1. Introduction
MicroRNAs (miRNAs) are small (21–22 nt), non-coding RNA fragments found in many organisms, from plants to humans, which function to negatively regulate gene expression. In humans, roles for miRNA have been identified in metabolism, growth, and development. Dicer, a protein critical for miRNA processing, has been associated with proper development of the limbs, lungs, and hair follicles and in T-cell differentiation [
1,
2]. Dysregulation of human miRNA has been implicated in the pathogenesis of many cancers [
3]. As negative regulators of gene expression, miRNAs can function as tumor suppressors that, if dysregulated, can augment the chance of cancer development [
4]. For example, a chromosomal deletion associated with the development of chronic lymphocytic leukaemia (CLL) causes a loss of intrinsic miRNA tumor suppressors [
5]. Currently, over 800 miRNAs have been identified in the human genome with diverse functions. miRNAs function by binding to partially complementary sites in the messenger RNA (mRNA) of other genes, inhibiting the production of the proteins synthesized from these genes. A single miRNA can interact with hundreds and thousands of genes in regulating various physiological processes, thus adding to the complex regulation of the human genome.
Viruses are known to subvert and manipulate the human gene machinery, and miRNAs are no exception. Host miRNAs are dysregulated by viruses, and both DNA and RNA viruses have been found to encode and express miRNAs in the infected host, demonstrating the importance of the role of host- and virus-encoded miRNAs in host-virus interactions. The miRNA expression profile of cells is altered in both clinical PBMC samples and cultured cells. Cells transfected with an infectious HIV-1 strain have significantly altered miRNA profiles compared to control cells [
6]. In 2008, Houzet
et al. showed that the vast majority of host miRNAs are downregulated in HIV infection of peripheral blood mononuclear cells (PBMCs). When individuals were separated into groups based on their T-cell counts and viral loads (VLs), specific miRNA “profiles” could be seen for each of the classes. Furthermore, many miRNA changes in patient cells could not be accounted for by infection alone, indicating a complex role for miRNA in gene regulation [
7].
In 2007, Huang
et al. showed overexpression of host miRNAs in resting T-cells that target sequences in the 3′ end of HIV-1 RNA, silencing viral mRNA and enforcing latency [
8]. Furthermore, Witwer
et al. showed that PBMC miRNA profiles could distinguish elite suppressors (ES) and uninfected controls from viremic HIV-1 infected patients [
9]. Their results demonstrated correlations between miRNA expression, CD4+ T-cell count and viral load. Some miRNAs found to differ in expression have previously been shown to correlate with HIV-1 latency, including miR-29s, miR-125b, and miR-150. Their analysis identified several miRNAs that have not been previously described in association with HIV infection, including miR-31, which distinguishes controls and ES and regulates a protein with implications for T-cell differentiation. Although this study has also shown that HIV-1-positive ES are characterized by a PBMC miRNA profile that in general resembles that of uninfected individuals, they also reiterate that the ES, on the basis of miRNA expression, are a heterogeneous group. This suggests that different mechanisms, shaped or marked by different miRNA expression patterns, underlie sustained and durable control in therapy-naïve HIV-infected individuals. In a recent International AIDS Society (IAS) meeting, Zhu
et al. showed a set of 18 differentially expressed miRNAs, which could identify the outcome of HIV disease at the chronic stage more accurately. Six out of 18 miRNAs were significantly related to faster rate of CD4+ T-cell decline [
10]. Studies of larger cohorts of individuals are needed to address miRNA specific to different stages of HIV disease and explain the underlying genomic basis of natural control of HIV in therapy-naive ES.
Since all the studies to date have been performed on whole PBMC or tissue, we endeavored to address disease- and cell-type-specific miRNAs and their role in HIV pathogenesis. We have adopted a novel approach for this study, which simultaneously analyzes miRNAs from the CD4+ and CD8+ T-cells from viremic, aviremic BDL patients, and elite controllers. This study is unique in showing the HIV disease-stage and cell-type specificity of miRNA during HIV infection and its natural control in elite controllers.
3. Discussion
Along with their association with pathological and physiological pathways, host miRNAs play a significant role in many viral infections, including HIV. In this work, we describe the first direct examination of virus-mediated cellular miRNA expression manipulation in the CD4+ and CD8+ T-cell fractions taken from HIV+ patients at differing stages of HIV disease. This disease-stage stratification was primarily based on the detectable virus in the plasma of diseased individuals and the length of infection. The main rationale behind conducting a parallel analysis of CD4+ and CD8+ T-cells for miRNAome was based on the roles of these cells in HIV infection. CD4+ T-cells are the primary targets of HIV [
11], whereas the CD8+ T-cells mediate antiviral control replication through cytotoxic and non-cytotoxic mechanisms. The effectiveness of this response directly correlated with prognosis [
12,
13]. It is evident that the HIV infection of host cells leads to an alteration of the global RNA interference machinery [
14]. Although it is possible that HIV-induced changes in cellular miRNA expression results from combinatorial molecular interactions among proteins, transcripts, and genomes, the mechanisms behind these changes during host-virus interactions remain to be understood. Here, we demonstrate the first evidence for cell-, disease-stage- and HIV-specific host miRNA during HIV disease stages.
Previous studies have shown unique and variable global modifications in microRNA expressions exhibited by different host cell types and cell lines in response to HIV-1 infection [
6,
7,
15,
16,
17]. These findings showed a consistent and prominent trend of downregulation of cellular miRNAs in HIV-infected individuals. For instance, in 2008, Houzet
et al. showed a prominent trend towards downregulation of miRNA in HIV+ individuals, with 59 of 62 DE miRNAs downregulated in HIV+ PBMCs [
7]. This finding is in agreement with other studies performed on cell lines [
6,
15], which found that when HELA cell lines are transfected with HIV-1 in a pNL4-3 vector, >43% of cellular miRNAs were downregulated, with the majority of the remainder unchanged. Our study concurs with these findings and further demonstrates that the downregulation of cellular miRNAs was systematic across both CD4+ and CD8+ T-cells and diverse disease groups. This trend of cellular miRNA downregulation was confirmed in our qRT-PCR expression trends, suggesting the functional significance of downregulation of cellular miRNAs during HIV infection and guiding the disease course.
A large body of work on miRNAs has been done on cell lines rather than primary patient cells. It is important to iterate that miRNA signatures of PBMCs from HIV+ patient blood are more than 50% discordant with those of
in vitro-infected PBMCs [
7]. This implies that a significant portion of the miRNA signature changes seen in HIV+ patients cannot be accounted for by infection alone. These changes could be attributed to
in vivo interaction between cell types and other combinatorial interactions, which together play a functional role in determining the dysregulation of host miRNAs during HIV infection. Thus, the analysis of primary CD4+ and CD8+ T-cells we have performed from diverse HIV disease groups carry considerable functional relevance in the context of HIV infection
in vivo.
To obtain differentially and statistically significant miRNA candidates, we employed the Benjamini–Hochberg FDR corrective testing with a p-value cut-off of <0.05 throughout. The Benjamini–Hochberg algorithm is designed to sort through a list of p-values and determine the likelihood of a false rejection of the null hypothesis (a false positive) and so “adjust” the p-value to control the likelihood of the false positive. Employing this algorithm provided data of greater integrity, and this is the data we have made use of in interpreting our DE dataset. Due to the stringency of this method of analysis, we failed to detect differential expression of miRNAs in some comparisons, while the DE miRNAs detected for other groups were statistically strong. To clarify any possible occlusion of biologically relevant miRNAs, we also analyzed the dataset without FDR corrective testing. To minimize false positives in the absence of FDR correction, the p-value cut-off was then set at <0.01. This allowed the detection of other DE miRNAs for the groups we failed to detect DE miRNAs using FDR corrective testing. Our hypothesis for using this rationale was that the statistical stringency possibly occludes biologically relevant miRNAs, as several of these miRNAs identified at p < 0.01 are not only of functional relevance, but are also known in the literature.
