very early without triggering extensive inflammation. If commercial killed IAV-S vaccines consistently included the predominant circulating North American strains, they would likely be more effective at preventing IAV-S outbreaks. The annual strain selection process for human seasonal influenza vaccines aims for this ideal. The human seasonal vaccines are reformulated most years, based on the results of global surveillance and identification of new strains that emerged through antigenic drift or shift [
148,
149]. This updating process, while imperfect, keeps the human inactivated virus vaccines approximately up-to-date with the evolution of A/H1N1, A/H3N2, and B viruses, so that vaccinated persons are more likely to develop antibodies that can neutralize the predominant circulating strains. A similar system of strain updating to support IAV-S vaccines for the United States has been advocated [
150].
For IAV-S, maintaining close antigenic matches in the commercial inactivated virus vaccines is more difficult. First, the evolution of swine H1 and H3 viruses in North America has resulted in co-circulating strains that belong to about seven antigenically distinct clusters within the two subtypes (see
Section 2.4) [
51]. Second, IAV-S surveillance to monitor the evolution of strains within the United States is still a young program, and the geographical distribution of IAV-S genotypes across North America is far from homogenous [
151]. Third, manufacturers of IAV-S vaccines make independent decisions about the strains contained in their products, and typically do not share those strains or publish their HA gene sequences.
Table 3.
Licensed influenza vaccines for swine in the United States.
Table 3.
Licensed influenza vaccines for swine in the United States.
| Vaccine Name | Manufacturer | IAV-S Lineages Included | Specific Strain Names (If Available) | Adjuvant | Combination Products |
|---|
| FluSure XP® | Zoetis | Delta-1 H1N2
Delta-2 H1N1
Gamma H1N1
Cluster IV H3N2 | A/Sw/Oklahoma/0726H/2008 (H1N2)
A/Sw/North Carolina/031/2005 (H1N1)
A/Sw/Iowa/110600/2000 (H1N1)
A/Sw/Missouri/069/2005 (H3N2) | Amphigen® | FluSure XP/RespiSure®
FluSure XP/FarrowSure GOLD®
FluSure XP/FarrowSure GOLD B®
FluSure XP/RespiSureONE®
FluSure XP/RespirSureONE/ER Bac Plus® |
| FluSure® Pandemic | Zoetis | Pandemic H1N1 | A/California/04/2009 (H1N1) | Amphigen® | |
| MaxiVac Excell® 5.0 | Merck Animal Health | Beta H1N1
Delta-2 H1N1
Gamma H1N1
Cluster I H3N2
Cluster IV H3N2 | | EMUNADE® | |
| PneumoStar® SIV | Novartis Animal Health | Alpha H1N1
Cluster I H3N2 | | ImmunSTAR® | |
| Swine Influenza Vaccine, RNA | Harrisvaccines | Cluster IV H3N2 | | none | |
Until 2007, updating strains in an IAV-S vaccine required a new licensure process for the product, including efficacy and field safety testing, which was recognized as a barrier to manufacturers making timely strain changes. In 2007, USDA-CVB introduced a new policy to allow manufacturers flexibility to change strains under an existing license, by either adding or replacing viruses, and demonstrating immunogenicity equivalent to that of the prior strains [
152]. Despite this move to foster better matching between vaccines and the evolving IAV-S subtypes, strain updates over the following years have not been common. Reasons for this might include lack of confidence by firms that existing IAV-S surveillance data can identify better strains, and the time and expense required for regulatory approval, even under the 2007 revised policy. Recent serological data strongly suggest that antigenic drift among the Cluster IV H3N2 viruses has been substantial enough to reduce efficacy of the commercial vaccines [
153]. With the strengthening of North American IAV-S surveillance in recent years [
51,
57], data are available to support more frequent and collaborative vaccine strain decisions.
7.2. Polyvalent IAV-S Vaccines Containing Multiple H1 and H3 Clusters
Several past and present IAV-S vaccines marketed in the US contained two strains of the H1 and H3 subtypes (End-FLUence2, Intervet Inc., Millsboro, DE, USA; FluSure, Pfizer Inc., New York, NY, USA; PneumoSTAR, Novartis Animal Health, Greensboro, NC, USA). Two of these were replaced several years ago with new products containing 4–5 strains of IAV-S (MaxiVac Excell 5.0, Merck Animal Health, Summit, NJ, USA; FluSure XP, Zoetis, Florham Park, NJ, USA) (
Table 3). The greater valency of the newer products was a response to the emergence of antigenically distinct clusters within the H1 and H3 subtypes. This was an important development. Three of the earlier commercial vaccines that contained only a cluster I H3N2 strain were insufficient to reduce shedding of a cluster III H3N2 virus after challenge infection, whereas an experimental vaccine containing a cluster III virus conferred full protection [
154]. A more recent experimental challenge study gave similar evidence that polyvalent commercial vaccines containing cluster IV H3N2 can protect against a drifted contemporary cluster IV strain better than a commercial vaccine containing only cluster I H3N2 [
155]. Although antigenic drift is likely diminishing the quality of protection from the newer polyvalent vaccines [
153,
156,
157], it is reasonable to assume these will protect herds more reliably than bivalent vaccines that contain even older IAV-S strains.
7.4. Keys to Better-Matched and Innovative IAV-S Vaccines in the Future
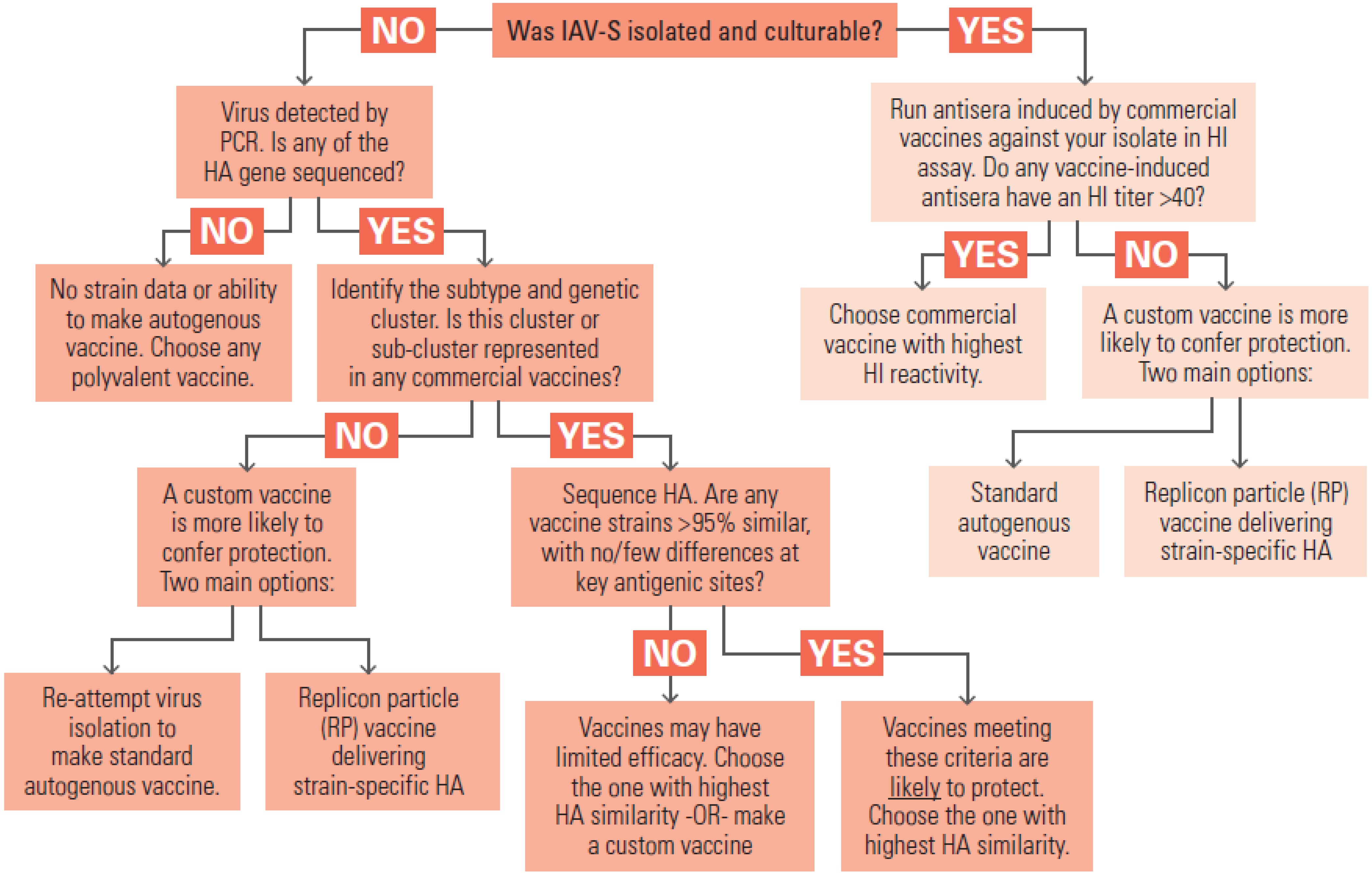
Significant improvement in IAV-S vaccines in the longer term will require coordinated efforts by the various parties participating in field surveillance, strain selection, manufacturing, and product licensure. If IAV-S strains in the conventional inactivated vaccines could be updated more rapidly, in response to new trends in the surveillance data, the vaccines would have more consistent efficacy in the field. In
Section 7.1 we discussed some practical factors that make it difficult to simply copy the human seasonal influenza vaccine strain selection program that is guided by the World Health Organization. However, veterinary vaccine experts have outlined long-range visions of a more flexible, coordinated, and data-driven program to select IAV-S strains [
150,
159]. Multi-agency cooperation would need to include manufacturers, NAHLN laboratories, and the relevant agencies within USDA. Cooperation could be focused on several aims:
Continuous evaluation of circulating strains and their antigenic similarity to current vaccines. The USDA’s program for influenza surveillance in swine should continue to monitor contemporary viruses identified in cases of respiratory disease outbreak, tracking the frequency of subtypes and the emergence of new variants. Representative viruses from prevalent subtypes or genotypes should be evaluated at an antigenic level by serologic cross-reactivity with vaccine anti-sera. Challenge experiments in pigs could be conducted to test for loss of vaccine efficacy when antigenic changes are identified.
Strain update working group. Manufacturers currently perform independent serology and in vivo challenge experiments, at significant cost, to assess if their polyvalent inactivated vaccines provide enough protection against circulating IAV-S strains. If the serology, bioinformatics analysis, and challenge experiments were performed under USDA leadership, it should be a more efficient use of resources.
Engineering of versatile “donor virus” strain(s) that confer superior manufacturing properties.Some IAV-S strains with desired antigenic properties are limited by poor growth properties, making them difficult to manufacture. As discussed elsewhere in the paper, antibody targets are mainly in HA and NA genes while efficient replication and protein expression in culture tend to be controlled by the internal proteins. Traditional gene reassortment or modern reverse genetics techniques can be used to combine HA and NA genes of a new strain of interest with internal genes of a high-growth strain, making an optimal seed virus for manufacturing. If a donor strain could be qualified in advance, receive regulatory approval, and be made available to all manufacturers, it could speed the process of making strain updates.
Revise regulatory policies to accommodate new vaccine platforms, such as LAIV and recombinant viral vectored products.
