Application of Near-Infrared Hyperspectral Imaging to Detect Sulfur Dioxide Residual in the Fritillaria thunbergii Bulbus Treated by Sulfur Fumigation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Hyperspectral Image Acquisition
2.3. Measurement of SO2 Residuals
2.4. Multivariate Analysis
2.4.1. Spectral Preprocessing and Outlier Detection
2.4.2. Calibration Model
2.4.3. Optimal Wavelength Selection Methods
2.4.4. Image Visualization
2.4.5. Software and Model
3. Results and Discussion
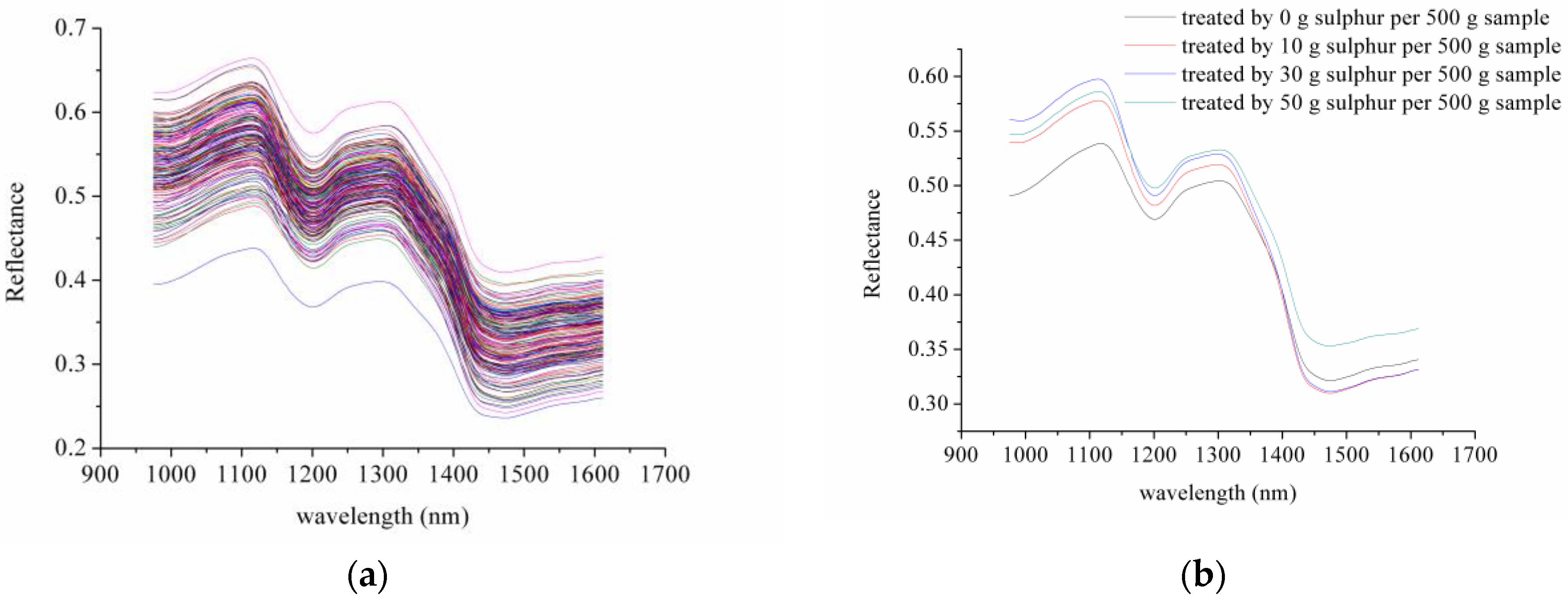
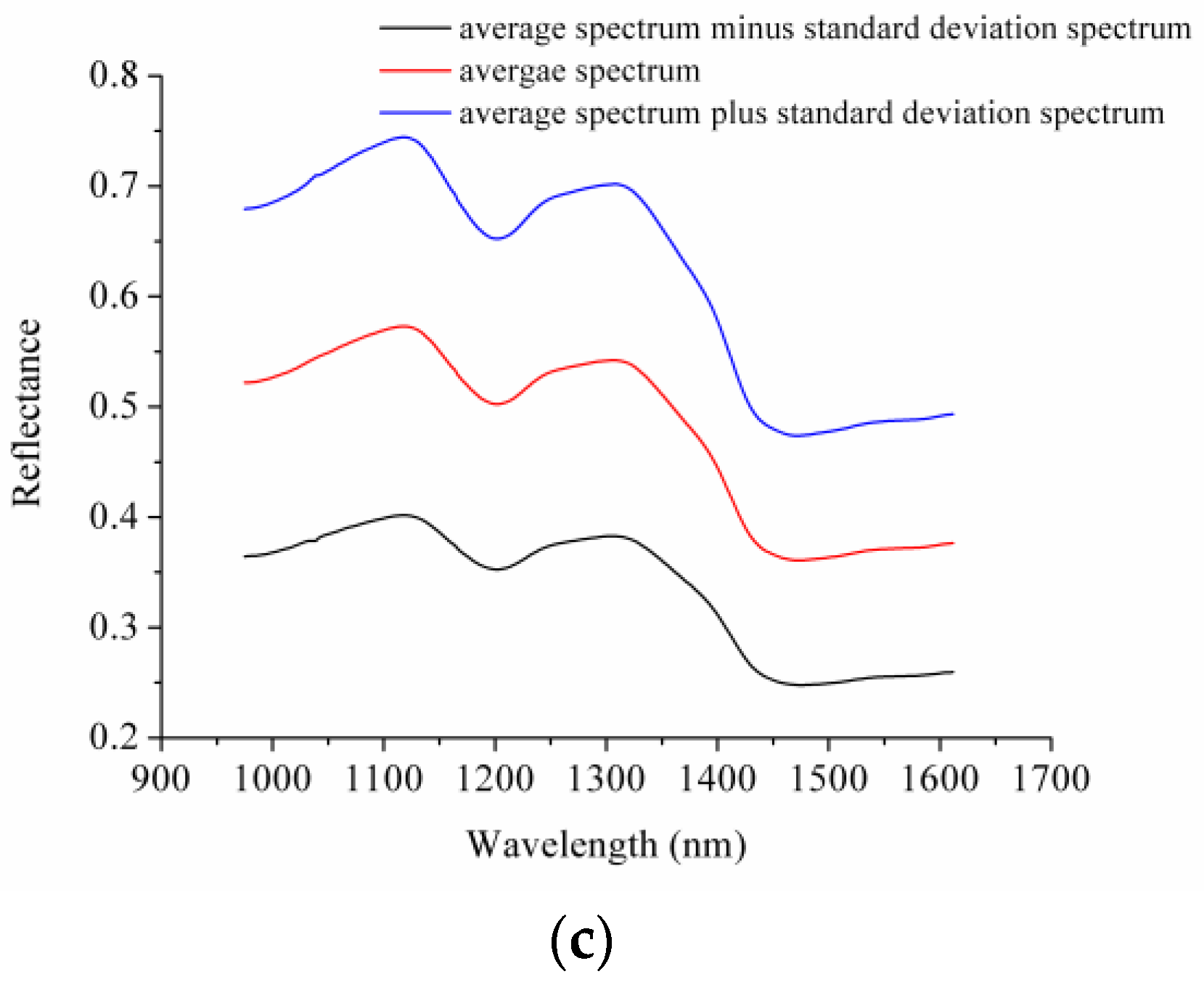
3.1. Spectral Profiles
3.2. Statistical Analysis of SO2 Residual Content
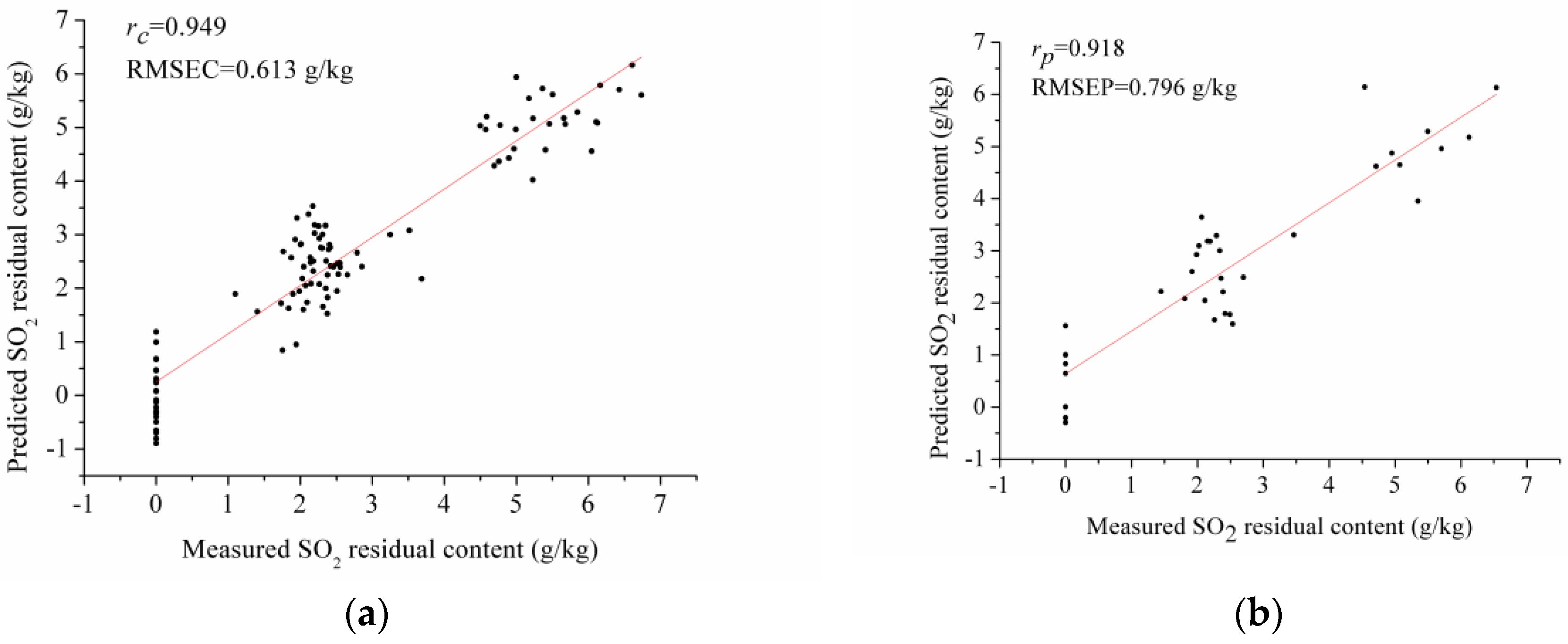
3.3. PLSR Model Using Full Spectra
3.4. Optimal Wavelength Selection
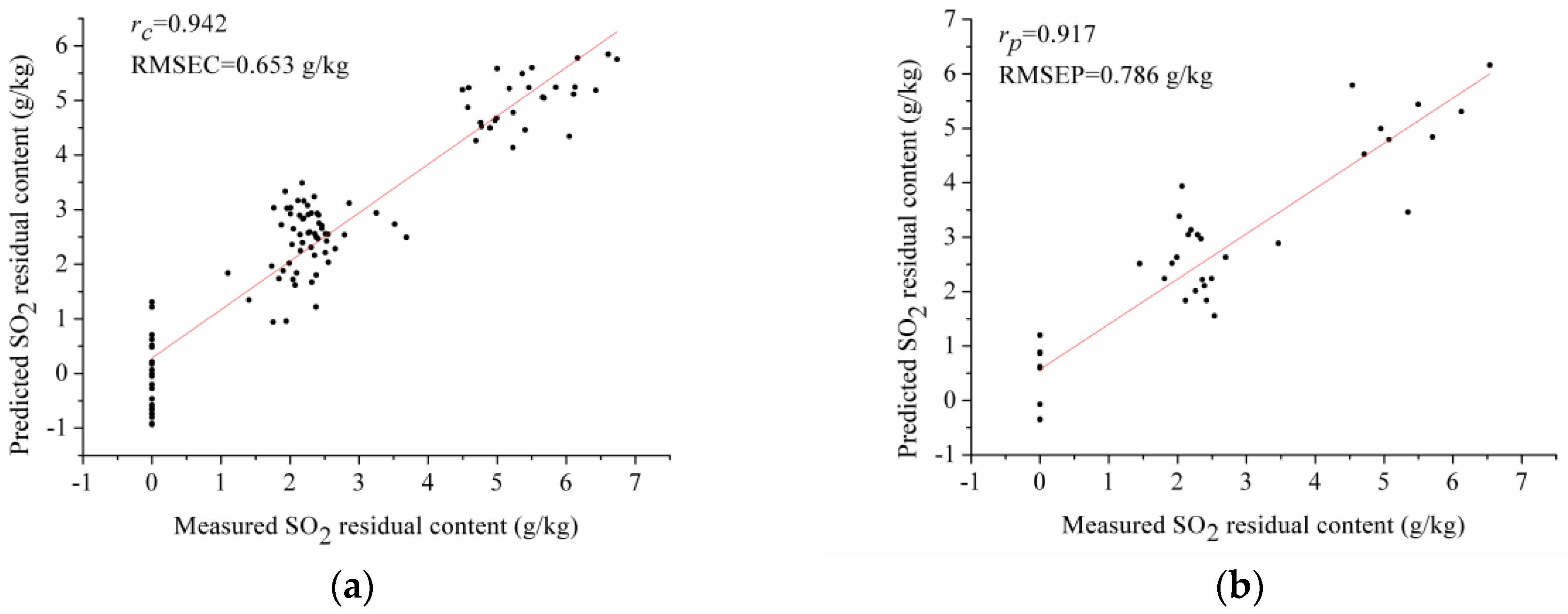
3.5. PLSR Model Using Optimal Wavelengths
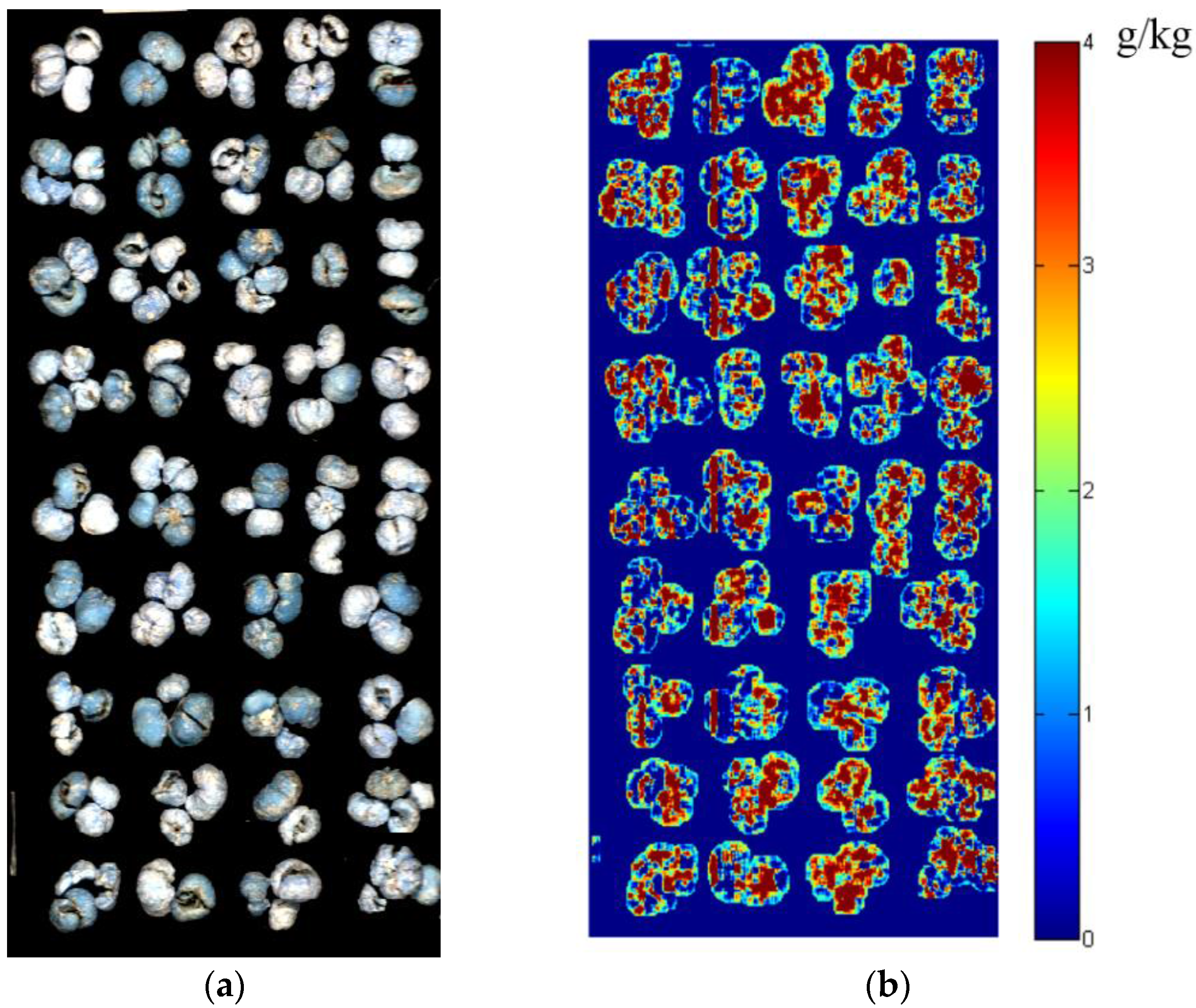
3.6. Visualization of SO2 Residual Content Distribution
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Duan, B.; Huang, L.; Chen, S. Study on the destructive effect to inherent quality of Fritillaria thunbergii Miq. (Zhebeimu) by sulfur-fumigated process using chromatographic fingerprinting