3.1. Signature miRNA in HIV Disease Class Prediction
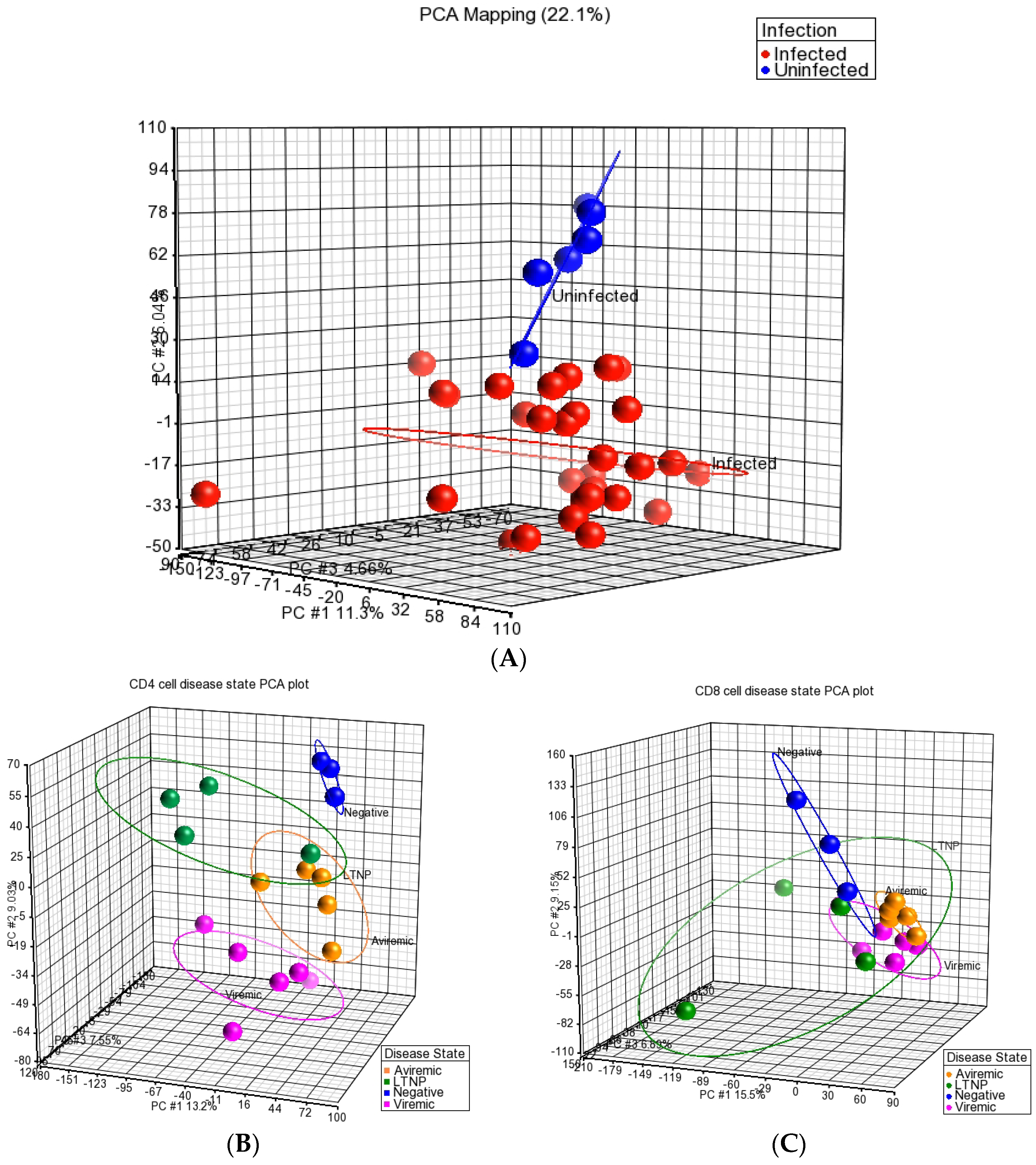
Through the use of PCA, we demonstrated that the miRNA profiles of HIV+ CD4+ and CD8+ T-cells were distinct from T-cells of HIV– individuals as all HIV-infected groups (viremic, aviremic, and LTNPs) were more closely related as a group and segregated from the uninfected group. PCA also highlighted that the individual samples from the same disease stages cluster together, and the sample classes with the greater similarities were more closely associated. Despite the close relationship of all HIV+ samples, PCA could also distinguish between HIV disease sub-groups, with the LTNP, aviremic, and HIV groups showing a common miRNA signature specific to them. This is plausible, as these three groups have undetectable levels of plasma viremia. Our results can be likened to the results shown by Witwer et al. on PBMCs, where the miRNA signatures of LTNP HIV+ individuals with low viral load and good CD4+ T-cell counts showed greater similarities to uninfected controls than HAART-treated HIV+ patients.
In support of these findings, several previous papers have suggested that a broad statistical analysis of the miRNAs that are differentially expressed in HIV+ individuals distinguish patients according to their CD4+ T-cell counts, viremia, or HIV exposure level [
7,
9,
15]. In 2008, Houzet
et al. showed that HIV+ patients could be categorized into disease groups based on DE miRNAs present in the PBMC fraction of their blood. Further, the miRNA signature of CD4+ T-cells from multiply exposed uninfected (MEU) individuals can distinguish them from therapy-naïve infected CD4+ T-cells [
15]. Bignami
et al. also showed that miRNA signatures of LTNPs, MEUs, and therapy-naïve HIV+ patient CD4+ T-cells cluster based on their exposure level and disease state [
15]. As mentioned, Witwer
et al. showed that, through examination of DE miRNAs, LTNPs with low CD4+ T-cell counts and high plasma viremia correlate more closely with HIV+ viremic individuals, and those with high CD4+ T-cell counts correlate with HIV− controls [
9]. Thus, the characteristics of an miRNA signature allowed accurate assignment of samples into disease states using a variety of predictive programs [
9], which is what our analysis has shown at both cell-type and disease-stage levels.