8. Autogenous Influenza Vaccines for Swine
Autogenous vaccines are intended for use in situations where commercial vaccines are ineffective or unavailable [
160]. Before using an autogenous vaccine, two questions to consider are whether commercial vaccines have been employed and found ineffective, and whether other barriers to vaccine efficacy, such as management factors, exist in the herd and could be contributing to the apparent lack of efficacy [
161]. A significant number of U.S. swine farms vaccinate their animals with autogenous IAV-S vaccines. These vaccines were used by 20% of the U.S. farms that vaccinated breeding sows in 2006 [
39], and in 2008, they accounted for more than half of all IAV-S vaccine doses released for sale (personal communication in [
162]). The primary reason for their use is thought to be the diversity of IAV-S variants circulating among pigs in North America, combined with the limited number of IAV-S strains available in commercial vaccines [
39].
8.1. Regulatory Requirements for Autogenous Vaccines
In the U.S., autogenous vaccines are regulated by the USDA Center for Veterinary Biologics (CVB). These vaccines must be inactivated, nontoxic, and used only by a veterinarian under a veterinary client/patient relationship [
163]. Ordinarily, the vaccine organism(s) must come from the herd in which the vaccine is to be used [
163]. Multiple isolates from the herd are commonly used in an autogenous vaccine. In addition to vaccine components from other pathogens, autogenous IAV-S vaccines may contain various combinations of H1 and H3 viruses [
162].
An autogenous vaccine is only permitted for use in the herd of origin, unless there is epidemiological evidence that adjacent or nonadjacent herds are at risk [
163]. In this case, authorization can be secured from CVB, through a routine process consistent with 9 CFR 113.113.The State Veterinarian, or equivalent state official, must be informed when an authorized vaccine is used in such herds. Autogenous vaccines are tested for safety in laboratory animals (mice or guinea pigs) and for purity [
163]. Purity testing includes testing for the absence of viable bacteria and fungi. Autogenous influenza vaccines must also meet the general requirements for the production of viral vaccines [
163,
164]. These requirements [
164] stipulate that tests must be incorporated into the manufacturing process to ensure complete viral inactivation. However, testing is overall less rigorous than required of commercial IAV-S vaccines [
162]. In particular, manufacturers of autogenous vaccines do not need to conduct potency and efficacy testing. As a result, some vaccines might contain antigen doses that are suboptimal for that particular viral strain. The expiration date for formulated autogenous vaccines is a maximum of 18 months after harvest [
163].
A microorganism can be used for autogenous vaccine manufacturing until 15 months after it was first isolated, or for 12 months after the date of harvest for the first serial [
163]. Extensions to 24 months are now permitted without CVB review, provided the firm maintains records documenting, among other things, that the vaccine was beneficial and that the microorganism remains associated with disease in the herd [
163,
165]. With additional tests and CVB authorization, additional serials may be produced after 24 months has passed since virus isolation [
163].
8.2. Selection and Use of Autogenous Vaccines
Advantages of autogenous vaccines include the ability to customize the vaccine to the specific virus(es) in the herd, to include multiple agents, and to customize the adjuvant [
166]. When selecting a multi-agent autogenous vaccine, it should be noted that there is no requirement to test for potential interference between vaccine antigens or adjuvant components [
166]. Given the less rigorous testing requirements, an autogenous vaccine for a new viral strain can be produced and delivered much more rapidly than an updated commercial vaccine. There is nevertheless a significant lag period to isolate and characterize the agent and manufacture the vaccine, compared to ordering a commercial vaccine that is already available. While the absence of extensive testing may also lower the price of autogenous vaccines, this comes at the cost of potential uncertainties in potency and efficacy. In addition, there is an increased risk that adventitious agents might be present in a vaccine prepared from field viruses and not tested extensively [
166]. In addition, not all IAV-S isolates adapt well to cell culture, or grow to high titers for vaccine production.
When selecting an autogenous vaccine, consideration should be given to the choice of manufacturer, as the products produced by different companies may vary. Companies that produce USDA-approved commercial biologicals will most likely use similar quality control/quality assurance production standards for their autogenous vaccines [
166]. A complete economic evaluation, including performance, morbidity and mortality, can help determine the cost savings or loss from vaccine use; the selection of a vaccine should not be based solely on price [
166]. A vaccine trial can help evaluate whether the commercial or autogenous vaccine is more effective in the herd [
166]. It is highly recommended that post-vaccination sera be collected to test against the herd IAV-S strains contained in the autogenous vaccine for serologic evidence to support vaccine efficacy.
The efficacy of autogenous vaccines against IAV-S has not been evaluated and reported to a significant extent in the literature. One recent experiment assessed the efficacy of a bivalent autogenous vaccine containing β- and γ-cluster H1 IAV-S, as well as two commercial vaccines, against pandemic H1N1 viruses [
162]. All three vaccines provided partial protection against this virus, although all were less effective than an experimental vaccine containing a virus identical to the challenge strain. It is likely that autogenous and commercial vaccine efficacy will differ between herds, depending on the specific viruses that affect those herds.
9. Vaccine Associated Enhanced Respiratory Disease (VAERD)
Certain vaccines for other mammalian viruses have been shown to induce immune responses that later exacerbate the severity of infection [
167]. With influenza viruses, vaccine failure has usually been equated with little or no protective immunity and lack of prevention of virus transmission in the herd rather than disease exacerbation. However,
in vivo experiments in recent years have revealed a potentially important phenomenon of vaccine associated enhanced respiratory disease (VAERD) in IAV-S infected pigs. VAERD has been observed in pigs immunized with an adjuvanted, whole inactivated, monovalent IAV-S vaccine and later challenged with an antigenically divergent strain still of the same subtype [
38,
83,
147].
9.1. VAERD Pathogenesis
The best studied experimental model of VAERD is in weanling piglets that are vaccinated with δ-1 cluster H1N2 virus and then challenged with 2009 pandemic H1N1 virus [
38], or vice-versa [
168]. Serum IgG antibodies induced by the vaccine bind to the heterologous challenge virus strain, but provide no significant cross-neutralization. Pigs affected by VAERD sometimes present with more pronounced clinical signs (e.g., fever, dyspnea, coughing, lethargy) compared with non-vaccinated control pigs challenged with the same virus [
38,
169], but clinical signs are not always significantly exacerbated [
83]. Macroscopic pneumonia is more extensive in VAERD pigs’ lungs than in non-vaccinates. Histological examination of VAERD-affected lung reveals more prominent bronchiolitis, peribronchiolar lymphocytic cuffing, and interstitial pneumonia [
38,
169]. IAV-S may also be more widely distributed in the lungs of pigs with VAERD than in non-vaccinated, challenged animals [
83].
VAERD may be the result of multifactorial mechanisms, including dysregulation of proinflammatory cytokines and immune cell types. However, one consistent component seems to be vaccine-induced cross-reactive IgG antibodies in the absence of HI or neutralizing antibodies [
38,
83,
169]. Such cross-reactive IgG antibodies might activate inflammatory immune mechanisms that damage tissues without efficient control of virus replication.
In vitro assays with serum from VAERD-affected pigs showed evidence that these IgG antibodies bind to the heterologous strain but fail to neutralize it and instead promote virus infectivity and fusion, thus leading to elevated replication [
170].
9.2. Alternate Immunization Methods and VAERD
Based on experimental data, initial exposure to a live virus does not promote VAERD. In one study, pigs that were inoculated with live, wild-type virus were partially protected from heterologous challenge infection, whereas the group that received a whole inactivated virus vaccine of the same strain went on to develop VAERD after challenge [
83]. A different study tested whether a live attenuated influenza virus (LAIV) vaccine presenting the same H1N2 surface antigens as the whole inactivated virus vaccine would also potentiate VAERD after heterologous 2009 pandemic H1N1 challenge [
171]. In contrast, the LAIV vaccine conferred significant cross-protection against viral replication and disease. The pronounced difference between inactivated and live vaccines likely results from superior induction of cell-mediated and mucosal immunity with the LAIV vaccine. This further supports the idea that LAIV vaccines could be an effective tool to defend against diverse IAV-S strains.
An additional recent study tested whether VAERD would occur in piglets that received maternal antibodies from whole inactivated virus (WIV)-vaccinated sows, prior to challenge with the heterologous virus [
172]. Mismatched antibodies from the mother were sufficient to prime these piglets for VAERD. This result points to IgG antibodies as the immune mediator mainly responsible for triggering VAERD (as opposed to T lymphocytes), and also raises a note of caution about immunizing breeding sows with WIV vaccines if antigenic relevance of vaccine strains is in doubt.
VAERD-like disease has also been reported in pigs immunized with one experimental DNA vaccine and challenged with an H1N1 IAV-S strain. These vaccines encoded M2e (the highly conserved, extracellular domain of the M2 protein) and induced non-neutralizing antibodies against it [
32]. The relevance of this experiment to VAERD induced by inactivated virus vaccines is uncertain, although it is possible that the underlying mechanisms causing lung lesions are similar.
9.3. Potential for VAERD in Vaccinated Herds
To our knowledge, VAERD has not yet been definitively demonstrated in IAV-S-infected swine herds, although it is inherently difficult to make such a determination under field conditions. Conventionally licensed and autogenous vaccines used in the field contain inactivated viruses with adjuvant, and many circulating IAV-S strains are antigenically distinct from the vaccines, so it is a realistic possibility. On the other hand, vaccines in the VAERD experiments were monovalent, whereas polyvalent commercial vaccines contain antigens from multiple H1 and H3 clusters. Since VAERD pathogenesis requires a major antigenic difference between vaccine and challenge virus (
i.e., two viruses of different clusters), pigs receiving a polyvalent commercial vaccine may not be as vulnerable to VAERD. Data from two studies where pigs were vaccinated with multivalent commercial vaccines before challenge with 2009 pandemic H1N1 virus, with no report of VAERD, support this idea [
162,
173].
Natural infection also differs from the experimental setting in the route of exposure: droplet transmission from animal to animal, versus intratracheal or intranasal inoculation. The impact this might have on VAERD is not known. Thus the VAERD studies, like most animal experiments, simplify some of the complex conditions found in the field to make the outcomes more reproducible. It is nevertheless prudent to take vaccine-enhanced disease into account as a possible hazard to herd health, and consider ways to lower that risk.
9.4. Concepts to Limit the Potential for VAERD
With the imperfect choices of commercial and autogenous IAV-S vaccines available today, what strategies could reduce the risk of VAERD in vaccinated swine herds? The preference for a vaccine with the best-possible antigenic match, even if it is not a full match, holds true with respect to avoiding VAERD. Ideas mentioned above for optimal protection included comparing gene sequence of field isolates and vaccine seed viruses, or developing antisera to commercial vaccines that would enable serological tests against field strains (
Section 7.4). Both of these would be useful ways to identify the most relevant vaccine option and reduce the risk of VAERD. Also, a polyvalent vaccine that contains representatives of multiple H1 and H3 clusters would be less likely to potentiate VAERD than a bivalent vaccine with only one older, antigenically distant strain of each subtype.
Autogenous vaccines provide assurance that the currently circulating strain will be well matched to the vaccine and unlikely to cause VAERD. The drawback is that a new strain belonging to an antigenically divergent cluster could emerge or co-circulate, in which case antibodies already induced by the autogenous vaccine might predispose pigs to VAERD. Perhaps the most stringent way to supply protection and prevent VAERD is to immunize with an autogenous vaccine to control the locally predominant virus plus multivalent commercial vaccine to control other strains that may enter the herd. Finally, experimental data indicate that LAIV vaccines could one day be an excellent tool to provide cross-protection, and a minimal risk of VAERD, during outbreaks with diverse IAV-S strains.
