The Interaction of the Gut Microbiota with the Mucus Barrier in Health and Disease in Human
Abstract
:1. Introduction
2. The Structure of the Mucus Barrier
3. The Mucin Gene Family and Their Role in the Gut
4. Bacterial Species in the Human Gastrointestinal Tract
5. Mucin Glycosylation and the Sugar Code
5.1. Bulk Properties—Gel Formation and Viscoelasticity
5.2. Mucin Glycans; Sequence, Topography and Mucosal Interactions
6. Mucin Glycans as Biological Arrays Linked to Function
7. Screening for Mucin Glycans and Mucin Glycan Engineering
8. Metabolism of Mucin Glycans
9. Glycan Expression When the Gastrointestinal Microbiota Is Removed
10. Aberrant Mucin Glycosylation and Disease in the Gastrointestinal Tract
10.1. Bacterial Strains and Gastrointestinal Disease
10.2. Necrotising Enterocolitis (NEC)
10.3. Inflammatory Bowel Disease (IBD)
10.4. Colorectal Cancer
11. Mucin Glycans; Future Prospects
- The attachment of O-glycans to either serine or threonine residues in mucin tandem repeat domains has been identified as a regulated biological event. The significance of this biochemical configuration deserves wider exploration. Methods to carry out such screening are available.
- Why does fecal diversion trigger changes that are also seen in cancer?
- Nucleotide sugar metabolism is fundamental to normal glycosylation processes. However, the links with catabolic pathways and their integration with biosynthesis are still poorly understood and should be examined more closely at cell, tissue and organ levels.
- Different populations of O-acetylated sialic acids in colonic mucins are evident in the colorectal mucosa as detected by the PR3A5 and 6G4 antibodies. These patterns should be reexamined using the nidovirus probes reported recently in order to obtain more precise information regarding their biological functions. In addition, the identification and biological significance of non O-acetylators should be examined in order to understand the importance of sialic acid O-acetylation.
- The Sda antigen is implicated as a major sialoglycan present in colonic mucins, which is normally O-acetylated, but is lost in colorectal cancer and in fecal diversion. It is clear that bacteria talk to this mucin glycan. Does the Sda antigen represent a discrete group of O-acetylated sialic acids?
- Mucin sulphation is poorly studied. A combination of chemical and immunohistological studies is needed to address the expression patterns in normal health and the implications of specific deletions in disease.
- In IBD changes relating to the age of patients, their ethnicity, disease severity deserve further investigation. In addition, the composition of the microbiota during development should be examined for IBD patients.
Funding
Acknowledgments
Conflicts of Interest
Abbreviations Used in the Text Tables and Figures
Symbols for Individual Monosaccharides Used in Figures
 |  |  |  |
| d-glucose; | d-galactose; | d-mannose; | N-acetyl-d-glucosamine; |
 |  |  |
| N-acetyl-d-galactosamine; | l-fucose; | N-acetylneuraminic acid. |
References
- Lauc, G.; Kristic, J.; Zoldos, V. Glycans–the third revolution in evolution. Front. Genet. 2014, 5, 145. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Desseyn, J.L.; Aubert, J.P.; Porchet, N.; Laine, A. Evolution of the large secreted gel-forming mucins. Mol. Biol. Evol. 2000, 17, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Hansson, G.C.; Samuelsson, T. Gel-forming mucins appeared early in metazoan evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 16209–16214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corfield, A.P. Mucins: A biologically relevant glycan barrier in mucosal protection. Biochim. Biophys. Acta 2015, 1850, 236–252. [Google Scholar] [CrossRef] [PubMed]
- McGuckin, M.A.; Linden, S.K.; Sutton, P.; Florin, T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 2011, 9, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Bafna, S.; Kaur, S.; Batra, S.K. Membrane-bound mucins: The mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene 2010, 29, 2893–2904. [Google Scholar] [CrossRef] [PubMed]
- Ambort, D.; Johansson, M.E.; Gustafsson, J.K.; Ermund, A.; Hansson, G.C. Perspectives on mucus properties and formation–lessons from the biochemical world. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Gabius, H.J.; Roth, J. An introduction to the sugar code. Histochem. Cell Biol. 2017, 147, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Ouwerkerk, J.P.; de Vos, W.M.; Belzer, C. Glycobiome: Bacteria and mucus at the epithelial interface. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 25–38. [Google Scholar] [CrossRef] [PubMed]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. 2013, 11, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Gordon, J.I. Glycans as legislators of host-microbial interactions: Spanning the spectrum from symbiosis to pathogenicity. Glycobiology 2001, 11, 1R–10R. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.; Tharmalingam, T.; Rudd, P.M. Glycans as cancer biomarkers. Biochim. Biophys. Acta 2012, 1820, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Campos, D.; Freitas, D.; Gomes, J.; Magalhaes, A.; Steentoft, C.; Gomes, C.; Vester-Christensen, M.B.; Ferreira, J.A.; Afonso, L.P.; Santos, L.L.; et al. Probing the O-glycoproteome of gastric cancer cell lines for biomarker discovery. Mol. Cell. Proteom. 2015, 14, 1616–1629. [Google Scholar] [CrossRef] [PubMed]