3.2. DE miRNAs in CD8+ T-Cell Predict HIV Disease Stage Better than CD4+ T-Cells
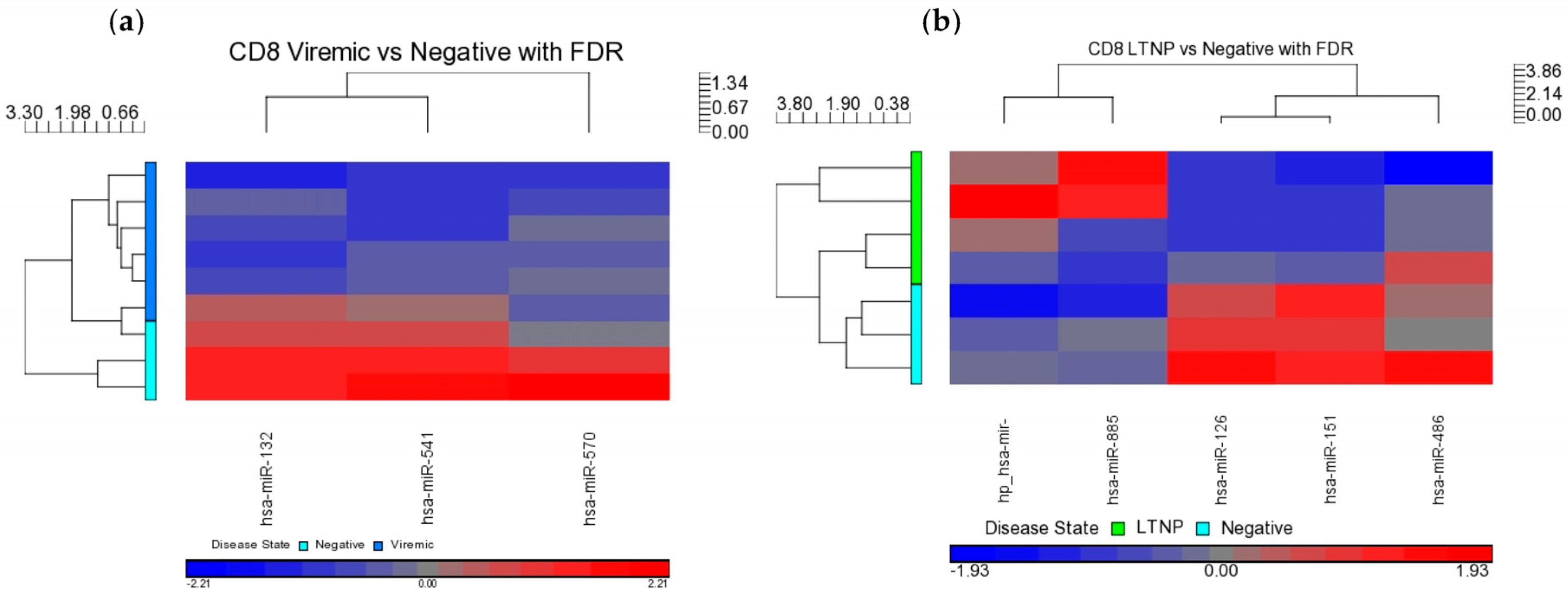
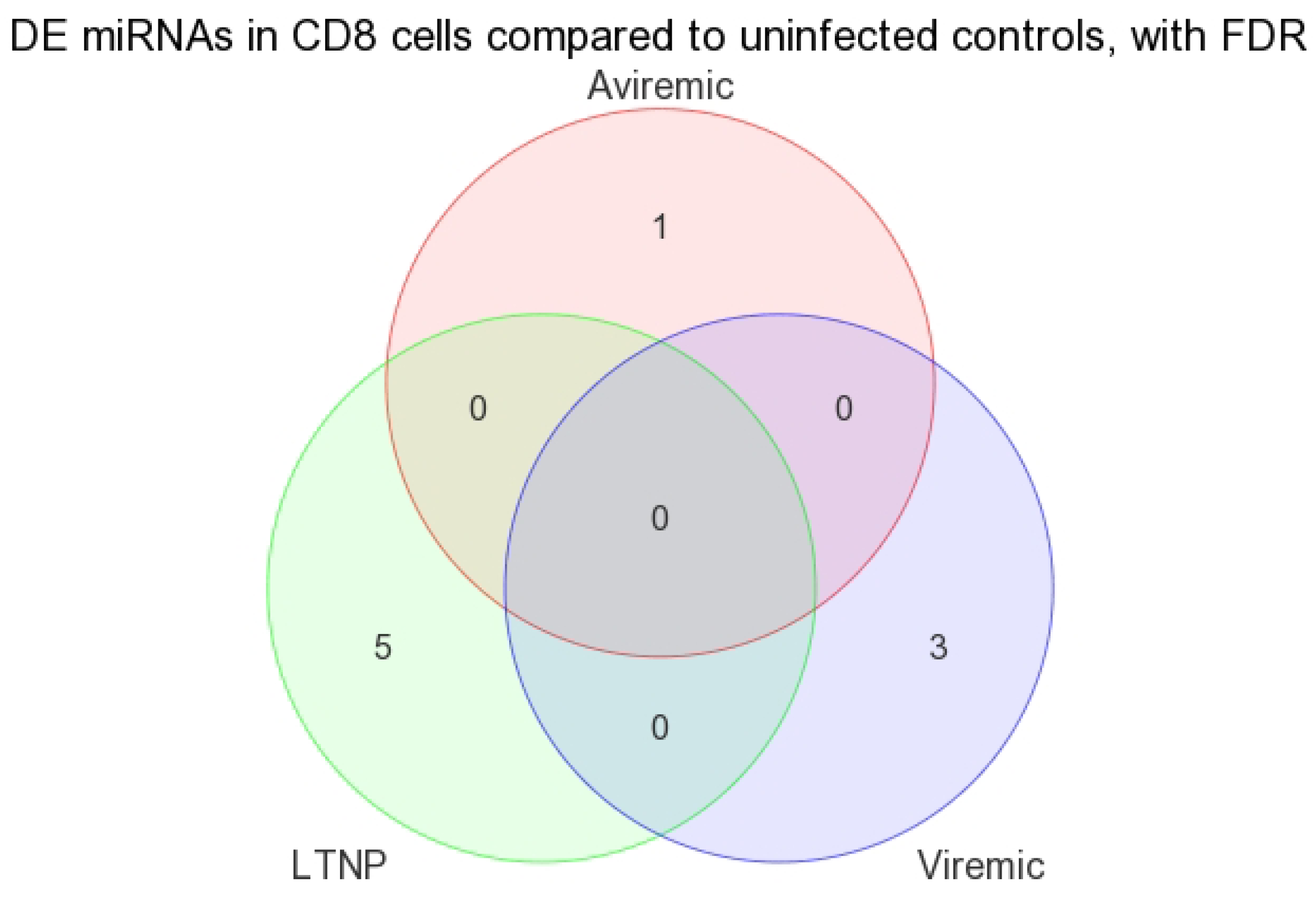
When DE miRNAs were analyzed in the CD8+ T-cells of HIV+ individuals from each of the disease groups (viremic, aviremic, and LTNP), it was shown that some miRNAs were highly specific for these disease groups when compared with negative controls. Namely, miR-1323, -541, and -570 were associated with the CD8+ viremic group, miR-1298 was associated with the CD8+ aviremic group, and the miRNA-572, -126, 151-5p, -486-5p, and -885-3p with the CD8+ LTNP group. This is the first direct evidence of HIV disease-stage-specific miRNAs in CD8+ T-cells, which is consistent with the role of CD8+ T-cells in containing viremia. A clear segregation of DE miRNA in each comparison demonstrates that the CD8+ T-cells are better suited to predicting HIV disease stage and cell-specific miRNAs than their CD4+ T-cell counterparts.