analysis. Phytomedicine 2012, 19, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Huang, L.F.; Zheng, S.H.; Chen, S.L. Sulfur fumigation, a better or worse choice in preservation of traditional Chinese medicine. Phytomedicine 2013, 20, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Liao, Q.; Li, J.; Lin, F.; Liu, E.; Jiang, X.; Deng, L. Chronic exposure to sulfur dioxide enhances airway hyperresponsiveness only in ovalbumin-sensitized rats. Toxicol. Lett. 2012, 214, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tian, J.; Bai, Y.; Yang, Z.; Zhang, H.; Meng, Z. Effects of sulfur dioxide and its derivatives on the functions of rat hearts and their mechanisms. Procedia Environ. Sci. 2013, 18, 43–50. [Google Scholar] [CrossRef]
- Ding, K.; Cao, G.; Xu, Z.; Chen, X. Quantitative analysis coupled with toxic evaluation to investigate the influence of sulfur-fumigation on the quality of Chrysanthemum morifolium. Nat. Prod. Commun. 2014, 9, 1357–1358. [Google Scholar] [PubMed]
- Chinese Pharmacopoeia Commission. Chinese Pharmacopoeia; China Medical Science Press: Beijing, China, 2015. [Google Scholar]
- Wu, X.Y.; Chao, Z.M.; Sun, W.; Wang, C. Qualitative and quantitative research on sulfur fumigation of Angelicae Dahuricae Radix (Baizhi) by near-infrared spectroscopy. China J. Chin. Mater. Med. 2014, 39, 1863–1868. [Google Scholar]
- Zhang, H.; Wu, T.; Zhang, L.; Zhang, P. Development of a Portable Field Imaging Spectrometer: Application for the Identification of Sun-Dried and Sulfur-Fumigated Chinese Herbals. Appl. Spectrosc. 2016, 70, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Araújo, M.C.U.; Saldanha, T.C.B.; Galvao, R.K.H.; Yoneyama, T.; Chame, H.C.; Visani, V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemom. Intell. Lab. 2001, 57, 65–73. [Google Scholar] [CrossRef]
- ElMasry, G.; Sun, D.W.; Allen, P. Near-infrared hyperspectral imaging for predicting colour, pH and tenderness of fresh beef. J. Food. Eng. 2012, 110, 127–140. [Google Scholar] [CrossRef]
- Li, H.D.; Xu, Q.S.; Liang, Y.Z. Random frog: An efficient reversible jump Markov chain Monte Carlo-like approach for variable selection with applications to gene selection and disease classification. Anal. Chim. Acta 2012, 740, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liang, Y.; Xu, Q.; Cao, D. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Cope, M.; Delpy, D.T.; Wray, J.S.; Reynolds, E.O.R. A CCD spectrophotometer to quantitate the concentration of chromophores in living tissue utilising the absorption peak of water at 975 nm. In Oxygen Transport to Tissue XI; Springer: New York, NY, USA, 1989; pp. 33–40. [Google Scholar]
- Sonnenschein, R.; Jarmer, T.; Vohland, M.; Werner, W. Spectral determination of plant water content of wheat canopies. In Proceedings of the 4th EARSel Workshop of Imaging Spectroscopy; New Quality in Environmental Studies; Warsaw University: Warsaw, Poland, 2005; pp. 727–737. [Google Scholar]
- Kawamura, K.; Watanabe, N.; Sakanoue, S.; Inoue, Y. Estimating forage biomass and quality in a mixed sown pasture based on partial least squares regression with waveband selection. Grassl. Sci. 2008, 54, 131–145. [Google Scholar] [CrossRef]
- Lee, M.H.; Cavinato, A.G.; Mayes, D.M.; Rasco, B.A. Noninvasive short-wavelength near-infrared spectroscopic method to estimate the crude lipid content in the muscle of intact rainbow trout. J. Agric. Food Chem. 1992, 40, 2176–2181. [Google Scholar] [CrossRef]