10. New Vaccine Technologies for IAV-S
10.1. Replicon Particle Vectored HA
Alphavirus-like replicon particles (RP) are the first viral vector vaccine technology approved by USDA as a vaccine for IAV-S (“Swine Influenza Vaccine, RNA”; Harrisvaccines, Ames, IA, USA) [
174]. The vector is a modified form of an attenuated Venezuelan equine encephalitis virus, from which structural genes have been deleted, thus preventing multi-cycle replication of the virus (reviewed by Vander Veen
et al. [
175]). In place of deleted genes, it is possible to package RNA that encodes a gene of interest, such as the HA gene of an IAV-S strain. The licensed RP IAV-S vaccine includes a sequence encoding the HA of a cluster IV H3N2 virus. Formation of the propagation-defective particles requires separate strands of RNA (helper RNAs) that are engineered to express the structural genes for only that initial cycle. A recombinant alphavirus RP then has the capacity to bind host cells, deliver genetic cargo to the cytoplasm, and drive protein expression of the inserted gene(s), without producing any progeny virus [
175]. Alphavirus RPs also have inherent adjuvant properties [
176], which likely contributes to effectiveness as a vaccine platform.
In vivo testing of RP-HA vaccines in pigs, using H1 or H3 genes of multiple IAV-S strains, demonstrated their ability to induce robust titers of HA inhibiting (HI) antibodies [
177,
178,
179]. Pigs vaccinated with two doses of RP-HA vaccine (H3 subtype) were protected against homologous H3N2 challenge infection, in terms of nasal shedding of virus, gross lung lesions, and histological lung scores [
177,
180]. Anti-vector immunity against alphavirus RPs is considered to be limited, due to the deleted structural genes, which suggests booster doses can be administered without interference by circulating antibodies [
181]. One might also predict that RP-HA vaccines can resist interference from prior IAV-S immunity, including maternal antibodies, but a study in maternal antibody-positive piglets showed clear interference with immune responses to RP-HA vaccination [
177]. Since HA is the only influenza gene encoded by this vaccine, there is no antibody response directed against other influenza proteins, such as NA, NP, or M1, which may diminish the partial immunity afforded by whole virus vaccines in the absence of a well-matched neutralizing response to the HA. However, the vaccine platform is well-suited to diagnostic strategies of differentiating infected from vaccinated animals (DIVA). Another advantage of the RP-HA vaccine system is its adaptability for making rapid IAV-S strain updates. By routine modern methods, the HA gene from any emerging or newly dominant strain can be cloned (or synthesized from scratch, based on published sequence) and inserted into the RP vector [
179]. This enables rapid establishment of new replicons for vaccine manufacturing.
The replicon particle vaccine platform that was the basis for licensure of “Swine Influenza Vaccine RNA” vaccine, identified as SirraVax™ technology (Harrisvaccines, Inc., Ames, IA, USA), is more frequently being used by Harrisvaccines to generate custom IAV-S vaccines. Each custom RP HA IAV-S vaccine is designed based on HA gene sequence(s) submitted in an electronic format by a veterinarian. A nucleic acid molecule is synthesized to encode the same HA protein sequence. Vero cell cultures are transfected with the recombinant replicon RNA and helper RNA, using electroporation, and incubated for a designated period of time. Harvested RP-HA is then purified by affinity chromatography and formulated, without an adjuvant. Pork producers that have identified multiple distinct strains may submit several HA sequences, enabling the production of polyvalent custom RP-HA vaccines. Combination products blending the RP-HA with custom RP-vectored antigens of other establishment-specific viruses, such as rotavirus variants, are also possible. The custom manufacturing process typically takes 4–6 weeks. These custom vaccines function essentially in the same way as autogenous vaccines. To date, a USDA autogenous vaccine license has not been issued for SirraVax™-based custom RP-HA influenza vaccines. Harrisvaccines produces and sells these products without a USDA license, citing the veterinary practitioner exemption (9 CFR, Part 107.1).
10.2. Adenovirus Vectored IAV-S Genes
Replication-defective human adenovirus serotype 5 (Ad5) is a viral vector strategy with a significant experimental track record in swine. Deletion of a portion of this virus’s genome created a replication defective phenotype, as well as space in the genome to insert foreign genes. Replication defective Ad5 can only be grown in specialized cell lines with genes that compensate for the deleted gene region of the virus.
The HA and NP genes of an H3N2 IAV-S strain were both cloned into Ad5 to generate separate recombinant viruses, which were then tested, together or individually for immunogenicity and vaccine efficacy in pigs [
182]. Single-dose intramuscular delivery of Ad5-HA, with or without the Ad5-NP construct, elicited HI antibodies. Pigs given Ad5-HA alone were protected moderately well against heterologous H3N2 virus challenge, while the combination of Ad5-HA and Ad5-NP supplied more complete protection, in terms of viral replication and lung lesion severity. One of the subsequent studies with this vaccine model showed that the Ad5-HA plus Ad5-NP antigens could be delivered to muscle tissue via a needle-free system, with similar results to intramuscular injection [
183].
It was also shown that the Ad5-vectored IAV-S vaccine can be used effectively in piglets with influenza-specific maternal antibodies. The combined Ad5-HA and Ad5-NP antigens, administered intramuscularly in the face of H3N2-specific maternal antibodies, triggered an active HI antibody response [
184]. A boosting dose with commercial inactivated IAV-S vaccine, given 4 weeks later, produced a clear anamnestic antibody response. This group of pigs then showed superior protection against H3N2 challenge infection compared to pigs that received only the inactivated IAV-S vaccine.
Most recently, a recombinant Ad5 encoding HA of the 2009 pandemic H1N1 virus was used to immunize pigs by a single intranasal dose [
168]. This immunization induced IAV-S-specific T cells and nasal IgA antibodies, while providing solid protection against homologous challenge. Immune mediators elicited by the vaccine were partially cross-reactive with the heterologous H1N2 challenge virus (δ-1 cluster), supplying a modest level of cross-protection.
Collectively, these data show that the adenovirus vector is a potentially useful system for immunizing pigs against IAV-S. The ability to generate IAV-S immunity in piglets with IAV-S specific maternal antibodies is important, but perhaps partly offset by difficulty boosting animals with Ad5-HA after they have already been primed with the same vaccine [
185]. Other desirable factors with Ad5 are the safety of a non-replicating vector and the versatility to respond rapidly to a new epidemic or pandemic by inserting a new, matching HA gene [
186]. Commercial development of this strategy is not underway at this time, to our knowledge, but the significant body of data published in the past suggests it could be a valuable vaccine system, perhaps for use in combination with conventional IAV-S antigens.
10.3. Live-Attenuated (Modified-Live) Virus
Over the past 10 years, experimental studies of live-attenuated influenza virus (LAIV) vaccines for swine have shown great promise. (LAIV is equivalent to modified-live virus (MLV).) The rationale for LAIV vaccines is that since they are administered by the intranasal route, the attenuated viruses replicate moderately in the upper respiratory tract and induce a balanced mucosal and systemic immune response. Mucosal antibodies in the respiratory tract lining are very desirable in the context of controlling airborne influenza virus infection. The potential for replicating LAIV to induce a robust cell-mediated immune response is also an attractive mechanism for defense against IAV-S because most T cells recognize epitopes in the conserved internal proteins, making them broadly reactive against many strains.
Three different research teams developed LAIV vaccines for IAV-S, each using distinct strategies of viral gene manipulation to cause an attenuated phenotype [
36,
187,
188]. All three designs were made possible with reverse genetics technology, which starts by inserting DNA copies of the eight viral RNA gene segments into separate plasmids that are manipulated using standard molecular biology techniques [
189]. A specific reassortant virus can be generated by combining gene segments from two or more viral strains, such as substituting the HA and NA from a contemporary circulating strain onto a lab adapted vaccine backbone strain. Attenuating mutations can be made in one or more gene segments, which then can be combined with HA and NA genes from a vaccine strain, thus yielding a virus with the desired surface antigens plus attenuating properties encoded elsewhere. One of the attenuation approaches was to handicap the virus’s ability to resist host cell type I interferon (IFN), by reducing the length of itsNS1 protein [
35,
187]. A second approach was to make the virus cold-adapted and temperature sensitive by changing amino acid residues in two of the viral polymerase genes, very similarly to the licensed human LAIV vaccine (FluMist) [
36]. A third approach modified the cleavage site in HA to require elastase enzyme; elastase can be added as a supplement during virus cultivation, but it is too scarce in the host animal to support significant replication of the virus [
188].
Independent studies tested all of these vaccine platforms in pigs, using two intranasal doses, for immunogenicity and efficacy against infection. Summarizing briefly, the LAIV vaccines proved immunogenic in terms of eliciting respiratory tract mucosal antibodies, systemic cell-mediated immune responses, and modest levels of systemic neutralizing antibodies [
34,
35,
36,
190]. In terms of efficacy, all of the experimental LAIV vaccines have shown robust protection against challenge with homologous virus, plus measurable and sometimes robust cross-protection against antigenic variant virus. An LAIV vaccine proved efficacious against infection with an antigenically distinct challenge strain, even if the vaccine was administered to piglets with high levels of maternal antibodies [
191].
A key factor in terms of the cost and convenience of vaccine application in the field is whether two doses are required (two doses are required with conventional inactivated vaccines). In one study, piglets receiving a single dose of NS1 mutant LAIV developed cross-protective immunity comparable to piglets that received two doses [
191]. Because sow vaccination represents a large part of the IAV-S vaccine market, another study explored the use of temperature sensitive H1N1pdm09 LAIV in sows [
172]. Piglets from litters that suckled colostrum from LAIV-vaccinated mothers received partial protection against homologous challenge, but not against heterologous δ1-H1N2 challenge.
These studies point to LAIV vaccines as a platform with potential to address two of the critical problems in IAV-S immunization: antigenic variation and interference from circulating maternal antibodies. Another problem that has been linked to whole inactivated virus vaccines, at least in experimental challenge studies, is VAERD. When LAIV and inactivated vaccines of identical strains were administered to piglets, followed by heterologous challenge, LAIV vaccinees showed none of the VAERD phenomenon present in WIV vaccinees, but were in fact partially cross-protected [
171]. Therefore, it can be envisioned that an LAIV vaccine may be successfully developed and licensed, as an equivalent multivalent product has been licensed for humans since 2003 [
192]. Regulatory concerns (e.g., safety and environmental impact) and designing a practical mechanism for intranasal mass-vaccination are among the hurdles that would have to be overcome.
10.4. DNA Vaccines
DNA plasmid vaccines that encode protective antigens are an attractive concept for veterinary medicine, including IAV-S immunization. A DNA vaccine offers a way to induce expression of proteins of interest in the host animal’s cells, which is a key for inducing robust cell-mediated immunity, yet without exposing the host to any pathogens. The platform enables versatile combination of antigens from multiple strains and simple substitution of one strain for another. Maternal antibodies are considered less likely to impede DNA vaccination, compared with injections of protein antigens or attenuated virus.