3.3. Unique miRNAs in Non-Progressive HIV Disease
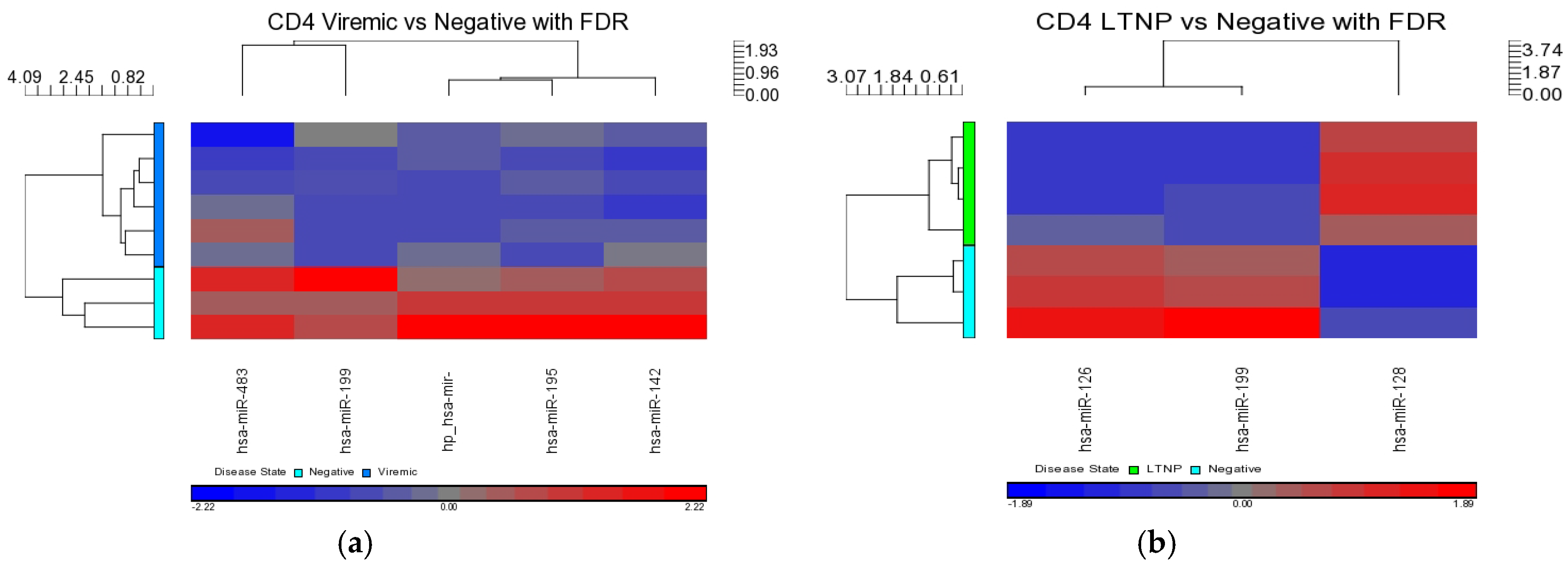
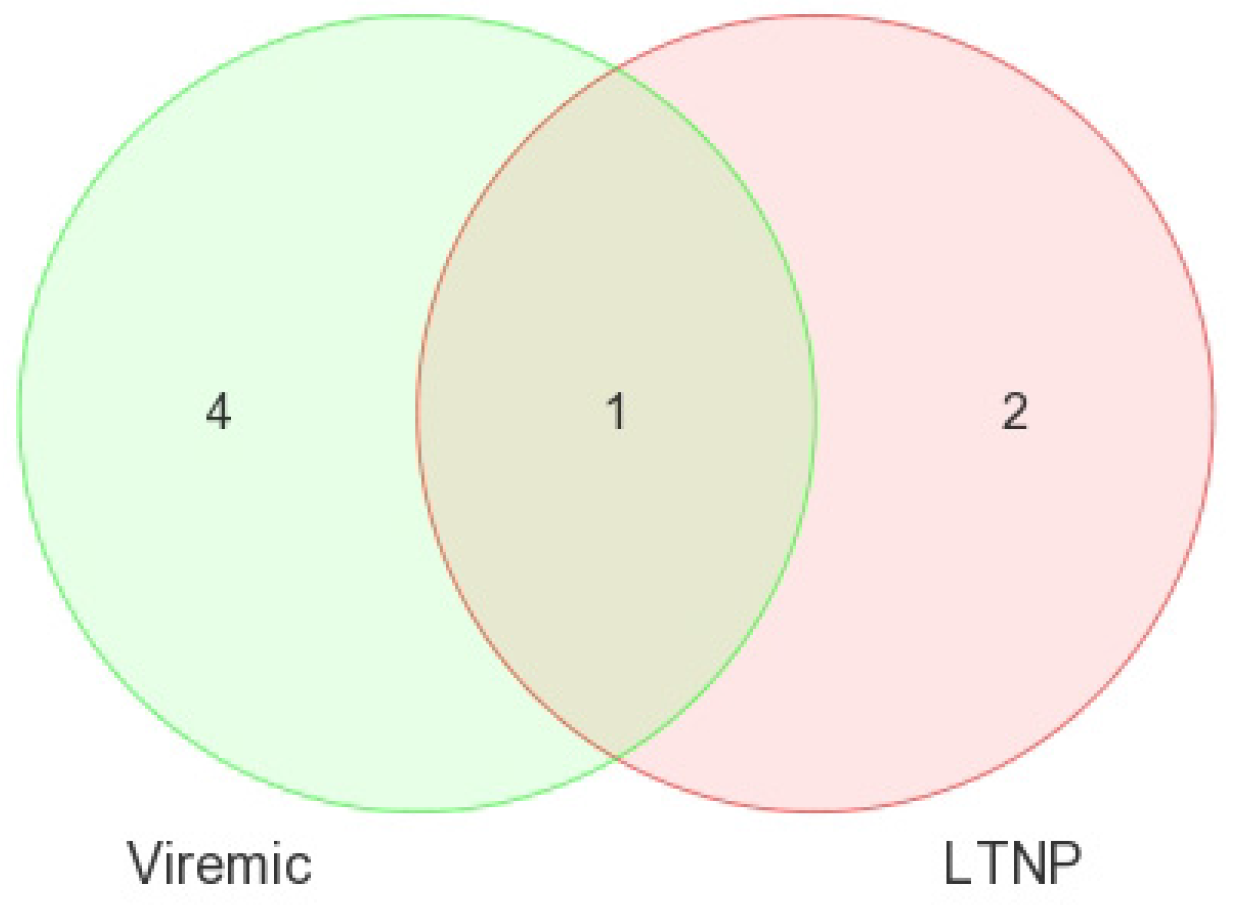
Our analysis performed in CD4+ and CD8+ T-cell types suggests that miRNAs-199a-5p, -1280, and -126 were differentially expressed in the CD4+ T-cells of LTNPs when compared to negative controls, and the miRNAs-151-5p, -486-5p, -572, -885-3p, and -126 were differentially expressed in the CD8+ T-cells of LTNPs when compared to negative controls. MiR-126 was differentially expressed in both CD4+ and CD8+ T-cells in the LTNP disease state. Consistent expression trends between microarray and qRT-PCR for miR-126 suggests that this miRNA is significantly downregulated in both the CD4+ and CD8+ T-cells of LTNPs, suggesting that it is a possible biomarker for non-progressive HIV disease. This, to our knowledge, is the first report of a miRNA that correlates with the LTNP disease state and may be involved in the regulation of cellular immunity and possibly the natural control of HIV in LTNPs.