- Reis, C.A.; Osorio, H.; Silva, L.; Gomes, C.; David, L. Alterations in glycosylation as biomarkers for cancer detection. J. Clin. Pathol. 2010, 63, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Freeze, H.H.; Kinoshita, T.; Schnaar, R.L. Genetic Disorders of Glycan Degradation; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratoty Press: New York, NY, USA, 2017; pp. 553–568. [Google Scholar]
- Freeze, H.H.; Schachter, H.; Kinoshita, T. Genetic Disorders of Glycosylation. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 569–582. [Google Scholar]
- Varki, A.; Kannagi, R.; Toole, B.P.; Stanley, P. Glycosylation Changes in Cancer. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 597–609. [Google Scholar]
- Bergstrom, K.; Liu, X.; Zhao, Y.; Gao, N.; Wu, Q.; Song, K.; Cui, Y.; Li, Y.; McDaniel, J.M.; McGee, S.; et al. Defective Intestinal Mucin-Type O-Glycosylation Causes Spontaneous Colitis-Associated Cancer in Mice. Gastroenterology 2016, 151, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Brockhausen, I. Mucin-type O-glycans in human colon and breast cancer: Glycodynamics and functions. EMBO Rep. 2006, 7, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, M.N.; Chik, J.; Lee, L.; Anugraham, M.; Abrahams, J.L.; Packer, N.H. Cell surface protein glycosylation in cancer. Proteomics 2014, 14, 525–546. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.; Malagolini, N.; Trinchera, M.; Chiricolo, M. Mechanisms of cancer-associated glycosylation changes. Front. Biosci. (Landmark Ed.) 2012, 17, 670–699. [Google Scholar] [CrossRef] [PubMed]
- Groux-Degroote, S.; Wavelet, C.; Krzewinski-Recchi, M.A.; Portier, L.; Mortuaire, M.; Mihalache, A.; Trinchera, M.; Delannoy, P.; Malagolini, N.; Chiricolo, M.; et al. B4GALNT2 gene expression controls the biosynthesis of Sda and sialyl Lewis X antigens in healthy and cancer human gastrointestinal tract. Int. J. Biochem. Cell Biol. 2014, 53, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Robbe-Masselot, C.; Michalski, J.C.; Capon, C. Tumour associated antigens of mucin O-glycans. Clinical relevance of glycobiology in cancer and inflammatory diseases of the epithelium. In The Epithelial Mucins: Structure/Function Roles in Cancer and Inflammation; Van Seuningen, I., Ed.; Research Signpost: Kerala, India, 2008; pp. 55–73. [Google Scholar]
- Sheng, Y.H.; Hasnain, S.Z.; Florin, T.H.; McGuckin, M.A. Mucins in inflammatory bowel diseases and colorectal cancer. J. Gastroenterol. Hepatol. 2012, 27, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Watt, F.M. Defining Adult Stem Cells by Function, Not by Phenotype. Annu. Rev. Biochem. 2018, 87, 13.1–13.13. [Google Scholar] [CrossRef] [PubMed]
- Birchenough, G.M.; Johansson, M.E.; Gustafsson, J.K.; Bergstrom, J.H.; Hansson, G.C. New developments in goblet cell mucus secretion and function. Mucosal Immunol. 2015, 8, 712–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, M.E.; Hansson, G.C. Mucus and the goblet cell. Dig. Dis. (Basel Switz.) 2013, 31, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Pelaseyed, T.; Bergstrom, J.H.; Gustafsson, J.K.; Ermund, A.; Birchenough, G.M.; Schutte, A.; van der Post, S.; Svensson, F.; Rodriguez-Pineiro, A.M.; Nystrom, E.E.; et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014, 260, 8–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCauley, H.A.; Guasch, G. Three cheers for the goblet cell: Maintaining homeostasis in mucosal epithelia. Trends Mol. Med. 2015, 21, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Birchenough, G.M.; Nystrom, E.E.; Johansson, M.E.; Hansson, G.C. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 2016, 352, 1535–1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ermund, A.; Meiss, L.N.; Rodriguez-Pineiro, A.M.; Bahr, A.; Nilsson, H.E.; Trillo-Muyo, S.; Ridley, C.; Thornton, D.J.; Wine, J.J.; Hebert, H.; et al. The normal trachea is cleaned by MUC5B mucin bundles from the submucosal glands coated with the MUC5AC mucin. Biochem. Biophys. Res. Commun. 2017, 492, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Ostedgaard, L.S.; Moninger, T.O.; McMenimen, J.D.; Sawin, N.M.; Parker, C.P.; Thornell, I.M.; Powers, L.S.; Gansemer, N.D.; Bouzek, D.C.; Cook, D.P.; et al. Gel-forming mucins form distinct morphologic structures in airways. Proc. Natl. Acad. Sci. USA 2017, 114, 6842–6847. [Google Scholar] [CrossRef] [PubMed]