DNA vaccines against several swine viral pathogens have been tested in experimental settings. A number of these studies have demonstrated immune responses and protection against challenge infection, including IAV-S [
193,
194,
195]. HA-encoding DNA vaccines for IAV-S were administered successfully by intramuscular needle injection as well as a needle-free subcutaneous/intramuscular injection method [
196]. The major drawback with these proof-of-concept studies is that large doses of DNA were required, often in a series of three or more doses. Because materials and labor costs to administer these doses would be high, DNA vaccines for IAV-S are probably not commercially viable, with current methods.
10.5. Subunit Vaccines
Viral subunit vaccines contain protein antigens that are either fractionated from whole virus or expressed individually in a recombinant system. A subunit vaccine consists of a simpler protein mixture than whole inactivated virus, which allows the target proteins (such as HA) to be formulated at a greater concentration and purity. Most human seasonal influenza vaccines in North America and Europe are subunit vaccines derived by treating virus with detergent and isolating the membrane proteins (mainly HA and NA) [
149].
One recently licensed human seasonal influenza vaccine is a subunit product called FluBlok, which is comprised of recombinant HA (rHA) proteins expressed from a recombinant baculovirus vector [
197]. The rHA proteins of different strains are produced in insect cell culture, purified, and blended to form a trivalent final product. The baculovirus-vectored expression system should be equally capable of generating IAV-S vaccine antigens. In addition, the alphavirus RP vector system can be utilized to express high levels of rHA protein in mammalian cell culture [
179,
198]. (This differs from the alphavirus RP vaccine approach discussed in
Section 10.1, which delivers recombinant RP-HA into the animal, leading to
in vivo expression of the rHA protein.) Other culture systems that have been engineered to express immunogenic rHA include
E. coli, yeast, and plants. Subunit vaccines for IAV-S might offer advantages in terms of production efficiency and the flexibility to substitute strains. It is unlikely that subunit IAV-S vaccines would differ very much from conventional inactivated virus vaccines in terms of efficacy, but they would be suitable for developing DIVA diagnostics.
11. Conclusions
IAV-S became a much more costly and complicated problem for the US pork industry after the triple reassortant virus lineage emerged in the late 1990s and began undergoing gene reassortment with various other swine and human influenza viruses. Since that time the antigenic diversity of circulating strains has increased and the field efficacy of conventional IAV-S vaccine formulations has been increasingly questioned. The conventional IAV-S vaccines, which contain inactivated viruses representing up to 5 strains, are designed to induce robust titers of serum neutralizing antibodies. However, the degree of cross-neutralization against antigenically drifted strains is often very limited. Some experimental results suggest that vaccine-induced antibodies can even enhance the severity of subsequent infection with a poorly matched strain.
The efficacy of conventional vaccines would be greater if IAV-S strains were updated more frequently to improve the likelihood of antibodies matching the prevalent circulating strains. The USDA IAV-S surveillance program that was launched in 2010 generates data that could support a coordinated system of vaccine strain selection. Practitioners seeking to identify the best-matched vaccine for an IAV-S strain in a client’s herd can utilize gene sequencing and/or serology to analyze the similarity between that field virus and the strains used in each of the commercial vaccines. Resulting data may indicate there are no closely matching commercial vaccines, and that an autogenous or custom vaccine is a better strategy. Experimental research shows promising results for new forms of IAV-S vaccines, such as live-attenuated or recombinant viral vectors, which may at some point offer a more broadly-protective method of immunizing pigs. Vaccines that reliably prevent IAV-S outbreaks in swine would be expected to improve the efficiency of pork production while reducing the danger of virus transmission to humans.
Acknowledgments
This document was produced with funding provided through a cooperative agreement between Iowa State University and the United States Department of Agriculture, Animal and Plant Health Inspection Services, Veterinary Services. The authors wish to thank the following individuals for helpful comments: Gayle Brown, Marie Culhane, Brian Erdahl, Mark Hammer, Paul Hauer, John K. Johnson, Michael Kuhn, Dave Pyburn, Byron Rippke and Amy Vincent.
Author Contributions
Matthew R. Sandbulte, Anna R. Spickler, Pamela K. Zaabel, and James A. Roth all participated in writing the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Holtkamp, D.; Rotto, H.; Garcia, R. The economic cost of major health challenges in large US swine production systems. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, Orlando, FL, USA, 3–6 March 2007.
- Donovan, T.S. The role of influenza on growing pig performance. In Proceedings of the Allen D. Leman Swine Conference, Saint Paul, MN, USA, 17–20 September 2005.
- Myers, K.P.; Olsen, C.W.; Gray, G.C. Cases of swine influenza in humans: A review of the literature. Clin. Infect. Dis. 2007, 44, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Bowman, A.S.; Nelson, S.W.; Page, S.L.; Nolting, J.M.; Killian, M.L.; Sreevatsan, S.; Slemons, R.D. Swine-to-Human Transmission of Influenza A(H3N2) Virus at Agricultural Fairs, Ohio, USA, 2012. Emerg. Infect. Dis. 2014, 20, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Pappaioanou, M.; Gramer, M. Lessons from pandemic H1N1 2009 to improve prevention, detection, and response to influenza pandemics from a One Health perspective. ILAR J. 2010, 51, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.F. Control of influenza A viruses in large production systems. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, San Diego, CA, USA, 8–11 March 2008.
- Corzo, C.A.; Gramer, M.R.; Kuhn, M.; Mohr, M.; Morrison, R. Observations Regarding Influenza A Virus Shedding in a Swine Breeding Farm After Mass Vaccination. J. Swine Health Prod. 2012, 20, 283–289. [Google Scholar]
- Romagosa, A.; Allerson, M.; Gramer, M.; Joo, H.S.; Deen, J.; Detmer, S.; Torremorell, M. Vaccination of influenza a virus decreases transmission rates in pigs. Vet. Res. 2011. [Google Scholar] [CrossRef]
- Brown, I.H. Influenza Virus Infections of Pigs. Part 1: Swine, Avian & Human Influenza Viruses. Available online: http://www.pighealth.com/influenza.htm (accessed on 22 May 2014).
- Fouchier, R.A.; Munster, V.J. Epidemiology of low pathogenic avian influenza viruses in wild birds. Rev. Sci. Tech. 2009, 28, 49–58. [Google Scholar] [PubMed]
- Heinen, P. Swine influenza: A zoonosis. 9-15-2003. In Vet Sci Tomorrow [Serial Online]; Utrecht University: Utrecht, The Netherlands, 2004. [Google Scholar]
- Reid, A.H.; Taubenberger, J.K. The origin of the 1918 pandemic influenza virus: A continuing enigma. J. Gen. Virol. 2003, 84, 2285–2292. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Webby, R.; Lam, T.T.; Smith, D.K.; Peiris, J.S.; Guan, Y. History of swine influenza viruses in Asia. Curr. Top. Microbiol. Immunol. 2013, 370, 57–68. [Google Scholar] [PubMed]
- Ma, W.; Richt, J.A. Swine influenza vaccines: Current status and future perspectives. Anim. Health Res. Rev. 2010, 11, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Samji, T. Influenza A: Understanding the viral life cycle. Yale J. Biol. Med. 2009, 82, 153–159. [Google Scholar] [PubMed]
- Chakrabarti, A.K.; Pasricha, G. An insight into the PB1F2 protein and its multifunctional role in enhancing the pathogenicity of the influenza A viruses. Virology 2013, 440, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Huber, V.C.; Peltola, V.; Iverson, A.R.; McCullers, J.A. Contribution of vaccine-induced immunity toward either the HA or the NA component of influenza viruses limits secondary bacterial complications. J. Virol. 2010, 84, 4105–4108. [Google Scholar] [CrossRef] [PubMed]
- Kilbourne, E.D.; Laver, W.G.; Schulman, J.L.; Webster, R.G. Antiviral activity of antiserum specific for an influenza virus neuraminidase. J. Virol. 1968, 2, 281–288. [Google Scholar] [PubMed]
- Kilbourne, E.D.; Pokorny, B.A.; Johansson, B.; Brett, I.; Milev, Y.; Matthews, J.T. Protection of mice with recombinant influenza virus neuraminidase. J. Infect. Dis. 2004, 189, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Bosch, B.J.; Bodewes, R.; de Vries, R.P.; Kreijtz, J.H.; Bartelink, W.; van Amerongen, G.; Rimmelzwaan, G.F.; de Haan, C.A.; Osterhaus, A.D.; Rottier, P.J. Recombinant soluble, multimeric HA and NA exhibit distinctive types of protection against pandemic swine-origin 2009 A(H1N1) influenza virus infection in ferrets. J. Virol. 2010, 84, 10366–10374. [Google Scholar] [CrossRef] [PubMed]
- Heinen, P.P.; Rijsewijk, F.A.; de Boer-Luijtze, E.A.; Bianchi, A.T. Vaccination of pigs with a DNA construct expressing an influenza virus M2-nucleoprotein fusion protein exacerbates disease after challenge with influenza A virus. J. Gen. Virol. 2002, 83, 1851–1859. [Google Scholar] [PubMed]
- Kitikoon, P.; Strait, E.L.; Thacker, E.L. The antibody responses to swine influenza virus (SIV) recombinant matrix 1 (rM1), matrix 2 (M2), and hemagglutinin (HA) proteins in pigs with different SIV exposure. Vet. Microbiol. 2008, 126, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Kappes, M.A.; Sandbulte, M.R.; Platt, R.; Wang, C.; Lager, K.M.; Henningson, J.N.; Lorusso, A.; Vincent, A.L.; Loving, C.L.; Roth, J.A.; et al. Vaccination with NS1-truncated H3N2 swine influenza virus primes T cells and confers cross-protection against an H1N1 heterosubtypic challenge in pigs. Vaccine 2012, 30, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Janke, B.H.; Webby, R.J.; Garcia-Sastre, A.; Richt, J.A. Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 2007, 25, 7999–8009. [Google Scholar] [CrossRef] [PubMed]
- Pena, L.; Vincent, A.L.; Ye, J.; Ciacci-Zanella, J.R.; Angel, M.; Lorusso, A.; Gauger, P.C.; Janke, B.H.; Loving, C.L.; Perez, D.R. Modifications in the polymerase genes of a swine-like triple-reassortant influenza virus to generate live attenuated vaccines against 2009 pandemic H1N1 viruses. J. Virol. 2011, 85, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.; Stech, J.; Klenk, H.D. Influenza receptors, polymerase and host range. Rev. Sci. Tech. 2009, 28, 203–217. [Google Scholar] [PubMed]
- Nicholls, J.M.; Chan, R.W.; Russell, R.J.; Air, G.M.; Peiris, J.S. Evolving complexities of influenza virus and its receptors. Trends Microbiol. 2008, 16, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Trebbien, R.; Larsen, L.E.; Viuff, B.M. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol. J. 2011. [Google Scholar] [CrossRef]