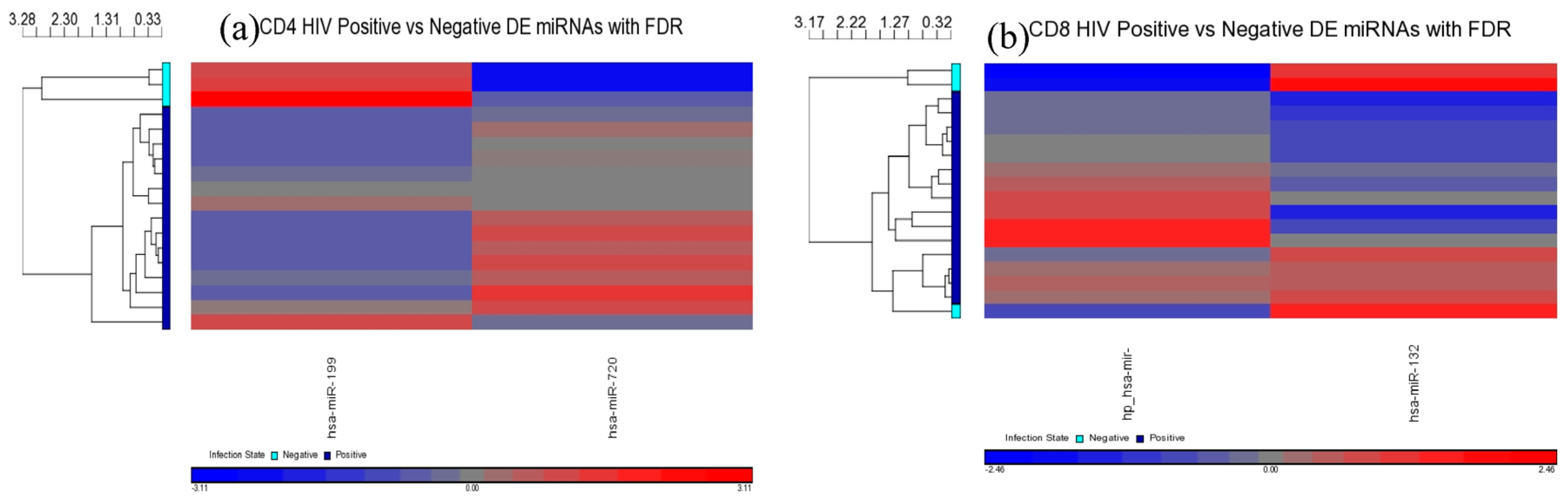
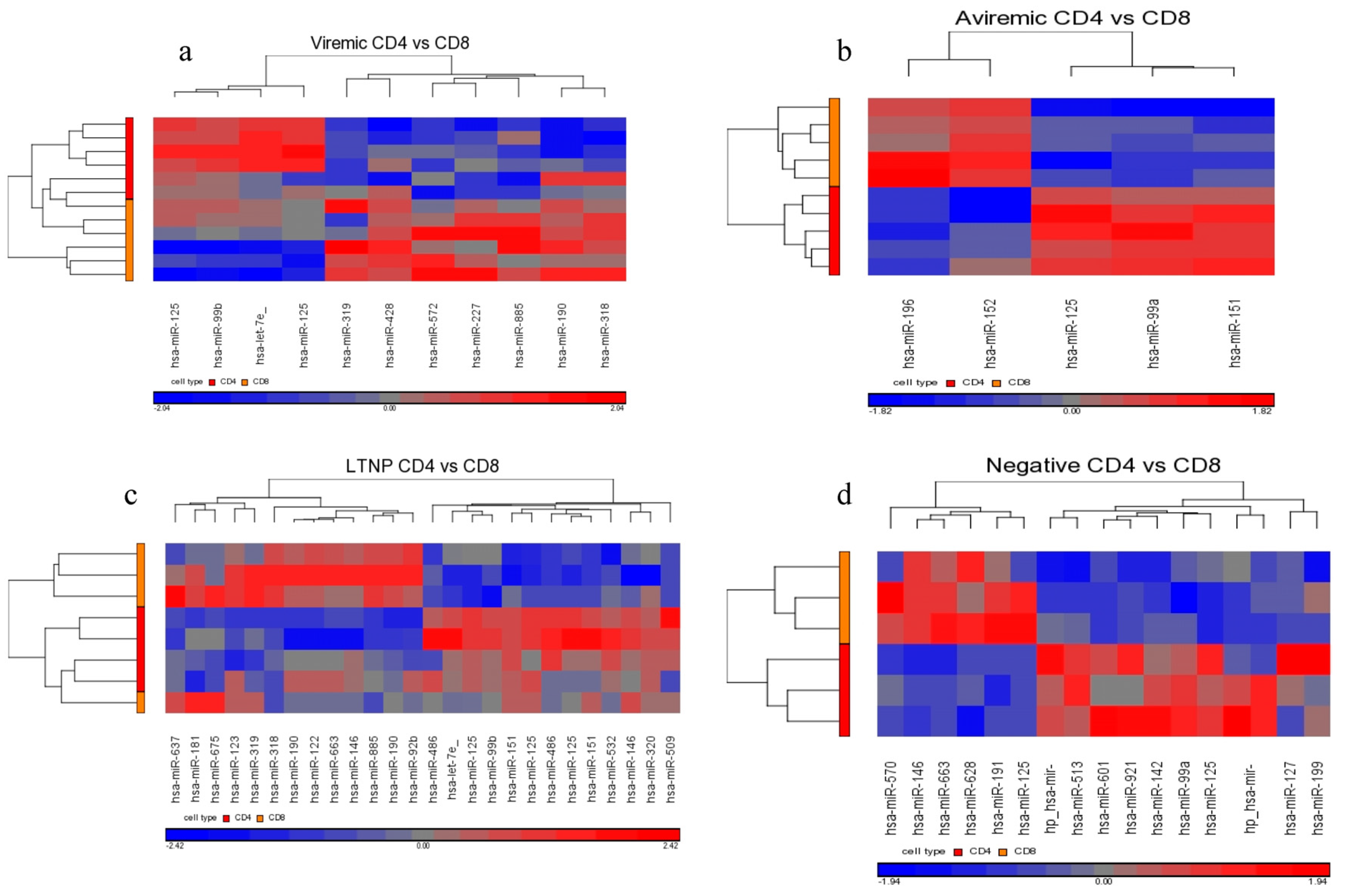
MiR-199a-5p was differentially expressed in HIV+ CD4+ T-cells belonging to all HIV+ groups analyzed (viremic, aviremic, and LTNPs). Stronger downregulation was observed in LTNP vs. negative disease group than in either the viremic or aviremic CD4+ T-cell comparisons against HIV− individuals. Thus, it is likely that miR-199a-5p plays a role in predicting HIV+ status based on CD4+ T-cells. We hypothesize that, although it is expressed and downregulated in the CD4+ T-cells of all HIV+ groups, the degree of downregulation of miR199a-5p in different groups imparts susceptibility or protection, as evident from the CD4+ T-cells from the LTNPs that showed the highest degree of downmodulation of miRNA199-5p, which may play a protective role against the virus. Supporting this conclusion, it has previously been shown that miR-199a (along with miRNAs-143, -303-3p, and -335) is differentially expressed in the PBMCs of class 1 (HIV+, high CD4+ T-cell count, low viral load) individuals, which agrees with our work with miRNA 199a showing the strongest downregulation in the LTNPs group. Our analysis using three phenotypically different HIV+ groups clarifies and unambiguously demonstrates a clear association of miR-199a with HIV positivity based uniquely on CD4+ T-cells, a trend which was further confirmed by qRT-PCR analysis, implicating miR-199a to be an HIV-specific miRNA.
About other differentially expressed miRNAs, miRNA-155 in HIV+ CD4+ T-cells was recently observed to distinguish LTNPs from therapy-naive HIV+ patients [
15]. They suggest that high miR-155 in CD4+ T-cells may correlate with HIV-1 pathogenesis. Our analysis showed similar trends to those of studies by Bignami
et al. and Witwer
et al.