- Atuma, C.; Strugala, V.; Allen, A.; Holm, L. The adherent gastrointestinal mucus gel layer: Thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 280, G922–G999. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, J.K.; Ermund, A.; Johansson, M.E.; Schutte, A.; Hansson, G.C.; Sjovall, H. An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G430–G438. [Google Scholar] [CrossRef] [PubMed]
- Kamphuis, J.B.J.; Mercier-Bonin, M.; Eutamene, H.; Theodorou, V. Mucus organisation is shaped by colonic content: A new view. Sci. Rep. 2017, 7, 8527. [Google Scholar] [CrossRef] [PubMed]
- Andrianifahanana, M.; Moniaux, N.; Batra, S.K. Regulation of mucin expression: Mechanistic aspects and implications for cancer and inflammatory diseases. Biochim. Biophys. Acta 2006, 1765, 189–222. [Google Scholar] [CrossRef] [PubMed]
- Desseyn, J.-L.; Gouyer, V.; Tetaert, D. Architecture of the gel forming mucins. In The Epithelial Mucins: Structure/Function. Roles in Cancer and Inflammatory Diseases; Van Seuningen, I., Ed.; Research Signpost: Kerala, India, 2008; pp. 1–16. [Google Scholar]
- Hattrup, C.L.; Gendler, S.J. Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol. 2008, 70, 431–457. [Google Scholar] [CrossRef] [PubMed]
- Thornton, D.J.; Rousseau, K.; McGuckin, M.A. Structure and Function of the Polymeric Mucins in Airways Mucus. Annu. Rev. Physiol. 2008, 70, 459–486. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Sjovall, H.; Hansson, G.C. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 352–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desseyn, J.L. Mucin CYS domains are ancient and highly conserved modules that evolved in concert. Mol. Phylogenet. Evol. 2009, 52, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Desseyn, J.L.; Tetaert, D.; Gouyer, V. Architecture of the large membrane-bound mucins. Gene 2008, 410, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Linden, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the mucosal barrier to infection. Nat. Mucosal Immunol. 2008, 1, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Gendler, S.J.; Spicer, A.P.; Lalani, E.N.; Duhig, T.; Peat, N.; Burchell, J.; Pemberton, L.; Boshell, M.; Taylor-Papadimitriou, J. Structure and biology of a carcinoma-associated mucin, MUC1. Am. Rev. Respir. Dis. 1991, 144 Pt 2, S42–S47. [Google Scholar] [CrossRef] [PubMed]
- Thornton, D.J.; Davies, J.R.; Carlstedt, I.; Sheehan, J.K. Structure and Biochemistry of Respiratory Mucins. In Airway Mucus: Basic Mechanisms and Clinical Perspectives; Rogers, D.F., Lethem, M.I., Eds.; Birkhäuser Verlag: Basel, Switzerland, 1997. [Google Scholar]
- Lang, T.; Klasson, S.; Larsson, E.; Johansson, M.E.; Hansson, G.C.; Samuelsson, T. Searching the Evolutionary Origin of Epithelial Mucus Protein Components-Mucins and FCGBP. Mol. Biol. Evol. 2016, 33, 1921–1936. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Berry, M. Glycan variation and evolution in the eukaryotes. Trends Biochem. Sci. 2015, 40, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Angata, T.; Varki, A. Chemical diversity in the sialic acids and related alpha-keto acids: An evolutionary perspective. Chem. Rev. 2002, 102, 439–469. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.R.; Gagneux, P. Evolution of carbohydrate antigens–microbial forces shaping host glycomes? Glycobiology 2007, 17, 23R–34R. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Nothing in glycobiology makes sense, except in the light of evolution. Cell 2006, 126, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Dell, A.; Galadari, A.; Sastre, F.; Hitchen, P. Similarities and differences in the glycosylation mechanisms in prokaryotes and eukaryotes. Int. J. Microbiol. 2010, 2010, 148178. [Google Scholar] [CrossRef] [PubMed]
- Gipson, I.K.; Spurr-Michaud, S.J.; Tisdale, A.S.; Kublin, C.; Cintron, C.; Keutmann, H. Stratified squamous epithelia produce mucin-like glycoproteins. Tissue Cell 1995, 27, 397–404. [Google Scholar] [CrossRef]
- Hijikata, M.; Matsushita, I.; Tanaka, G.; Tsuchiya, T.; Ito, H.; Tokunaga, K.; Ohashi, J.; Homma, S.; Kobashi, Y.; Taguchi, Y.; et al. Molecular cloning of two novel mucin-like genes in the disease-susceptibility locus for diffuse panbronchiolitis. Hum. Genet. 2011, 129, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Ihrke, G.; Gray, S.R.; Luzio, J.P. Endolyn is a mucin-like type I membrane protein targeted to lysosomes by its cytoplasmic tail. Biochem. J. 2000, 345 Pt 2, 287–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsh, P.D.; Percival, R.S. The oral microflora--friend or foe? Can we decide? Int. Dent. J. 2006, 56 (Suppl. 1), 233–239. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.Q.; Zhang, X.; Liu, S.; Zheng, L.; Ma, F.; Chen, T.; Xu, B. Exploring the oral microflora of preschool children. J. Microbiol. 