- Ito, T.; Couceiro, J.N.; Kelm, S.; Baum, L.G.; Krauss, S.; Castrucci, M.R.; Donatelli, I.; Kida, H.; Paulson, J.C.; Webster, R.G.; et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998, 72, 7367–7373. [Google Scholar]
- Nelli, R.K.; Kuchipudi, S.V.; White, G.A.; Perez, B.B.; Dunham, S.P.; Chang, K.C. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet. Res. 2010. [Google Scholar] [CrossRef]
- Van Poucke, S.G.; Nicholls, J.M.; Nauwynck, H.J.; van Reeth, K. Replication of avian, human and swine influenza viruses in porcine respiratory explants and association with sialic acid distribution. Virol. J. 2010. [Google Scholar] [CrossRef] [Green Version]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 2004, 78, 12665–12667. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Saif, Y.M. Avian influenza virus. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Dundon, W.G.; Milani, A.; Cattoli, G.; Capua, I. Progressive truncation of the Non-Structural 1 gene of H7N1 avian influenza viruses following extensive circulation in poultry. Virus Res. 2006, 119, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Tumpey, T.M.; Alvarez, R.; Swayne, D.E.; Suarez, D.L. Diagnostic approach for differentiating infected from vaccinated poultry on the basis of antibodies to NS1, the nonstructural protein of influenza A virus. J. Clin. Microbiol. 2005, 43, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Pena, L.; Vincent, A.L.; Loving, C.L.; Henningson, J.N.; Lager, K.M.; Li, W.; Perez, D.R. Strain-dependent effects of PB1-F2 of triple-reassortant H3N2 influenza viruses in swine. J. Gen. Virol. 2012, 93, 2204–2214. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.; Chen, L.M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269–4274. [Google Scholar] [CrossRef] [PubMed]
- Gauger, P.C.; Vincent, A.L.; Loving, C.L.; Lager, K.M.; Janke, B.H.; Kehrli, M.E., Jr.; Roth, J.A. Enhanced pneumonia and disease in pigs vaccinated with an inactivated human-like (delta-cluster) H1N2 vaccine and challenged with pandemic 2009 H1N1 influenza virus. Vaccine 2011, 29, 2712–2719. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Janke, B.H.; Richt, J.A. Swine influenza viruses a North American perspective. Adv. Virus Res. 2008, 72, 127–154. [Google Scholar] [PubMed]
- Brown, I.H. History and epidemiology of swine influenza in Europe. Curr. Top. Microbiol. Immunol. 2013, 370, 133–146. [Google Scholar] [PubMed]
- Shope, R.E. Immunization experiments with swine influenza virus. J. Exp. Med. 1936, 64, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Kanegae, Y.; Sugita, S.; Shortridge, K.F.; Yoshioka, Y.; Nerome, K. Origin and evolutionary pathways of the H1 hemagglutinin gene of avian, swine and human influenza viruses: Cocirculation of two distinct lineages of swine virus. Arch. Virol. 1994, 134, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Karasin, A.I.; Carman, S.; Olsen, C.W. Identification of human H1N2 and human-swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003 to 2005). J. Clin. Microbiol. 2006, 44, 1123–1126. [Google Scholar] [CrossRef] [PubMed]
- Karasin, A.I.; Schutten, M.M.; Cooper, L.A.; Smith, C.B.; Subbarao, K.; Anderson, G.A.; Carman, S.; Olsen, C.W. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977–1999: Evidence for wholly human and reassortant virus genotypes. Virus Res. 2000, 68, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.N.; Senne, D.A.; Landgraf, J.S.; Swenson, S.L.; Erickson, G.; Rossow, K.; Liu, L.; Yoon, K.J.; Krauss, S.; Webster, R.G. Emergence of H3N2 reassortant influenza A viruses in North American pigs. Vet. Microbiol. 2000, 74, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Poljak, Z.; Friendship, R.M.; Carman, S.; McNab, W.B.; Dewey, C.E. Investigation of exposure to swine influenza viruses in Ontario (Canada) finisher herds in 2004 and 2005. Prev. Vet. Med. 2008, 83, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, C.A.; Spearman, G.; Hamel, A.; Godson, D.L.; Fortin, A.; Fontaine, G.; Tremblay, D. Characterization of a Canadian mink H3N2 influenza A virus isolate genetically related to triple reassortant swine influenza virus. J. Clin. Microbiol. 2009, 47, 796–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsen, C.W.; Karasin, A.I.; Carman, S.; Li, Y.; Bastien, N.; Ojkic, D.; Alves, D.; Charbonneau, G.; Henning, B.M.; Low, D.E.; et al. Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg. Infect. Dis. 2006, 12, 1132–1135. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.L.; Swenson, S.L.; Lager, K.M.; Gauger, P.C.; Loiacono, C.M.; Zhang, Y. Characterization of an influenza A virus isolated from pigs during an outbreak of respiratory disease in swine and people during a county fair in the United States. Vet. Microbiol. 2009, 137, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Gramer, M.R.; Richt, J.A.; Janke, B.H. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 2009, 39, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Anderson, T.K.; Nelson, M.I.; Kitikoon, P.; Swenson, S.L.; Korslund, J.A.; Vincent, A.L. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir. Viruses 2013, 7, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Vincent, A.L.; Lager, K.M.; Janke, B.H.; Henry, S.C.; Rowland, R.R.; Hesse, R.A.; Richt, J.A. Identification and characterization of a highly virulent triple reassortant H1N1 swine influenza virus in the United States. Virus Genes 2010, 40, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, A.; Vincent, A.L.; Harland, M.L.; Alt, D.; Bayles, D.O.; Swenson, S.L.; Gramer, M.R.; Russell, C.A.; Smith, D.J.; Lager, K.M.; et al. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J. Gen. Virol. 2011, 92, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.I.; Vincent, A.L.; Kitikoon, P.; Holmes, E.C.; Gramer, M.R. Evolution of novel reassortant A/H3N2 influenza viruses in North American swine and humans, 2009–2011. J. Virol. 2012, 86, 8872–8878. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V.; et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009, 325, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Corzo, C.A.; Culhane, M.; Juleen, K.; Stigger-Rosser, E.; Ducatez, M.F.; Webby, R.J.; Lowe, J.F. Active surveillance for influenza A virus among swine, midwestern United States, 2009–2011. Emerg. Infect. Dis. 2013, 19, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Ducatez, M.F.; Hause, B.; Stigger-Rosser, E.; Darnell, D.; Corzo, C.; Juleen, K.; Simonson, R.; Brockwell-Staats, C.; Rubrum, A.; Wang, D.; et al. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg. Infect. Dis. 2011, 17, 1624–1629. [Google Scholar] [CrossRef] [PubMed]
- Nfon, C.K.; Berhane, Y.; Hisanaga, T.; Zhang, S.; Handel, K.; Kehler, H.; Labrecque, O.; Lewis, N.S.; Vincent, A.L.; Copps, J.; et al. Characterization of H1N1 swine influenza viruses circulating in Canadian pigs in 2009. J. Virol. 2011, 85, 8667–8679. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Khatri, M.; Wang, L.; Saif, Y.M.; Lee, C.W. Identification of swine H1N2/pandemic H1N1 reassortant influenza virus in pigs, United States. Vet. Microbiol. 2012, 158, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Ji, J.; Chang, S.; Xue, C.; Ma, J.; Bi, Y.; Xie, Q. Phylogenetic diversity and genotypic complexity of H1N1 subtype swine influenza viruses isolated in mainland China. Virol. J. 2012. [Google Scholar] [CrossRef]
- Tremblay, D.; Allard, V.; Doyon, J.F.; Bellehumeur, C.; Spearman, J.G.; Harel, J.; Gagnon, C.A. Emergence of a new swine H3N2 and pandemic (H1N1) 2009 influenza A virus reassortant in two Canadian animal populations, mink and swine. J. Clin. Microbiol. 2011, 49, 4386–4390. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Vincent, A.L.; Gramer, M.R.; Brockwell, C.B.; Lager, K.M.; Janke, B.H.; Gauger, P.C.; Patnayak, D.P.; Webby, R.J.; Richt, J.A. Identification of H2N3 influenza A viruses from swine in the United States. Proc. Natl. Acad. Sci. USA 2007, 104, 20949–20954. [Google Scholar] [CrossRef] [PubMed]
- Killian, M.L.; Zhang, Y.; Panigrahy, B.; Trampel, D.; Yoon, K.J. Identification and characterization of H2N3 avian influenza virus from backyard poultry and comparison with novel H2N3 swine influenza virus. Avian Dis. 2011, 55, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Lekcharoensuk, P.; Lager, K.M.; Vemulapalli, R.; Woodruff, M.; Vincent, A.L.; Richt, J.A. Novel swine influenza virus subtype H3N1, United States. Emerg. Infect. Dis. 2006, 12, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Gramer, M.; Rossow, K.; Yoon, K.J. Isolation and genetic characterization of new reassortant H3N1 swine influenza virus from pigs in the midwestern United States. J. Virol. 2006, 80, 5092–5096. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.Y.; Song, M.S.; Lee, E.H.; Lee, Y.M.; Kim, S.Y.; Kim, H.K.; Choi, J.K.; Kim, C.J.; Webby, R.J.; Choi, Y.K. Isolation and characterization of novel H3N1 swine influenza viruses from pigs with respiratory diseases in Korea. J. Clin. Microbiol. 2006, 44, 3923–3927. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Barbieri, I.; Sozzi, E.; Luppi, A.; Lelli, D.; Lombardi, G.; Zanoni, M.G.; Cordioli, P. Novel swine influenza virus subtype H3N1 in Italy. Vet. Microbiol. 2009, 138, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Cappuccio, J.A.; Pena, L.; Dibarbora, M.; Rimondi, A.; Pineyro, P.; Insarralde, L.; Quiroga, M.A.; Machuca, M.; Craig, M.I.; Olivera, V.; et al. Outbreak of swine influenza in Argentina reveals a non-contemporary human H3N2 virus highly transmissible among pigs. J. Gen. Virol. 2011, 92, 2871–2878. [Google Scholar] [CrossRef] [PubMed]
- Meseko, C.; Olaleye, D.; Capua, I.; Cattoli, G. Swine influenza in sub-saharan Africa—Current knowledge and emerging insights. Zoonoses Public Health 2014, 61, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Waffarn, E.E.; Baumgarth, N. Protective B cell responses to flu—No fluke! J. Immunol. 2011, 186, 3823–3829. [Google Scholar] [CrossRef] [PubMed]
- Kreijtz, J.H.; Fouchier, R.A.; Rimmelzwaan, G.F. Immune responses to influenza virus infection. Virus Res. 2011, 162, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Braciale, T.J.; Sun, J.; Taeg, S.K. Regulating the adaptive immune response to respiratory virus infection. Nat. Rev. Immunol. 2012, 12, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Achdout, H.; Meningher, T.; Hirsh, S.; Glasner, A.; Bar-On, Y.; Gur, C.; Porgador, A.; Mendelson, M.; Mandelboim, M.; Mandelboim, O. Killing of avian and swine influenza virus by natural killer cells. J. Virol. 2010, 84, 3993–4001. [Google Scholar] [CrossRef] [PubMed]