3.4. The miR-125 Family Is Important in the Maintenance of Latency and Aviremia
The 3′ ends of HIV-1 mRNAs (Nef) are targeted by cellular miR-125b (along with miR-28, miR-150, miR-223, and miR-382), all of which are enriched in resting CD4+ T-cells compared to active CD4+ T-cells [
8]. These miRNAs have inhibitory effects and enforce viral latency in resting HIV+ CD4+ lymphocytes by reducing protein translation and viral production [
8]. Moreover, miR-125b has been identified as one of the constitutive miRNA signatures of naïve CD4+ T-cells [
18]. MiR-125a-3p, along with miR-23 and miR-155, is shown to be upregulated in HIV+ therapy-naïve CD4+ T-cells, normal in LTNP CD4+ T-cells, and downregulated in the CD4+ T-cells of MEU samples [
15]. A recent study involving PBMC miRNA profiling [
9] did not find miR-125b correlating with replication control and latency; however, this study involved only the whole PBMCs and not the individual cell subsets [
18,
19], which may harbor unique miRNA characteristics shown in our study.
Interestingly, our results show that the family of miR-125 was consistently upregulated in the comparison of diseased CD4+ T- and CD8+ T-cells using a non-FDR statistical analysis. This upregulation was markedly higher in the CD4+ T-cells of LTNPs than those of viremic or aviremic individuals, followed by its upregulation in aviremic individuals, suggesting a possible role for this family of miRNAs in aviremia and natural control and possibly viral latency. Our results agree in part with the findings by Huang
et al., and we concur that miR-125 family members are potential candidates associated with latency in CD4+ T-cells [
8]. More work is needed to prove the biological relevance of this miRNA. It is important to iterate that this differential expression was only achieved when less stringent corrective testing was applied. Nonetheless, the fold changes and
p-values of miRNA candidates with the disease stage added reliability and confidence to these conclusions.
3.5. Possible Role of miR-29 Family in Viral Replication and Management of Viremia during HAART Treatment
miR-29 targets the viral Nef 3′UTR and downregulates Nef protein expression and interferes with HIV-1 replication, thereby inhibiting viral production and infectivity in HIV-infected T lymphocytes [
7]. Low levels of miR-29b are associated with disease progression and viremia [
9,
10], whereas high expression of miR-29b positively correlates with CD4+ T-cell count. Furthermore, miR-29a specifically targets the HIV-1 3′UTR [
10,
20,
21]. This implies a role for miR-29 members in prohibiting viremia and disease progression [
9]. Recently, it has been discovered that miR-29 suppresses HIV-1 replication and binds directly to HIV-1 Nef mRNAs [
22]. An indirect role for miR-29 has also been suggested in the modulation of cyclins in HIV infection [
23].
Although our results did not immediately suggest a role for miR-29 in managing viremia and disease progression, it became clear upon examination that miR-29 was indeed upregulated in aviremic CD4+ T-cells, when compared to both viremic and LTNP CD4+ T-cells. Although it is peculiar that the miR-29s did not show association with the LTNP disease state, this is not unexpected. The biggest difference between HIV+ aviremic and LTNP individuals is the preservation of immune system strength and natural control of HIV in LTNP individuals, where aviremic individuals require HAART to control HIV replication. It is likely that miRNAs may have some role in maintaining below detectable levels of plasma viremia while on HAART therapy. On another note, it is important to iterate that we also found miR-1298 specific to the CD8+ aviremic group (with respect to negative control), showing 2-fold downregulation. This was obtained using the FDR corrective testing, and we believe it is the first miRNA specific to CD8+ T-cells of HIV patients that may be involved in control HIV during HAART therapy and may offer some biological significance. Thus, these two miRNAs may have some functional relevance during HAART therapy, and whether they come into play when therapy is effective in controlling plasma viremia remains unknown. Nonetheless, the two miRNAs in some ways predict the efficacy of HAART and should be explored in the context.
Similarly, in aviremic CD8+ T-cells, miR-29b/c were upregulated when compared to negative controls, whereas the miR-29a/b/c were upregulated in aviremic individuals when compared to LTNPs. Taken together, these data suggest that the expression of the cellular miR-29 family is triggered by viral replication and implies a potential role for miR-29 family members in both CD4+ and CD8+ T-cell subsets in HIV infection, although perhaps not in the management of viremia in the LTNP group. Determining whether miR-1298 and the miR-29 family have some functional synergy between them requires future functional validation.
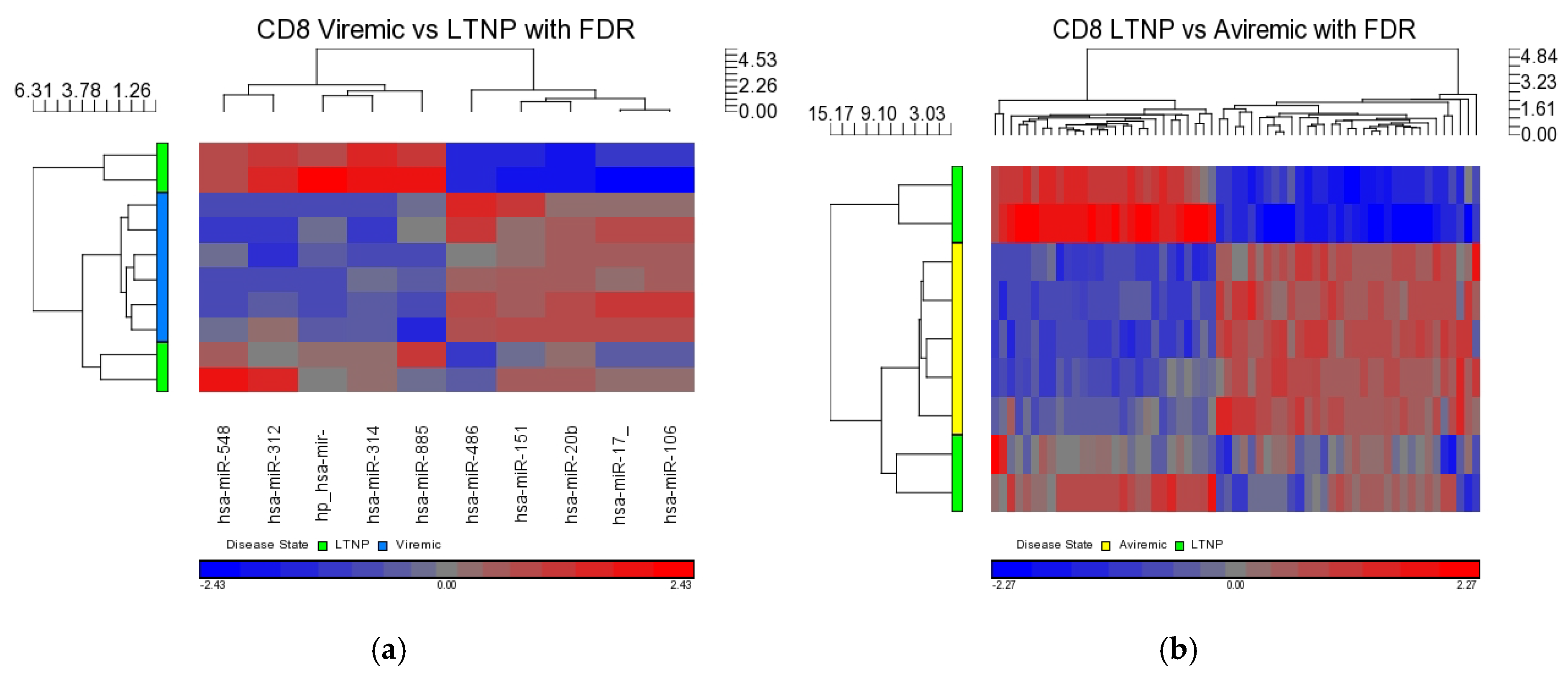
3.6. miRNA-17/20 Are Downregulated in CD8+ T Cells in LTNPs: A Tentative Link to Their Role in Viral Replication
miR-17/20 target 3′UTR of PCAF acetyltransferase and are known to indirectly inhibit HIV infection [
7,
8]. It has been shown that the transfection of miR-17-5p and -20 into the PBMCs of HIV-1 infected donors leads to reduced HIV-1 production, and that transfection of antisense inhibitor enhances PCAF expression and HIV-1 production [
24]. PCAF1 is an important cofactor for tat in HIV-1 gene expression and has potential sites for these miRNAs in its 3′UTR. Recently, miR-17a has been associated with enhanced viral production and predicted to bind cellular factors that indirectly influence HIV-1 replication [
22].