2017, 55, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Rosen, R.; Amirault, J.; Liu, H.; Mitchell, P.; Hu, L.; Khatwa, U.; Onderdonk, A. Changes in gastric and lung microflora with acid suppression: Acid suppression and bacterial growth. JAMA Pediatr. 2014, 168, 932–937. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Mahbub, R.; Fox, J.G. Xenobiotics: Interaction with the Intestinal Microflora. ILAR J. 2015, 56, 218–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, H.J.; O’Toole, P.W.; Walker, A.W. Special issue: The Human Intestinal Microbiota. Microbiology 2010, 156 Pt 11, 3203–3204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirinian, G.; Shimizu, T.; Sugar, C.; Slots, J.; Chen, C. Periodontopathic bacteria in young healthy subjects of different ethnic backgrounds in Los Angeles. J. Periodontol. 2002, 73, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Wagner, S.A.; Clamp, J.R.; Kriaris, M.S.; Hoskins, L.C. Mucin degradation in the human colon: Production of sialidase, sialate O-acetyl esterase, N-acetylneuraminate lyase, arylesterase and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 1992, 60, 3971–3978. [Google Scholar] [PubMed]
- Variyam, E.P.; Hoskins, L.C. In vitro degradation of gastric mucin. Carbohydrate side chains protect polypeptide core from pancreatic proteases. Gastroenterology 1983, 84, 533–537. [Google Scholar] [PubMed]
- Bottacini, F.; van Sinderen, D.; Ventura, M. Omics of bifidobacteria: Research and insights into their health-promoting activities. Biochem. J. 2017, 474, 4137–4152. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turroni, F.; Bottacini, F.; Foroni, E.; Mulder, I.; Kim, J.H.; Zomer, A.; Sanchez, B.; Bidossi, A.; Ferrarini, A.; Giubellini, V.; et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. USA 2010, 107, 19514–19519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, R.; Sharma, N.; Chaudhry, R.; Deorari, A.; Paul, V.K.; Gewolb, I.H.; Panigrahi, P. Effects of oral Lactobacillus GG on enteric microflora in low-birth-weight neonates. J. Pediatr. Gastroenterol. Nutr. 2003, 36, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.; O’Connell Motherway, M.; Schoterman, M.H.; van Neerven, R.J.; Nauta, A.; van Sinderen, D. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J. Appl. Microbiol. 2013, 114, 1132–1146. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; van Sinderen, D.; O’Toole, P.W. The genus Lactobacillus—A genomic basis for understanding its diversity. FEMS Microbiol. Lett. 2007, 269, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.R.; Ahrne, S.; Hyde, L.; Wei, S.; Hollingsworth, M.A. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 2003, 52, 827–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, T.H.; Boudry, C.; Everaert, N.; Thewis, A.; Portetelle, D.; Daube, G.; Nezer, C.; Taminiau, B.; Bindelle, J. Adding mucins to an in vitro batch fermentation model of the large intestine induces changes in microbial population isolated from porcine feces depending on the substrate. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed]
- Dols, J.A.; Molenaar, D.; van der Helm, J.J.; Caspers, M.P.; de Kat Angelino-Bart, A.; Schuren, F.H.; Speksnijder, A.G.; Westerhoff, H.V.; Richardus, J.H.; Boon, M.E.; et al. Molecular assessment of bacterial vaginosis by Lactobacillus abundance and species diversity. BMC Infect. Dis. 2016, 16, 180. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4680–4687. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, G.S.; Carvalho, F.P.; Arantes, R.M.; Nunes, A.C.; Moreira, J.L.; Mendonca, M.; Almeida, R.B.; Farias, L.M.; Carvalho, M.A.; Nicoli, J.R. Characteristics of Lactobacillus and Gardnerella vaginalis from women with or without bacterial vaginosis and their relationships in gnotobiotic mice. J. Med. Microbiol. 2012, 61 Pt 8, 1074–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collado, M.C.; Derrien, M.; Isolauri, E.; de Vos, W.M.; Salminen, S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 2007, 73, 7767–7770. [Google Scholar] [CrossRef] [PubMed]
- De Vos, W.M. Microbe Profile: Akkermansia muciniphila: A conserved intestinal symbiont that acts as the gatekeeper of our mucosa. Microbiology 2017, 163, 646–648. [Google Scholar] [CrossRef] [PubMed]
- Belzer, C.; de Vos, W.M. Microbes inside–from diversity to function: The case of Akkermansia. ISME J. 2012, 6, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Reunanen, J.; Kainulainen, V.; Huuskonen, L.; Ottman, N.; Belzer, C.; Huhtinen, H.; de Vos, W.M.; Satokari, R. Akkermansia muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl. Environ. Microbiol. 2015, 81, 3655–3662. [Google Scholar] [CrossRef] [PubMed]