- Wu, D.; Chen, X.; Cao, F.; Sun, D.-W.; He, Y.; Jiang, Y. Comparison of infrared spectroscopy and nuclear magnetic resonance techniques in tandem with multivariable selection for rapid determination of ω-3 polyunsaturated fatty acids in fish oil. Food Bioprocess Tech. 2014, 7, 1555–1569. [Google Scholar] [CrossRef]
- Weyer, L.G.; Lo, S.C. Spectra– Structure Correlations in the Near-Infrared. In Handbook of Vibrational Spectroscopy; John Wiley & Sons, Inc.: New York, NY, USA, 2006; pp. 1817–1837. [Google Scholar]
- Rogel-Castillo, C.; Boulton, R.; Opastpongkarn, A.; Huang, G.; Mitchell, A.E. Use of near-infrared spectroscopy and chemometrics for the non-destructive identification of concealed damage in raw almonds (Prunus dulcis). J. Agric. Food Chem. 2016, 64, 5958–5962. [Google Scholar] [CrossRef] [PubMed]
- De la Roza-Delgado, B.; Soldado, A.; de Faria Oliveira, A.F.G.; Martínez-Fernández, A.; Argamentería, A. Assessing the value of a portable near infrared spectroscopy sensor for predicting pork meat quality traits of “Asturcelta autochthonous swine breed”. Food Anal. Methods 2014, 7, 151–156. [Google Scholar] [CrossRef]
- Workman, J.J., Jr. Interpretive spectroscopy for near infrared. Appl. Spectrosc. Rev. 1996, 31, 251–320. [Google Scholar] [CrossRef]