- Larsen, D.L.; Karasin, A.; Zuckermann, F.; Olsen, C.W. Systemic and mucosal immune responses to H1N1 influenza virus infection in pigs. Vet. Microbiol. 2000, 74, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.W.; Bey, R.F.; Baarsch, M.J.; Larson, M.E. Class specific antibody response to influenza A H1N1 infection in swine. Vet. Microbiol. 1995, 43, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Pomorska-Mol, M.; Markowska-Daniel, I.; Kwit, K. Immune and acute phase response in pigs experimentally infected with H1N2 swine influenza virus. FEMS Immunol. Med. Microbiol. 2012, 66, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Heinen, P.P.; van Nieuwstadt, A.P.; Pol, J.M.; de Boer-Luijtze, E.A.; van Oirschot, J.T.; Bianchi, A.T. Systemic and mucosal isotype-specific antibody responses in pigs to experimental influenza virus infection. Viral Immunol. 2000, 13, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Van Reeth, K.; Labarque, G.; Pensaert, M. Serological profiles after consecutive experimental infections of pigs with European H1N1, H3N2, and H1N2 swine influenza viruses. Viral Immunol. 2006, 19, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Grebe, K.M.; Yewdell, J.W.; Bennink, J.R. Heterosubtypic immunity to influenza A virus: Where do we stand? Microbes Infect. 2008, 10, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Khatri, M.; Dwivedi, V.; Krakowka, S.; Manickam, C.; Ali, A.; Wang, L.; Qin, Z.; Renukaradhya, G.J.; Lee, C.W. Swine influenza H1N1 virus induces acute inflammatory immune responses in pig lungs: A potential animal model for human H1N1 influenza virus. J. Virol. 2010, 84, 11210–11218. [Google Scholar] [CrossRef] [PubMed]
- Bikour, M.H.; Cornaglia, E.; Elazhary, Y. Evaluation of a protective immunity induced by an inactivated influenza H3N2 vaccine after an intratracheal challenge of pigs. Can. J. Vet. Res. 1996, 60, 312–314. [Google Scholar] [PubMed]
- Vincent, A.L.; Lager, K.M.; Janke, B.H.; Gramer, M.R.; Richt, J.A. Failure of protection and enhanced pneumonia with a US H1N2 swine influenza virus in pigs vaccinated with an inactivated classical swine H1N1 vaccine. Vet. Microbiol. 2008, 126, 310–323. [Google Scholar] [CrossRef] [PubMed]
- Johansson, B.E.; Moran, T.M.; Kilbourne, E.D. Antigen-presenting B cells and helper T cells cooperatively mediate intravirionic antigenic competition between influenza A virus surface glycoproteins. Proc. Natl. Acad. Sci. USA 1987, 84, 6869–6873. [Google Scholar] [CrossRef] [PubMed]
- Van Reeth, K.; Gregory, V.; Hay, A.; Pensaert, M. Protection against a European H1N2 swine influenza virus in pigs previously infected with H1N1 and/or H3N2 subtypes. Vaccine 2003, 21, 1375–1381. [Google Scholar] [CrossRef] [PubMed]
- Marcelin, G.; Sandbulte, M.R.; Webby, R.J. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev. Med. Virol. 2012, 22, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Van Reeth, K.; Brown, I.; Essen, S.; Pensaert, M. Genetic relationships, serological cross-reaction and cross-protection between H1N2 and other influenza A virus subtypes endemic in European pigs. Virus Res. 2004, 103, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Gauger, P.C.; Vincent, A.L. Serum virus neutralization assay for detection and quantitation of serum-neutralizing antibodies to influenza A virus in swine. Methods Mol. Biol. 2014, 1161, 313–324. [Google Scholar] [PubMed]
- Kitikoon, P.; Vincent, A.L. Microneutralization assay for swine influenza virus in swine serum. Methods Mol. Biol. 2014, 1161, 325–335. [Google Scholar] [PubMed]
- Abed, Y.; Hardy, I.; Li, Y.; Boivin, G. Divergent evolution of hemagglutinin and neuraminidase genes in recent influenza A:H3N2 viruses isolated in Canada. J. Med. Virol. 2002, 67, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Kilbourne, E.D.; Johansson, B.E.; Grajower, B. Independent and disparate evolution in nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc. Natl. Acad. Sci. USA 1990, 87, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Westgeest, K.B.; de Graaf, M.; Fourment, M.; Bestebroer, T.M.; van Beek, R.; Spronken, M.I.; de Jong, J.C.; Rimmelzwaan, G.F.; Russell, C.A.; Osterhaus, A.D.; et al. Genetic evolution of the neuraminidase of influenza A (H3N2) viruses from 1968 to 2009 and its correspondence to haemagglutinin evolution. J. Gen. Virol. 2012, 93, 1996–2007. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.B.; Braciale, T.J. Resistance to and recovery from lethal influenza virus infection in B lymphocyte-deficient mice. J. Exp. Med. 1997, 186, 2063–2068. [Google Scholar] [CrossRef] [PubMed]
- Heinen, P.P.; de Boer-Luijtze, E.A.; Bianchi, A.T. Respiratory and systemic humoral and cellular immune responses of pigs to a heterosubtypic influenza A virus infection. J. Gen. Virol. 2001, 82, 2697–2707. [Google Scholar] [PubMed]
- Lange, E.; Kalthoff, D.; Blohm, U.; Teifke, J.P.; Breithaupt, A.; Maresch, C.; Starick, E.; Fereidouni, S.; Hoffmann, B.; Mettenleiter, T.C.; et al. Pathogenesis and transmission of the novel swine-origin influenza virus A/H1N1 after experimental infection of pigs. J. Gen. Virol. 2009, 90, 2119–2123. [Google Scholar] [CrossRef] [PubMed]
- McMichael, A.J.; Gotch, F.M.; Noble, G.R.; Beare, P.A. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 1983, 309, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Hillaire, M.L.; Rimmelzwaan, G.F.; Kreijtz, J.H. Clearance of influenza virus infections by T cells: Risk of collateral damage? Curr. Opin. Virol. 2013, 3, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Doherty, P.C.; Christensen, J.P. Accessing complexity: The dynamics of virus-specific T cell responses. Annu. Rev. Immunol. 2000, 18, 561–592. [Google Scholar] [CrossRef] [PubMed]
- Flynn, K.J.; Belz, G.T.; Altman, J.D.; Ahmed, R.; Woodland, D.L.; Doherty, P.C. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity 1998, 8, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Flynn, K.J.; Riberdy, J.M.; Christensen, J.P.; Altman, J.D.; Doherty, P.C. In vivo proliferation of naive and memory influenza-specific CD8+ T cells. Proc. Natl. Acad. Sci. USA 1999, 96, 8597–8602. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Braciale, T.J. Role of T cell immunity in recovery from influenza virus infection. Curr. Opin. Virol. 2013, 3, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Wrammert, J.; Koutsonanos, D.; Li, G.M.; Edupuganti, S.; Sui, J.; Morrissey, M.; McCausland, M.; Skountzou, I.; Hornig, M.; Lipkin, W.I.; et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 2011, 208, 181–193. [Google Scholar] [CrossRef] [PubMed]
- De Vleeschauwer, A.R.; van Poucke, S.G.; Karasin, A.I.; Olsen, C.W.; van Reeth, K. Cross-protection between antigenically distinct H1N1 swine influenza viruses from Europe and North America. Influenza Other Respir. Viruses 2011, 5, 115–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillaire, M.L.; van Trierum, S.E.; Kreijtz, J.H.; Bodewes, R.; Geelhoed-Mieras, M.M.; Nieuwkoop, N.J.; Fouchier, R.A.; Kuiken, T.; Osterhaus, A.D.; Rimmelzwaan, G.F. Cross-protective immunity against influenza pH1N1 2009 viruses induced by seasonal influenza A (H3N2) virus is mediated by virus-specific T-cells. J. Gen. Virol. 2011, 92, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Van Reeth, K.; Braeckmans, D.; Cox, E.; van Borm, S.; van den Berg, T.; Goddeeris, B.; de Vleeschauwer, A. Prior infection with an H1N1 swine influenza virus partially protects pigs against a low pathogenic H5N1 avian influenza virus. Vaccine 2009, 27, 6330–6339. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.; Vincent, A.L.; Gauger, P.C.; Loving, C.L.; Zanella, E.L.; Lager, K.M.; Kehrli, M.E., Jr.; Kimura, K.; Roth, J.A. Comparison of humoral and cellular immune responses to inactivated swine influenza virus vaccine in weaned pigs. Vet. Immunol. Immunopathol. 2011, 142, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.M.; Schram, B.R.; McGregor, M.W.; Sheoran, A.S.; Olsen, C.W.; Lunn, D.P. Local and systemic isotype-specific antibody responses to equine influenza virus infection versus conventional vaccination. Vaccine 1998, 16, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Heinen, P.P.; van Nieuwstadt, A.P.; de Boer-Luijtze, E.A.; Bianchi, A.T. Analysis of the quality of protection induced by a porcine influenza A vaccine to challenge with an H3N2 virus. Vet. Immunol. Immunopathol. 2001, 82, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Wang, Z.; Sreenivasan, C.C.; Hause, B.M.; Gourapura, R.; Li, F.; Francis, D.H.; Kaushik, R.S.; Khatri, M. Poly I:C adjuvanted inactivated swine influenza vaccine induces heterologous protective immunity in pigs. Vaccine 2014. [Google Scholar] [CrossRef]
- Salmon, H.; Berri, M.; Gerdts, V.; Meurens, F. Humoral and cellular factors of maternal immunity in swine. Dev. Comp. Immunol. 2009, 33, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Thacker, B. Vaccination strategies for swine influenza virus. In Proceedings of the Keeping pace with SIV: Proceedings from the Allen D. Leman Swine Conference, Minneapolis, MN, USA; 2000. [Google Scholar]
- Renshaw, H.W. Influence of antibody-mediated immune suppression on clinical, viral, and immune responses to swine influenza infection. Am. J. Vet. Res. 1975, 36, 5–13. [Google Scholar] [PubMed]
- Blaskovic, D.; Rathova, V.; Kociskova, D.; Kaplan, M.M.; Jamrichova, O.; Sadecky, E. Experimental infection of weanling pigs with A-swine influenza virus. 3. Immunity in piglets farrowed by antibody-bearing dams experimentally infected a year earlier. Bull. World Health Organ. 1970, 42, 771–777. [Google Scholar] [PubMed]
- Mensik, J.; Valicek, L.; Pospisil, Z. Pathogenesis of swine influenza infection produced experimentally in suckling piglets. 3. Multiplication of virus in the respiratory tract of suckling piglets in the presence of colostrum-derived specific antibody in their blood stream. Zentralbl. Veterinarmed. B 1971, 18, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Mensik, J.; Pokorny, J. Development of antibody response to swine influenza virus in pigs. I. The influence of experimental infection of pregnant sows on serum antibody production by their progeny during postnatal development. Zentralbl. Veterinarmed. B 1971, 18, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Goyal, S.M.; Joo, H.S. Evaluation of transmission of swine influenza type A subtype H1N2 virus in seropositive pigs. Am. J. Vet. Res. 2004, 65, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Loeffen, W.L.; Heinen, P.P.; Bianchi, A.T.; Hunneman, W.A.; Verheijden, J.H. Effect of maternally derived antibodies on the clinical signs and immune response in pigs after primary and secondary infection with an influenza H1N1 virus. Vet. Immunol. Immunopathol. 2003, 92, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Markowska-Daniel, I.; Pomorska-Mol, M.; Pejsak, Z. The influence of age and maternal antibodies on the postvaccinal response against swine influenza viruses in pigs. Vet. Immunol. Immunopathol. 2011, 142, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Kitikoon, P.; Nilubol, D.; Erickson, B.J.; Janke, B.H.; Hoover, T.C.; Sornsen, S.A.; Thacker, E.L. The immune response and maternal antibody interference to a heterologous H1N1 swine influenza virus infection following vaccination. Vet. Immunol. Immunopathol. 2006, 112, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Allerson, M.W.; Davies, P.R.; Gramer, M.R.; Torremorell, M. Infection dynamics of pandemic 2009 H1N1 influenza virus in a two-site swine herd. Transbound. Emerg. Dis. 2014, 61, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Larsen, L.E.; Christian, N.K.; Aakerblom, S.; Hjulsager, C.K.; Nielsen, J.P.; Stege, H.H.; Kristensen, C.S.; Elvstrom, A.; Lau, L. Dynamics of swine influenza infections in the farrowing unit of a Danish sow herd. In Proceedings of the 21st IPVS Congress, Vancouver, BC, Canada, 18–21 July 2010; p. 80.