- Van Herreweghen, F.; Van den Abbeele, P.; De Mulder, T.; De Weirdt, R.; Geirnaert, A.; Hernandez-Sanabria, E.; Vilchez-Vargas, R.; Jauregui, R.; Pieper, D.H.; Belzer, C.; et al. In vitro colonisation of the distal colon by Akkermansia muciniphila is largely mucin and pH dependent. Benef. Microbes 2017, 8, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Wang, M.M.; Kulinich, A.; Yao, H.L.; Ma, H.Y.; Martinez, J.E.; Duan, X.C.; Chen, H.; Cai, Z.P.; Flitsch, S.L.; et al. Biochemical characterisation of the neuraminidase pool of the human gut symbiont Akkermansia muciniphila. Carbohydr. Res. 2015, 415, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Ozcan, E.; Milani, C.; Mancabelli, L.; Viappiani, A.; van Sinderen, D.; Sela, D.A.; Ventura, M. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front. Microbiol. 2015, 6, 1030. [Google Scholar] [CrossRef] [PubMed]
- Belenguer, A.; Duncan, S.H.; Calder, A.G.; Holtrop, G.; Louis, P.; Lobley, G.E.; Flint, H.J. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 2006, 72, 3593–3599. [Google Scholar] [CrossRef] [PubMed]
- Rios-Covian, D.; Gueimonde, M.; Duncan, S.H.; Flint, H.J.; de Los Reyes-Gavilan, C.G. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.L.; Versalovic, J. The human microbiome and its potential importance to pediatrics. Pediatrics 2012, 129, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, C.; Reigstad, C.S.; Backhed, F. Intestinal microbiota during infancy and its implications for obesity. J. Pediatr. Gastroenterol. Nutr. 2009, 48, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Charbonneau, M.R.; Blanton, L.V.; DiGiulio, D.B.; Relman, D.A.; Lebrilla, C.B.; Mills, D.A.; Gordon, J.I. A microbial perspective of human developmental biology. Nature 2016, 535, 48–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, D.T.; Ten Hagen, K.G. Mucin-type O-glycosylation during development. J. Biol. Chem. 2013, 288, 6921–6929. [Google Scholar] [CrossRef] [PubMed]
- Ten Hagen, K.G. Developmental glycobiology. Semin. Cell Dev. Biol. 2010, 21, 599. [Google Scholar] [CrossRef] [PubMed]
- Tian, E.; Hagen, K.G. O-linked glycan expression during Drosophila development. Glycobiology 2007, 17, 820–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, E.; Hoffman, M.P.; Ten Hagen, K.G. O-glycosylation modulates integrin and FGF signalling by influencing the secretion of basement membrane components. Nat. Commun. 2012, 3, 869. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T.A.; Ten Hagen, K.G.; Jamison, O. Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase ortholog pairs. Glycobiology 2008, 18, 861–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buisine, M.P.; Devisme, L.; Savidge, T.C.; Gespach, C.; Gosselin, B.; Porchet, N.; Aubert, J.P. Mucin gene expression in human embryonic and fetal intestine. Gut 1998, 43, 519–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corfield, A.P.; Carroll, D.; Myerscough, N.; Probert, C.S. Mucins in the gastrointestinal tract in health and disease. Front. Biosci. 2001, 6, D1321–D1357. [Google Scholar] [CrossRef] [PubMed]
- Reid, P.E.; Owen, D.A.; Magee, F.; Park, C.M. Histochemical studies of intestinal epithelial goblet cell glycoproteins during the development of the human foetus. Histochem. J. 1990, 22, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Lev, R.; Orlic, D. Histochemical and radio autographic studies of normal human foetal colon. Histochemistry 1974, 39, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Muchmore, E.A.; Varki, N.M.; Fukuda, M.; Varki, A. Developmental regulation of sialic acid modifications in rat and human colon. FASEB J. 1987, 1, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Robbe-Masselot, C.; Maes, E.; Rousset, M.; Michalski, J.C.; Capon, C. Glycosylation of human fetal mucins: A similar repertoire of O-glycans along the intestinal tract. Glycoconj. J. 2009, 26, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Fiat, A.M.; Chevan, J.; Jolles, P.; De Waard, P.; Vliegenthart, J.F.; Piller, F.; Cartron, J.P. Structural variability of the neutral carbohydrate moiety of cow colostrum kappa-casein as a function of time after parturition. Identification of a tetrasaccharide with blood group I specificity. Eur. J. Biochem. 1988, 173, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Fiat, A.M.; Jolles, P. Caseins of various origins and biologically active casein peptides and oligosaccharides: Structural and physiological aspects. Mol. Cell. Biochem. 1989, 87, 5–30. [Google Scholar] [CrossRef] [PubMed]
- Barboza, M.; Pinzon, J.; Wickramasinghe, S.; Froehlich, J.W.; Moeller, I.; Smilowitz, J.T.; Ruhaak, L.R.; Huang, J.; Lonnerdal, B.; German, J.B.; et al. Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria-host interactions. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Froehlich, J.W.; Dodds, E.D.; Barboza, M.; McJimpsey, E.L.; Seipert, R.R.; Francis, J.; An, H.J.; Freeman, S.; German, J.B.; Lebrilla, C.B. Glycoprotein expression in human milk during lactation. J. Agric. Food Chem. 2010, 58, 6440–6448. [Google Scholar] [CrossRef] [PubMed]