- Stuart, B.H. Infrared Spectroscopy: Fundamentals and Applications; John Wiley & Sons Ltd.: Chichester, UK, 2004. [Google Scholar]

| Sample Set | Number | Range (g/kg) | Means (g/kg) | Standard Deviation (g/kg) |
|---|---|---|---|---|
| Calibration set | 111 | 0–6.736 | 2.502 | 1.950 |
| Prediction set | 37 | 0–6.536 | 2.472 | 1.963 |
| Method | Number | Wavelength (nm) |
|---|---|---|
| SPA | 15 | 1581.51, 1611.96, 1483.46, 1318.09, 1345.0601, 1281.01, 1372.05, 1213.65, 1159.8108, 1112.72, 1554.45, 1436.1801, 1237.22, 975.01, 1055.59 |
| Bw | 21 | 975.01, 995.15, 1018.65, 1028.72, 1052.23, 1075.75, 1106, 1173.26, 1210.29, 1233.85, 1281.01, 1314.72, 1348.4399, 1388.92, 1422.67, 1449.6801, 1480.08, 1507.12, 1537.54, 1578.13, 1611.96 |
| Random frog | 24 | 1581.51, 1503.74, 1524.02, 1605.2, 1500.36, 1473.33, 1469.95, 1554.45, 1507.12, 1608.58, 1534.16, 1584.89, 975.01, 1520.64, 1527.4, 1601.8101, 1476.71, 1530.78, 1496.98, 1210.29, 1206.92, 1449.6801, 1466.5699, 1446.3101 |
| CARS | 26 | 975.01, 981.72, 1156.4399, 1163.9, 1206.92, 1230.49, 1233.85, 1247.33, 1250.6899, 1257.4301, 1311.35, 1338.3199, 1449.6801, 1463.1899, 1466.5699, 1469.95, 1500.36, 1503.74, 1524.02, 1527.4, 1530.78, 1534.16, 1554.45, 1581.51, 1605.2, 1608.58 |