- Allerson, M.; Deen, J.; Detmer, S.E.; Gramer, M.R.; Joo, H.S.; Romagosa, A.; Torremorell, M. The impact of maternally derived immunity on influenza A virus transmission in neonatal pig populations. Vaccine 2013, 31, 500–505. [Google Scholar] [CrossRef] [PubMed]
- USDA Swine Influenza Surveillance Update; USDA Animal and Plant Health Inspection Service. Available online: http://www.aphis.usda.gov/wps/portal/aphis/ourfocus/animalhealth/sa_animal_disease_information/sa_swine_health/ct_siv_surveillance/ (accessed on 15 October 2014).
- Influenza Surveillance in Swine: Procedures Manual; USDA Animal and Plant Health Inspection Service. Available online: http://www.aphis.usda.gov/animal_health/animal_dis_spec/swine/downloads/influenza_surv_swine_proc_manual.pdf (accessed on 22 January 2014).
- National Surveillance Plan for Swine Influenza Virus in Pigs, Version 3.2; USDA Animal and Plant Health Inspection Service. Available online: http://www.aphis.usda.gov/animal_health/animal_dis_spec/swine/downloads/appendix_a_national_surv_plan.pdf (accessed on 22 January 2014).
- Centers for Disease Control and Prevention. Overview of Influenza Surveillance in the United States. 6 October 2014. Available online: http://www.cdc.gov/flu/pdf/weekly/overview.pdf (accessed on 3 April 2015). [Google Scholar]
- USDA Swine Program Initiatives and Updates: Collaboration to Achieve Results; Presentation by USDA-APHIS-Veterinary Services: Ames, IA, USA, 1 April 2014.
- Nelson, M.I.; Gramer, M.R.; Vincent, A.L.; Holmes, E.C. Global transmission of influenza viruses from humans to swine. J. Gen. Virol. 2012, 93, 2195–2203. [Google Scholar] [CrossRef] [PubMed]
- Janke, B.H. Diagnosis of swine influenza. Swine Health Prod. 2000, 8, 79–84. [Google Scholar]
- Swenson, S.L.; Vincent, L.L.; Lute, B.M.; Janke, B.H.; Lechtenberg, K.E.; Landgraf, J.G.; Schmitt, B.J.; Kinker, D.R.; McMillen, J.K. A comparison of diagnostic assays for the detection of type A swine influenza virus from nasal swabs and lungs. J. Vet. Diagn Investig. 2001, 13, 36–42. [Google Scholar] [CrossRef]
- Hill, C.; Torremorell, M. Evaluating Sampling Strategies to Diagnose Influenza Virus in Pre-Weaned Pigs. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, San Diego, CA, USA, 2–5 March 2013.
- Goodell, C.K.; Kittawornrat, A.; Panyasing, Y.; Olsen, C.; Zhou, F.; Overbay, T.; Rauh, R.; Nelson, W.M.; O’Connell, C.; Burrell, A.; et al. Influenza A Virus Detection from Oral Fluid and Nasal Swabs of IAV-Innoculated Pigs. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, San Diego, CA, USA, 2–5 March 2013.
- Dobesh, K.; Bretey, K.; Holtkamp, D. Utilizing Snout Wiping for PCR Detection and Virus Isolation of SIV in Nursery Pigs. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, San Diego, CA, USA, 2–5 March 2013.
- Swine Influenza. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; World Organisation for Animal Health (OIE): Paris, France, 2013; pp. 1–11.
- Janke, B.H. Influenza A virus infections in swine: Pathogenesis and diagnosis. Vet. Pathol. 2014, 51, 410–426. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.W.; Bey, R.F.; Baarsch, M.J.; Emery, D.A. Subtype specific ELISA for the detection of antibodies against influenza A H1N1 and H3N2 in swine. J. Virol. Methods 1993, 45, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Skibbe, D.; Zhou, E.M.; Janke, B.H. Comparison of a commercial enzyme-linked immunosorbent assay with hemagglutination inhibition assay for serodiagnosis of swine influenza virus (H1N1) infection. J. Virol. Methods 2004, 16, 86–89. [Google Scholar]
- Barbe, F.; Labarque, G.; Pensaert, M.; van Reeth, K. Performance of a commercial Swine influenza virus H1N1 and H3N2 antibody enzyme-linked immunosorbent assay in pigs experimentally infected with European influenza viruses. J. Vet. Diagn. Investig. 2009, 21, 88–96. [Google Scholar] [CrossRef]
- Ciacci-Zanella, J.R.; Vincent, A.L.; Prickett, J.R.; Zimmerman, S.M.; Zimmerman, J.J. Detection of anti-influenza A nucleoprotein antibodies in pigs using a commercial influenza epitope-blocking enzyme-linked immunosorbent assay developed for avian species. J. Vet. Diagn. Investig. 2010, 22, 3–9. [Google Scholar] [CrossRef]
- Donovan, T.S. Influenza isolate selection methodology for timely autogenous vaccine use. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, San Diego, CA, USA, 8–11 March 2008.
- Senne, D.A. Virus propagation in embryonating eggs. In A Laboratory Manual for the Isolation and Identification of Avian Pathogens, 4th ed.; Swayne, D.E., Glisson, J.R., Jackwood, M.W., Pearson, J.E., Reed, W.M., Eds.; American Association of Avian Pathologists, University of Pennsylvania: Kennett Square, PA, USA, 1998; pp. 235–240. [Google Scholar]
- Fouchier, R.A.; Bestebroer, T.M.; Herfst, S.; van der Kemp, L.; Rimmelzwaan, G.F.; Osterhaus, A.D. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 2000, 38, 4096–4101. [Google Scholar] [PubMed]
- Spackman, E.; Senne, D.A.; Myers, T.J.; Bulaga, L.L.; Garber, L.P.; Perdue, M.L.; Lohman, K.; Daum, L.T.; Suarez, D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002, 40, 3256–3260. [Google Scholar] [CrossRef] [PubMed]
- Landolt, G.A.; Karasin, A.I.; Hofer, C.; Mahaney, J.; Svaren, J.; Olsen, C.W. Use of real-time reverse transcriptase polymerase chain reaction assay and cell culture methods for detection of swine influenza A viruses. Am. J. Vet. Res. 2005, 66, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Vincent, L.L.; Janke, B.H.; Paul, P.S.; Halbur, P.G. A monoclonal-antibody-based immunohistochemical method for the detection of swine influenza virus in formalin-fixed, paraffin-embedded tissues. J. Vet. Diagn. Investig. 1997, 9, 191–195. [Google Scholar] [CrossRef]
- Veterinary Services Memorandum No. 800.203: General Licensing Considerations: Compatibility of Components; USDA Animal and Plant Health Inspection Service: Ames, IA, USA, 2007.
- Vincent, A.L.; Ma, W.; Lager, K.M.; Richt, J.A.; Janke, B.H.; Sandbulte, M.R.; Gauger, P.C.; Loving, C.L.; Webby, R.J.; Garcia-Sastre, A. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. J. Virol. 2012, 86, 10597–10605. [Google Scholar] [CrossRef] [PubMed]
- Russell, C.A.; Jones, T.C.; Barr, I.G.; Cox, N.J.; Garten, R.J.; Gregory, V.; Gust, I.D.; Hampson, A.W.; Hay, A.J.; Hurt, A.C.; et al. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine 2008, 26, D31–D34. [Google Scholar] [CrossRef] [PubMed]
- Stohr, K.; Bucher, D.; Colgate, T.; Wood, J. Influenza virus surveillance, vaccine strain selection, and manufacture. Methods Mol. Biol. 2012, 865, 147–162. [Google Scholar] [PubMed]
- AASV H1N1 Influenza Working Group. American Association of Swine Veterinarians Position Statement on Pandemic (H1N1) 2009 Influenza. Available online: https://www.aasv.org/aasv/position-pH1N1.pdf (accessed on 23 May 2014).
- Nelson, M.I.; Lemey, P.; Tan, Y.; Vincent, A.; Lam, T.T.; Detmer, S.; Viboud, C.; Suchard, M.A.; Rambaut, A.; Holmes, E.C.; et al. Spatial dynamics of human-origin H1 influenza A virus in North American swine. PLOS Pathog. 2011, 7, e1002077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Center for Veterinary Biologics Notice No. 07-17: Labeling of Equine Influenza and Swine Influenza Vaccines; USDA Animal and Plant Health Inspection Service: Ames, IA, USA, 2007.
- Kitikoon, P.; Gauger, P.C.; Anderson, T.K.; Culhane, M.R.; Swenson, S.; Loving, C.L.; Perez, D.R.; Vincent, A.L. Swine influenza virus vaccine serologic cross-reactivity to contemporary US swine H3N2 and efficacy in pigs infected with an H3N2 similar to 2011–2012 H3N2v. Influenza Other Respir. Viruses 2013, 7, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Gramer, M.R.; Joo, H.S. Efficacy of swine influenza A virus vaccines against an H3N2 virus variant. Can. J. Vet. Res. 2007, 71, 207–212. [Google Scholar] [PubMed]
- Loving, C.L.; Lager, K.M.; Vincent, A.L.; Brockmeier, S.L.; Gauger, P.C.; Anderson, T.K.; Kitikoon, P.; Perez, D.R.; Kehrli, M.E., Jr. Efficacy in pigs of inactivated and live attenuated influenza virus vaccines against infection and transmission of an emerging H3N2 similar to the 2011–2012 H3N2v. J. Virol. 2013, 87, 9895–9903. [Google Scholar] [CrossRef] [PubMed]
- Detmer, S.E.; Gramer, M.R.; King, V.L.; Mathur, S.; Rapp-Gabrielson, V.J. In vivo evaluation of vaccine efficacy against challenge with a contemporary field isolate from the alpha cluster of H1N1 swine influenza virus. Can. J. Vet. Res. 2013, 77, 24–32. [Google Scholar] [PubMed]
- Grgic, H.; Costa, M.; Friendship, R.M.; Carman, S.; Nagy, E.; Wideman, G.; Weese, S.; Poljak, Z. Molecular characterization of H3N2 influenza A viruses isolated from Ontario swine in 2011 and 2012. Virol. J. 2014, 11. [Google Scholar] [CrossRef] [PubMed]
- Lewis, N.S.; Anderson, T.K.; Kitikoon, P.; Skepner, E.; Burke, D.F.; Vincent, A.L. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. J. Virol. 2014, 88, 4752–4763. [Google Scholar] [CrossRef] [PubMed]
- Kehrli, M.E.; Vincent, A.L.; Loving, C.L.; Lager, K.M. Swine influenza virus vaccine research and technology: What does the future hold and what are our next steps? In Proceedings of the 21st Annual Swine Disease Conference for Swine Practioners, Ames, IA, USA; 2013. [Google Scholar]
- USDA-APHIS. Autogenous Biologics. Available online: http://www.aphis.usda.gov/animal_health/vet_biologics/publications/pel_4_16.pdf (accessed on 22 May 2014).