- Gabrielli, O.; Zampini, L.; Galeazzi, T.; Padella, L.; Santoro, L.; Peila, C.; Giuliani, F.; Bertino, E.; Fabris, C.; Coppa, G.V. Preterm milk oligosaccharides during the first month of lactation. Pediatrics 2011, 128, e1520–e1531. [Google Scholar] [CrossRef] [PubMed]
- Viverge, D.; Grimmonprez, L.; Cassanas, G.; Bardet, L.; Bonnet, H.; Solere, M. Variations of lactose and oligosaccharides in milk from women of blood types secretor A or H, secretor Lewis, and secretor H/nonsecretor Lewis during the course of lactation. Ann. Nutr. Metab. 1985, 29, 1–11. [Google Scholar] [CrossRef] [PubMed]
- De Leoz, M.L.; Kalanetra, K.M.; Bokulich, N.A.; Strum, J.S.; Underwood, M.A.; German, J.B.; Mills, D.A.; Lebrilla, C.B. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: A proof-of-concept study. J. Proteome Res. 2015, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Smilowitz, J.T.; Lebrilla, C.B.; Mills, D.A.; German, J.B.; Freeman, S.L. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu. Rev. Nutr. 2014, 34, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bode, L. The functional biology of human milk oligosaccharides. Early Hum. Dev. 2015, 91, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Armanini, F.; Turroni, F.; James, K.; Ferretti, P.; Gorfer, V.; Ferrario, C.; Milani, C.; et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 2017, 5, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burger-van Paassen, N.; Loonen, L.M.; Witte-Bouma, J.; Korteland-van Male, A.M.; de Bruijn, A.C.; van der Sluis, M.; Lu, P.; Van Goudoever, J.B.; Wells, J.M.; Dekker, J.; et al. Mucin Muc2 deficiency and weaning influences the expression of the innate defense genes Reg3beta, Reg3gamma and angiogenin-4. PLoS ONE 2012, 7, e38798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konstantinov, S.R.; Poznanski, E.; Fuentes, S.; Akkermans, A.D.; Smidt, H.; de Vos, W.M. Lactobacillus sobrius sp. nov. abundant in the intestine of weaning piglets. Int J. Syst. Evol. Microbiol. 2006, 56 Pt 1, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Sabharwal, H.; Sjoblad, S.; Lundblad, A. Sialylated oligosaccharides in human milk and feces of preterm, full-term, and weaning infants. J. Pediatr. Gastroenterol. Nutr. 1991, 12, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Denny, P.C.; Denny, P.A.; Klauser, D.K.; Hong, S.H.; Navazesh, M.; Tabak, L.A. Age-related changes in mucins from human whole saliva. J. Dent. Res. 1991, 70, 1320–1327. [Google Scholar] [CrossRef] [PubMed]
- Baughan, L.W.; Robertello, F.J.; Sarrett, D.C.; Denny, P.A.; Denny, P.C. Salivary mucin as related to oral Streptococcus mutans in elderly people. Oral Microbiol. Immunol. 2000, 15, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Biagi, E.; Nylund, L.; Candela, M.; Ostan, R.; Bucci, L.; Pini, E.; Nikkila, J.; Monti, D.; Satokari, R.; Franceschi, C.; et al. Through ageing, and beyond: Gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE 2010, 5, E10667. [Google Scholar] [CrossRef]
- Biagi, E.; Candela, M.; Franceschi, C.; Brigidi, P. The aging gut microbiota: New perspectives. Ageing Res. Rev. 2011, 10, 428–429. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Cusack, S.; O’Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G.; et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4586–4591. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Cucchiara, S.; Iebba, V.; Conte, M.P.; Schippa, S. The microbiota in inflammatory bowel disease in different age groups. Dig. Dis. 2009, 27, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, F.; van Sinderen, D.; O’Toole, P.W.; Stanton, C. Feeding the microbiota: Transducer of nutrient signals for the host. Gut 2017, 66, 1709–1717. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.C.; Lagstrom, H.; Suomalainen, T.; Salminen, S. Effect of probiotics on constipation, fecal azoreductase activity and fecal mucin content in the elderly. Ann. Nutr. Metab. 2002, 46, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Ticinesi, A.; Gerritsen, J.; Nouvenne, A.; Lugli, G.A.; Mancabelli, L.; Turroni, F.; Duranti, S.; Mangifesta, M.; Viappiani, A.; et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: A metagenomic study. Sci. Rep. 2016, 6, 25945. [Google Scholar] [CrossRef] [PubMed]
- Safe, A.F.; Warren, B.; Corfield, A.; McNulty, C.A.; Watson, B.; Mountford, R.A.; Read, A. Helicobacter pylori infection in elderly people: Correlation between histology and serology. Age Ageing 1993, 22, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Rampelli, S.; Candela, M.; Severgnini, M.; Biagi, E.; Turroni, S.; Roselli, M.; Carnevali, P.; Donini, L.; Brigidi, P. A probiotics-containing biscuit modulates the intestinal microbiota in the elderly. J. Nutr. Health Aging 2013, 17, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Demouveaux, B.; Gouyer, V.; Gottrand, F.; Narita, T.; Desseyn, J.L. Gel-forming mucin interactome drives mucus viscoelasticity. Adv. Colloid Interface Sci. 2018, 252, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.P.; Allen, A.; Hutton, D.A. Rheology of mucin. Methods Mol. Biol. 2000, 125, 99–109. [Google Scholar] [PubMed]