We found that miR-17 and miR-20a were both downregulated in the CD8+ T-cells of LTNPs when compared directly with the CD8+ T-cells from aviremic patients. MiR-17 was downregulated in CD8+ T-cells from the LTNPs when compared against the viremic patients. This upregulation of miRNAs could be related to the inhibition of HIV-1 in a more severe disease state (aviremic on HAART compared to LTNPs), rather than the downregulation of these miRNAs by HIV.
As can be noted in several sections of the discussion, the correlation of DE miRNAs observed in our study could only be compared with other studies when non-FDR corrective testing was used. Thus, it is important to reiterate that, since we performed a bimodal analysis of our dataset using FDR (p < 0.05) and non-FDR (p < 0.01) statistical analysis, we were able to observe both the strongly DE miRNAs responsible for clear segregation of disease groups using FDR analysis, as well as the gentler trends of miRNA expression between disease classes and cell types in the non-FDR analysis. Our observations regarding the two datasets raise two possibilities: that either most studies in the miRNA field are not as stringently controlled as our primary FDR analysis, or that the stringency we have employed in deriving significantly DE miRNA is too strict. If this is the case, high stringency analysis may occlude biologically significant miRNA candidates, as was apparent when functionally important miRNA deemed important in the literature were confirmed and supported by our non-FDR dataset.
In summary, the findings presented herein provide evidence that HIV infection plays a significant role in altering cellular miRNA expression and, conversely, that cell-specific miRNA expression may have implications for the control of HIV infection. We report the findings of a variety of miRNAs that are differentially expressed in a cell-, disease-stage- and HIV-infection-specific manner, and offer support to the findings of several other studies. This is the first evidence showing tight miRNA regulation during HIV disease in different cell types and disease stages, suggesting an essential role for miRNA-guided cell phenotype in HIV pathogenesis and the development of AIDS. A unique aspect of these findings is the evidence for miRNAs that are involved in HIV resistance, as seen in non-progression. Further study of the roles of these miRNAs, their cognate mRNAs, and the reason for their dysregulation in HIV infection may shed light on the mechanisms controlling the progression or non-progression of HIV disease, and open up a pathway to a new line of therapeutics for the effective treatment and control of HIV.
4. Materials and Methods
4.1. Fresh PBMC Isolation from Whole Blood
Up to 10 mL of freshly collected whole blood in EDTA was diluted to 35 mL with DPBS (Dulbecco’s phosphate buffered saline, Lonza BioWhittaker, Walkersville, MD, USA) and layered carefully onto 15 mL of Ficoll (Ficoll-Paque PLUS, 1.077 ± 0.001 g/mL, GE Healthcare, Uppsala, Sweden) in 50-mL tubes, without disrupting the Ficoll border. Separation of the PBMC buffy coat was achieved via centrifugation at 1800 rpm for 20 min at room temperature. The buffy coat containing peripheral blood mononuclear cells (PBMCs) was extracted to another 50-mL tube via transfer pipette. The separated PBMCs were washed twice with DPBS by centrifugation at 1500 rpm for 7 min at 4 °C, then once in 10 mL of cold (4 °C) magnet activated cell sorting method (MACS) buffer (DPBS pH 7.2, 2 mM EDTA (Amresco, Sydney, Australia), 0.5% FBS (Fetal Bovine Serum, SAFC Biosciences, Lenexa, KS, USA) via centrifugation at 1200 rpm for 10 min at 4 °C. The supernatant was aspirated retaining the PBMC pellet.
4.2. Processing of Cryopreserved PBMCs
PBMCs from the LTNPs used in this study were previously cryopreserved. These cells frozen in liquid nitrogen were raised to −80 °C for one day before revival. Cells were slowly thawed on ice and were transferred using a transfer pipette into 15-mL tubes containing 5 mL of RPMI 1640 (Lonza BioWhittaker) 10% FBS in 15-mL tubes. The cell suspension was centrifuged for 10 min at 1000 rpm at room temperature to pellet the revived cells. The supernatant was aspirated and decanted retaining the clean cell pellet in preparation for the CD4+ and CD8+ cell separation.
4.3. Macs Column Separation of CD4+ and CD8+ T Cells from Whole PBMC
The PBMCs were centrifuged at 1000g to remove cell debris. The cell pellet was further washed twice with phosphate buffered saline (PBS) to remove excess antigen. After aspirating all of the PBS, the cell pellet was further washed with 10ml of MACS buffer (50 mL PBS + 250 µL of Fetal Bovine Serum (FBS) + 200 µL of 2 mM EDTA) by centrifuging at 300 g for 10 min at 4 °C. Cells were washed with PBS, and MACS® Column Technology was used for separating PBMCs into different cell subsets. Using MACS® MicroBeads, CD4+ helper T-cells and CD8+ cytotoxic T-cells were extracted according to the manufacturer’s specifications using the positive selection MACS, Miltenyi Biotech (Marburg, Germany). Flow cytometric verification of cell purity of CD4+ and CD8+ T-cells was evaluated using flow cytometry, as previously described by us (Potter et al., 2006).
4.4. RNA Extraction and miRNA Enrichment
Both total RNA and miRNA were extracted from the brain cortex using the miRNeasy Mini Kit (Qiagen Pty Ltd., Clifton Hill, Victoria, Australia) with an integrated step of on-column DNase digestion as per the manufacturer’s protocol for the kit.
4.5. RNA Quality Check and Quantification
RNA quality was checked by RNA micro-electrophoresis on a 2100 Bioanalyzer apparatus (Agilent Technologies, Oxford, UK) and by using the RNA 6000 Nano Labchip Technology Kit. All reagents were equilibrated to room temperature for 30 min before use, and the quantity and quality of RNA were assessed based on the RNA integration numbers (RIN) for each sample. All RNA samples used in q-PCR and miRNA analysis had an RIN of >5, and anything <5 was deemed inappropriate for the miRNA chip, but were considered for the q-PCR.
4.6. FlashTag Biotin HSR RNA Labeling and Affymetrix GeneChip miRNA Arrays
MicroRNA was prepared for Affymetrix GeneChip arrays using the FlashTag® Biotin HSR RNA Labeling Kit for Affymetrix® GeneChip® miRNA Arrays (Genisphere, Hatfield, PA, USA). miRNA array preparation was conducted according to instructions provided by Genisphere for the Affymetrix GeneChip miRNA array procedure (FlashTag Biotin HSR RNA Labeling Kit for Affymetrix GeneChip miRNA Arrays), including the use of reagents from this kit (vials numbered). Hybridization of prepared samples, incubation, washing, staining, and scanning was completed by the sample submission service at the Ramaciotti Center for Gene Function Analysis, University of New South Wales, Sydney.