- Draayer, H.A. Autogenous vaccines: Determining product need and antigen selection, production and testing considerations. Dev. Biol. (Basel) 2004, 117, 43–47. [Google Scholar]
- Vincent, A.L.; Ciacci-Zanella, J.R.; Lorusso, A.; Gauger, P.C.; Zanella, E.L.; Kehrli, M.E., Jr.; Janke, B.H.; Lager, K.M. Efficacy of inactivated swine influenza virus vaccines against the 2009 A/H1N1 influenza virus in pigs. Vaccine 2010, 28, 2782–2787. [Google Scholar] [CrossRef] [PubMed]
- 9 CFR 113.113 Autogenous Biologics. In Code of Federal Regulations—Title 9: Animals and Animal Products; U.S. Government Publishing Office: Washington, DC. Available online: http://www.gpo.gov/fdsys/pkg/CFR-2012-title9-vol1/xml/CFR-2012-title9-vol1-sec113-113.xml (accessed on 23 May 2014).
- 9 CFR 113.200 General requirements for killed virus vaccines. In Code of Federal Regulations—Title 9: Animals and Animal Products; U.S. Government Publishing Office: Washington, DC. Available online: http://www.gpo.gov/fdsys/pkg/CFR-2012-title9-vol1/xml/CFR-2012-title9-vol1-sec113-200.xml (accessed on 23 May 2014).
- Veterinary Services Memorandum No. 800.69: Guidelines for Autogenous Biologics; USDA Animal and Plant Health Inspection Service: Ames, IA, USA, 2009.
- Chase, C.C. Autogenous vaccines: Current use in the field in the U.S. cattle and hog industry. Dev. Biol. (Basel) 2004, 117, 69–71. [Google Scholar]
- Huisman, W.; Martina, B.E.; Rimmelzwaan, G.F.; Gruters, R.A.; Osterhaus, A.D. Vaccine-induced enhancement of viral infections. Vaccine 2009, 27, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Braucher, D.R.; Henningson, J.N.; Loving, C.L.; Vincent, A.L.; Kim, E.; Steitz, J.; Gambotto, A.A.; Kehrli, M.E., Jr. Intranasal vaccination with replication-defective adenovirus type 5 encoding influenza virus hemagglutinin elicits protective immunity to homologous challenge and partial protection to heterologous challenge in pigs. Clin. Vaccine Immunol. 2012, 19, 1722–1729. [Google Scholar] [CrossRef] [PubMed]
- Gauger, P.C.; Vincent, A.L.; Loving, C.L.; Henningson, J.N.; Lager, K.M.; Janke, B.H.; Kehrli, M.E., Jr.; Roth, J.A. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet. Pathol. 2012, 49, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.; Loving, C.L.; Manischewitz, J.; King, L.R.; Gauger, P.C.; Henningson, J.; Vincent, A.L.; Golding, H. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci. Transl. Med. 2013, 5. [Google Scholar] [CrossRef]
- Gauger, P.C.; Loving, C.L.; Khurana, S.; Lorusso, A.; Perez, D.R.; Kehrli, M.E., Jr.; Roth, J.A.; Golding, H.; Vincent, A.L. Live attenuated influenza A virus vaccine protects against A(H1N1)pdm09 heterologous challenge without vaccine associated enhanced respiratory disease. Virology 2014, 471 473, 93–104. [Google Scholar] [CrossRef] [PubMed]
- RaJao, D.; Vincent, A.; Loving, C.; Gauger, P.; Sandbulte, M.; Kitikoon, P. Maternally Derived Antibodies Induce Vaccine-Associated Enhanced Respiratory Disease in Weaned Pigs Challenged with Heterologous Virus. In Proceedings of the American Association of Swine Veterinarians Annual Meeting, Dallas, TX, USA; 2014. [Google Scholar]
- Loeffen, W.L.; Stockhofe, N.; Weesendorp, E.; van Zoelen-Bos, D.; Heutink, R.; Quak, S.; Goovaerts, D.; Heldens, J.G.; Maas, R.; Moormann, R.J.; et al. Efficacy of a pandemic (H1N1) 2009 virus vaccine in pigs against the pandemic influenza virus is superior to commercially available swine influenza vaccines. Vet. Microbiol. 2011, 152, 304–314. [Google Scholar] [CrossRef] [PubMed]
- USDA. Veterinary Biological Products: Licensees and Permittees; USDA APHIS Center for Veterinary Biologics: Ames, IA, USA, 2014.
- Vander Veen, R.L.; Harris, D.L.; Kamrud, K.I. Alphavirus replicon vaccines. Anim. Health Res. Rev. 2012, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Carroll, T.D.; Matzinger, S.R.; Barro, M.; Fritts, L.; McChesney, M.B.; Miller, C.J.; Johnston, R.E. Alphavirus replicon-based adjuvants enhance the immunogenicity and effectiveness of Fluzone® in rhesus macaques. Vaccine 2011, 29, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Bosworth, B.; Erdman, M.M.; Stine, D.L.; Harris, I.; Irwin, C.; Jens, M.; Loynachan, A.; Kamrud, K.; Harris, D.L. Replicon particle vaccine protects swine against influenza. Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, e99–e103. [Google Scholar] [CrossRef] [PubMed]
- Erdman, M.M.; Kamrud, K.I.; Harris, D.L.; Smith, J. Alphavirus replicon particle vaccines developed for use in humans induce high levels of antibodies to influenza virus hemagglutinin in swine: Proof of concept. Vaccine 2010, 28, 594–596. [Google Scholar] [CrossRef] [PubMed]
- Vander Veen, R.; Kamrud, K.; Mogler, M.; Loynachan, A.T.; McVicker, J.; Berglund, P.; Owens, G.; Timberlake, S.; Lewis, W.; Smith, J.; et al. Rapid Development of an Efficacious Swine Vaccine for Novel H1N1. PLOS Curr. 2009. [Google Scholar] [CrossRef]
- Vander Veen, R.L.; Loynachan, A.T.; Mogler, M.A.; Russell, B.J.; Harris, D.L.; Kamrud, K.I. Safety, immunogenicity, and efficacy of an alphavirus replicon-based swine influenza virus hemagglutinin vaccine. Vaccine 2012, 30, 1944–1950. [Google Scholar] [CrossRef] [PubMed]
- Pushko, P.; Parker, M.; Ludwig, G.V.; Davis, N.L.; Johnston, R.E.; Smith, J.F. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: Expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 1997, 239, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Wesley, R.D.; Tang, M.; Lager, K.M. Protection of weaned pigs by vaccination with human adenovirus 5 recombinant viruses expressing the hemagglutinin and the nucleoprotein of H3N2 swine influenza virus. Vaccine 2004, 22, 3427–3434. [Google Scholar] [CrossRef] [PubMed]
- Wesley, R.D.; Lager, K.M. Evaluation of a recombinant human adenovirus-5 vaccine administered via needle-free device and intramuscular injection for vaccination of pigs against swine influenza virus. Am. J. Vet. Res. 2005, 66, 1943–1947. [Google Scholar] [CrossRef] [PubMed]
- Wesley, R.D.; Lager, K.M. Overcoming maternal antibody interference by vaccination with human adenovirus 5 recombinant viruses expressing the hemagglutinin and the nucleoprotein of swine influenza virus. Vet. Microbiol. 2006, 118, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Sumida, S.M.; Truitt, D.M.; Kishko, M.G.; Arthur, J.C.; Jackson, S.S.; Gorgone, D.A.; Lifton, M.A.; Koudstaal, W.; Pau, M.G.; Kostense, S.; et al. Neutralizing antibodies and CD8+ T lymphocytes both contribute to immunity to adenovirus serotype 5 vaccine vectors. J. Virol. 2004, 76, 2666–2673. [Google Scholar] [CrossRef]
- Steitz, J.; Barlow, P.G.; Hossain, J.; Kim, E.; Okada, K.; Kenniston, T.; Rea, S.; Donis, R.O.; Gambotto, A. A candidate H1N1 pandemic influenza vaccine elicits protective immunity in mice. PLOS ONE 2010, 5, e10492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solorzano, A.; Webby, R.J.; Lager, K.M.; Janke, B.H.; Garcia-Sastre, A.; Richt, J.A. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 2005, 79, 7535–7543. [Google Scholar] [CrossRef] [PubMed]
- Masic, A.; Babiuk, L.A.; Zhou, Y. Reverse genetics-generated elastase-dependent swine influenza viruses are attenuated in pigs. J. Gen. Virol. 2009, 90, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Neumann, G.; Kawaoka, Y.; Hobom, G.; Webster, R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2000, 97, 6108–6113. [Google Scholar] [CrossRef] [PubMed]
- Masic, A.; Lu, X.; Li, J.; Mutwiri, G.K.; Babiuk, L.A.; Brown, E.G.; Zhou, Y. Immunogenicity and protective efficacy of an elastase-dependent live attenuated swine influenza virus vaccine administered intranasally in pigs. Vaccine 2010, 28, 7098–7108. [Google Scholar] [CrossRef] [PubMed]
- Sandbulte, M.; Platt, R.; Roth, J.; Henningson, J.; Gibson, K.; RaJao, D.; Loving, C.; Vincent, A. Divergent immune responses and disease outcomes in piglets immunized with inactivated and attenuated H3N2 swine influenza vaccines in the presence of maternally-derived antibodies. Virology 2014, 464 465, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Carter, N.J.; Curran, M.P. Live attenuated influenza vaccine (FluMist(R); Fluenz): A review of its use in the prevention of seasonal influenza in children and adults. Drugs 2011, 71, 1591–1622. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.M.; Glansbeek, H.L.; Hilgers, L.A.; te Lintelo, E.G.; de Visser, Y.E.; Boersma, W.J.; Haagmans, B.L.; Bianchi, A.T. Protective antiviral immune responses to pseudorabies virus induced by DNA vaccination using dimethyldioctadecylammonium bromide as an adjuvant. J. Virol. 2002, 76, 10540–10545. [Google Scholar] [CrossRef] [PubMed]
- Ganges, L.; Barrera, M.; Nunez, J.I.; Blanco, I.; Frias, M.T.; Rodriguez, F.; Sobrino, F. A DNA vaccine expressing the E2 protein of classical swine fever virus elicits T cell responses that can prime for rapid antibody production and confer total protection upon viral challenge. Vaccine 2005, 23, 3741–3752. [Google Scholar] [CrossRef] [PubMed]
- Cedillo-Barron, L.; Foster-Cuevas, M.; Belsham, G.J.; Lefevre, F.; Parkhouse, R.M. Induction of a protective response in swine vaccinated with DNA encoding foot-and-mouth disease virus empty capsid proteins and the 3D RNA polymerase. J. Gen. Virol. 2001, 82, 1713–1724. [Google Scholar] [PubMed]
- Gorres, J.P.; Lager, K.M.; Kong, W.P.; Royals, M.; Todd, J.P.; Vincent, A.L.; Wei, C.J.; Loving, C.L.; Zanella, E.L.; Janke, B.; et al. DNA vaccination elicits protective immune responses against pandemic and classic swine influenza viruses in pigs. Clin. Vaccine Immunol. 2011, 18, 1987–1995. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.M.; Hollister, J.R. FluBlok, a next generation influenza vaccine manufactured in insect cells. Biologicals 2009, 37, 182–189. [Google Scholar] [CrossRef]
- Rayner, J.O.; Dryga, S.A.; Kamrud, K.I. Alphavirus vectors and vaccination. Rev. Med. Virol. 2002, 12, 279–296. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).