- Verdugo, P. Supramolecular dynamics of mucus. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.; Corfield, A.P.; McMaster, T.J. Mucins: A dynamic biology. Soft Matter 2013, 9, 1740–1743. [Google Scholar] [CrossRef]
- Allen, A.; Cunliffe, W.J.; Pearson, J.P.; Venables, C.W. The adherent gastric mucus gel barrier in man and changes in peptic ulceration. J. Intern. Med. 1990, 228 (Suppl. 1), 83–90. [Google Scholar] [CrossRef]
- Ermund, A.; Schutte, A.; Johansson, M.E.; Gustafsson, J.K.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G341–G347. [Google Scholar] [CrossRef] [PubMed]
- Hansson, G.C.; Johansson, M.E. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes 2010, 1, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, E.; Ota, H.; Hidaka, H.; Hayama, M.; Matsuzawa, K.; Akamatsu, T.; Nakayama, J.; Katsuyama, T. Helicobacter pylori and two ultrastructurally distinct layers of gastric mucous cell mucins in the surface mucous gel layer. Gut 2001, 49, 474–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, M.E.; Larsson, J.M.; Hansson, G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4659–4665. [Google Scholar] [CrossRef] [PubMed]
- Seeberger, P.H. Monosaccharide Diversity. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 19–30. [Google Scholar]
- Solis, D.; Bovin, N.V.; Davis, A.P.; Jimenez-Barbero, J.; Romero, A.; Roy, R.; Smetana, K., Jr.; Gabius, H.J. A guide into glycosciences: How chemistry, biochemistry and biology cooperate to crack the sugar code. Biochim. Biophys. Acta 2015, 1850, 186–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindler, B.; Barnes, L.; Renois, G.; Gray, C.; Chambert, S.; Fort, S.; Flitsch, S.; Loison, C.; Allouche, A.R.; Compagnon, I. Anomeric memory of the glycosidic bond upon fragmentation and its consequences for carbohydrate sequencing. Nat. Commun. 2017, 8, 973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madariaga, D.; Martinez-Saez, N.; Somovilla, V.J.; Garcia-Garcia, L.; Berbis, M.A.; Valero-Gonzalez, J.; Martin-Santamaria, S.; Hurtado-Guerrero, R.; Asensio, J.L.; Jimenez-Barbero, J.; et al. Serine versus threonine glycosylation with alpha-O-GalNAc: Unexpected selectivity in their molecular recognition with lectins. Chemistry 2014, 20, 12616–12627. [Google Scholar] [CrossRef] [PubMed]
- Bennett, E.P.; Mandel, U.; Clausen, H.; Gerken, T.A.; Fritz, T.A.; Tabak, L.A. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 2012, 22, 736–756. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T.A.; Jamison, O.; Perrine, C.L.; Collette, J.C.; Moinova, H.; Ravi, L.; Markowitz, S.D.; Shen, W.; Patel, H.; Tabak, L.A. Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J. Biol. Chem. 2011, 286, 14493–14507. [Google Scholar] [CrossRef] [PubMed]
- Ten Hagen, K.G.; Fritz, T.A.; Tabak, L.A. All in the family: The UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 2003, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Patsos, G.; Corfield, A. O-Glycosylation: Structural Diversity and Functions. In The Sugar Code. Fundamentals of Glycoscience; Gabius, H.-J., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2009; pp. 111–137. [Google Scholar]
- Patsos, G.; Corfield, A. Management of the Human Mucosal Defensive Barrier; Evidence for Glycan Legislation. Biol. Chem. 2009, 390, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Brockhausen, I. Glycodynamics of mucin biosynthesis in gastrointestinal tumor cells. Adv. Exp. Med. Biol. 2003, 535, 163–188. [Google Scholar] [PubMed]
- Corfield, A. Eukaryotic protein glycosylation: A primer for histochemists and cell biologists. Histochem. Cell Biol. 2017, 147, 119–147. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, M.A.; Swanson, B.J. Mucins in cancer: Protection and control of the cell surface. Nat. Rev. Cancer 2004, 4, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulos, G.; Carraway, K.L. Molecular signaling in the regulation of mucins. J. Cell. Biochem. 2007, 102, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 2013, 1833, 2430–2437. [Google Scholar] [CrossRef] [PubMed]
- Hofsteenge, J.; Muller, D.R.; de Beer, T.; Loffler, A.; Richter, W.J.; Vliegenthart, J.F. New type of linkage between a carbohydrate and a protein: C-glycosylation of a specific tryptophan residue in human RNase US. Biochemistry 1994, 33, 13524–13530. [Google Scholar] [CrossRef] [PubMed]