4.7. Analysis
4.7.1. Sample Classification and Criteria
Patients were classed into disease groups based on their HIV plasma VL determined by M3 Prep/Cobas TaqMan HIV-Version 2 kit (ROCHE, Inc., Pleasanton, CA, USA) to test HIV plasma VL, at the Westmead Hospital ICPMR (Institute of Clinical Pathology and Medical Research). Diverse HIV disease groups were analyzed, including HAART-experienced viremic (VL above 40 copies/mL plasma) and aviremic (below detectable levels of plasma viremia) patients. An LTNP group, comprising of HIV+ therapy-naïve individuals who showed high CD4+ and CD8+ T-cell counts and below detectable plasma viremia in the absence of HAART, was also used. All comparisons between disease groups were made against HIV– healthy individuals. Thus, the main criterion for analysis was the stratification of patients into viremic, aviremic, LTNP, and HIV– healthy groups, with the aim of defining disease-stage-specific and cell-specific (CD4+ and CD8+ T-cells) miRNA.
4.7.2. Normalization, QC, and PCA Analysis
The Analysis was performed in the Partek Genomics Suite using inbuilt data normalization for Affymetrix CEL files (consisting of background correction, quantile normalization, Log
2 transformation, and median polish summarization of probesets). The quality of the miRNA dataset and its integrity was assessed using PCA. PCA is a mathematical operation involving applied linear algebra, which transforms datasets into a new coordinate system such that the most variable components of the dataset are presented on axes where each component is independent of the last. This allows a complex dataset to be simplified and visualized by its most important gradients. In turn, the analysis also allows the determination of which factors are the most important in distinguishing one cluster from another. PCA plots were generated, to first test the integrity of infected samples in comparison to uninfected samples, and then PCA was used to inter-compare the disease states for each cell type and the cell types for each disease state (
Figure 1,
Figure 2 and
Figure 3). Frequency histograms were also generated for both the CD4+ and CD8+ T-cell subsets, showing the effectiveness of normalization applied by the Partek Genomics Suite and concordance of the datasets (
Figure 3).
4.7.3. Statistical Analysis
Samples were analyzed using the Partek Genomics Suite software, applying two forms of corrective and statistical testing. Primarily, the analysis of differential expression was completed using t-tests followed by Benjmini–Hochberg FDR corrective testing. The Benjamini–Hochberg algorithm sorts a list of p-values in ascending order and determines how likely value is to be a false rejection of the null hypothesis (a false positive) at each point in the list. In this way, it develops a list of corrected or adjusted p-values for the detected differentially expressed miRNAs. A fold change of 2 (up- or downregulation) was also applied to deem any miRNA to be biologically and statistically significant. Although not a part of the result section, data were also processed without FDR corrective testing to see the difference between two datasets. This was to determine whether the statistical manipulation of the data using the FDR method occluded biologically or functionally significant miRNAs. For non-FDR data, a stricter p < 0.01 significance threshold was applied for significant changes, along with the fold change of 2, to ensure biological significance. This method will herein be referred to as the non-FDR method and is used as a comparative set of data for the FDR results.
Notably, even though non-FDR testing showed a higher number of DE miRNAs than FDR-controlled corrective testing, all microRNAs deemed significant using strict FDR corrective testing were also picked up as highly significant miRNAs in the less stringent, non-FDR method, confirming the reliability of FDR corrective testing.
Comparisons of miRNA expression were made in an organized and structured manner. First, differential expression of miRNAs between HIV+ and HIV– individuals was considered. Then, comparisons were made between CD4+ and CD8+ T-cells before examining the differential expression between disease states within CD4+ and CD8+ T-cells. Lastly, results observed in independent CD4+ and CD8+ T-cell analyses were inter-compared. Analyses included clustering, the generation of heat maps, and intergroup comparisons through the generation of Venn diagrams.
4.7.4. Quantitative Real-Time PCR Validation of Selected and Significant DE miRNAs
Two miRNAs of significance and importance were selectively corroborated using quantitative real-time PCR (qRT-PCR) based on their adjusted
p-values, fold changes, and differential expression. cDNA was generated from RNA samples using the Qiagen miScript Reverse Transcription Kit (Qiagen, Sydney, Australia). A master mix containing 160 µL of buffer RT and 40 µL of miScript reverse transcriptase mix was produced, and 5 µL was added to each sample of 250-ng RNA. The samples were made with RNase free water (up to 20 µL) and incubated at 37 °C for 60 min, and then 95 °C for 5 min to inactivate the RT enzyme using the Hybaid Thermal Cycler for PCR. Expression levels of two differentially expressed miRNAs (hsa-miR-199a and miR-126) were validated by qRT-PCR using the Qiagen miScript PCR system (Qiagen, Valencia, CA, USA). Master mixes containing SYBR green PCR master-mix (10 µL/assay), miScript universal primer (2 µL/assay), and RNase-free water (5 µL/assay) was produced for each specific primer. Unique miScript primers specific for the miRNAs of interest were added (2 µL/assay). Data was analyzed using Microsoft Excel. RNU6B control was used to normalize data. All the experiments were performed in duplicate, and relative expression levels of these miRNAs were determined by the ΔΔ−C
T method (see
Table 9 for qRTPCR results).
5. Conclusions
MicroRNAs are the master regulators of gene expression; therefore, this analysis of differentially expressed miRNA in CD4+ and CD8+ T-cells at different stages of HIV infection provides highly valuable data on how these two T-cell subsets interact during HIV infection. Our analysis provides data on how miRNAs may guide disease progression: leading to gradual disruption and decline of the immune system in viremic individuals or providing possible resistance against HIV infection, as seen in LTNPs.
Determining the role of differentially expressed miRNAs in HIV infection is essential. We believe that an understanding of the dysregulated miRNAs in HIV infection will prove crucial for the development of miRNA-based therapies or the targeting of cellular miRNA as treatment for HIV infection. Before this is possible, a clear understanding of miRNA gene targets is needed. This is a challenge, as one miRNA can target thousands of genes, and HIV infection of cells can induce large changes in gene expression and overall cell state, meaning that the relationships between miRNAs and mRNAs in HIV infection may be very complex. As such, more work is needed to map precisely the mRNA and miRNA interactions that guide HIV pathogenesis. Such analysis will not only allow the development of new strategies for HIV control but may also lead to the identification of miRNA candidates that can be used as a new generation of therapeutic targets to control HIV. For example, a miR-122 agonist, miravirsen, is the first miRNA-targeted drug to be developed for the treatment of viral accumulation in hepatocytes during HCV infection [
25].
With an increase of research in the area of miRNA expression profiling and functional analysis, several confounding factors have emerged. Currently, a variety of papers in this area all used different cell types, different definitions of HIV disease states, different methods of analysis and statistical evaluation, and diverse microarray platforms [
7,
8,
9,
15,
21,
23]. With all these variances in methods used by different studies, it is no surprise that the majority of research in this area is not currently in agreement. A consensus on miRNAs that is relevant in HIV disease will provide a durable framework for future work involving functional assays and miRNA/mRNA interaction analysis. Further, these clarifications will provide a clear view to developing new generation therapeutics to treat HIV disease. What is significant is that these analyses show miRNA that is specific to the natural control of HIV disease in LTNPs, which may be of vital importance in designing a new generation of prognostics and diagnostics for HIV disease.