| Methods | Model | Par * | rc | RMSEC (g/kg) | rp | RMSEP (g/kg) |
|---|---|---|---|---|---|---|
| SPA | PLSR | 13 | 0.939 | 0.668 | 0.912 | 0.819 |
| Random frog | PLSR | 7 | 0.888 | 0.892 | 0.845 | 1.064 |
| Bw | PLSR | 13 | 0.942 | 0.653 | 0.917 | 0.786 |
| CARS | PLSR | 12 | 0.956 | 0.568 | 0.909 | 0.840 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Zhang, C.; He, Y. Application of Near-Infrared Hyperspectral Imaging to Detect Sulfur Dioxide Residual in the Fritillaria thunbergii Bulbus Treated by Sulfur Fumigation. Appl. Sci. 2017, 7, 77. https://doi.org/10.3390/app7010077
He J, Zhang C, He Y. Application of Near-Infrared Hyperspectral Imaging to Detect Sulfur Dioxide Residual in the Fritillaria thunbergii Bulbus Treated by Sulfur Fumigation. Applied Sciences. 2017; 7(1):77. https://doi.org/10.3390/app7010077
Chicago/Turabian StyleHe, Juan, Chu Zhang, and Yong He. 2017. "Application of Near-Infrared Hyperspectral Imaging to Detect Sulfur Dioxide Residual in the Fritillaria thunbergii Bulbus Treated by Sulfur Fumigation" Applied Sciences 7, no. 1: 77. https://doi.org/10.3390/app7010077