- Ambort, D.; van der Post, S.; Johansson, M.E.; Mackenzie, J.; Thomsson, E.; Krengel, U.; Hansson, G.C. Function of the CysD domain of the gel-forming MUC2 mucin. Biochem. J. 2011, 436, 61–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Vilar, J.; Randell, S.H.; Boucher, R.C. C-Mannosylation of MUC5AC and MUC5B cys subdomains. Glycobiology 2004, 14, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Hovenberg, H.W.; Davies, J.R.; Carlstedt, I. Different mucins are produced by the surface epithelium and the submucosa in human trachea: Identification of MUC5AC as a major mucin from the goblet cells. Biochem. J. 1996, 318, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Baldus, S.E.; Monig, S.P.; Arkenau, V.; Hanisch, F.G.; Schneider, P.M.; Thiele, J.; Holscher, A.H.; Dienes, H.P. Correlation of MUC5AC immunoreactivity with histopathological subtypes and prognosis of gastric carcinoma. Ann. Surg. Oncol. 2002, 9, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Nordman, H.; Davies, J.R.; Lindell, G.; De Bolos, C.; Real, F.; Carlstedt, I. Gastric MUC5AC and MUC6 are large oligomeric mucins that differ in size, glycosylation and tissue distribution. Biochem. J. 2002, 364 Pt 1, 191–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Kim, J.S.; Kim, K.O.; Park, K.H.; Yim, H.J.; Kim, J.Y.; Yeon, J.E.; Park, J.J.; Shim, J.J.; Byun, K.S.; et al. Expression of MUC3, MUC5AC, MUC6 and epidermal growth factor receptor in gallbladder epithelium according to gallstone composition. Korean J. Gastroenterol. 2003, 42, 330–336. [Google Scholar] [PubMed]
- Ellingham, R.B.; Berry, M.; Stevenson, D.; Corfield, A.P. Secreted human conjunctival mucus contains MUC5AC glycoforms. Glycobiology 1999, 9, 1181–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gipson, I.K. Distribution of mucins at the ocular surface. Exp. Eye Res. 2004, 78, 379–388. [Google Scholar] [CrossRef]
- Kerschner, J.E. Mucin gene expression in human middle ear epithelium. Laryngoscope 2007, 117, 1666–1676. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Tsuprun, V.; Kawano, H.; Paparella, M.M.; Zhang, Z.; Anway, R.; Ho, S.B. Characterization of mucins in human middle ear and Eustachian tube. Am. J. Physiol. Lung Cell Mol. Physiol. 2001, 280, L1157–L1167. [Google Scholar] [CrossRef] [PubMed]
- Cozzi, P.J.; Wang, J.; Delprado, W.; Perkins, A.C.; Allen, B.J.; Russell, P.J.; Li, Y. MUC1, MUC2, MUC4, MUC5AC and MUC6 expression in the progression of prostate cancer. Clin. Exp. Metastasis 2005, 22, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Gipson, I.K.; Ho, S.B.; Spurr-Michaud, S.J.; Tisdale, A.S.; Zhan, Q.; Torlakovic, E.; Pudney, J.; Anderson, D.J.; Toribara, N.W.; Hill, J.A., 3rd. Mucin genes expressed by human female reproductive tract epithelia. Biol. Reprod. 1997, 56, 999–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moncla, B.J.; Chappell, C.A.; Debo, B.M.; Meyn, L.A. The Effects of Hormones and Vaginal Microflora on the Glycome of the Female Genital Tract: Cervical-Vaginal Fluid. PLoS ONE 2016, 11, e0158687. [Google Scholar] [CrossRef] [PubMed]
- Robbe, C.; Capon, C.; Coddeville, B.; Michalski, J.C. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem. J. 2004, 384 Pt 2, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Robbe, C.; Capon, C.; Maes, E.; Rousset, M.; Zweibaum, A.; Zanetta, J.P.; Michalski, J.C. Evidence of regio-specific glycosylation in human intestinal mucins: Presence of an acidic gradient along the intestinal tract. J. Biol. Chem. 2003, 278, 46337–46348. [Google Scholar] [CrossRef] [PubMed]
- Larsson, J.M.; Karlsson, H.; Sjovall, H.; Hansson, G.C. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology 2009, 19, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Veerman, E.C.; Van Den Keijbus, P.A.; Nazmi, K.; Vos, W.; Van Der Wal, J.E.; Bloemena, E.; Bolscher, J.G.; Amerongen, A.V. Distinct localization of MUC5B glycoforms in the human salivary glands. Glycobiology 2003, 13, 363–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonckheere, N.; Skrypek, N.; Van Seuningen, I. Mucins and Pancreatic Cancer. Cancers 2010, 2, 1794–1812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argueso, P.; Gipson, I.K. Assessing mucin expression and function in human ocular surface epithelia in vivo and in vitro. Methods Mol. Biol. 2012, 842, 313–325. [Google Scholar] [PubMed]
- Thornton, D.J.; Howard, M.; Khan, N.; Sheehan, J.K. Identification of two glycoforms of the MUC5B mucin in human respiratory mucus–Evidence for a cysteine-rich sequence repeated within the molecule. J. Biol. Chem. 1997, 272, 9561–9566. [Google Scholar] [CrossRef] [PubMed]
- Tecle, E.; Gagneux, P. Sugar-coated sperm: Unraveling the functions of the mammalian sperm glycocalyx. Mol. Reprod. Dev. 2015, 82, 635–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilcoyne, M.; Gerlach, J.Q.; Gough, R.; Gallagher, M.E.; Kane, M.; Carrington, S.D.; Joshi, L. Construction of a natural mucin microarray and interrogation for biologically relevant glyco-epitopes. Anal. Chem. 2012, 84, 3330–3338. [Google Scholar] [CrossRef] [PubMed]
- Le Pendu, J. Histo-blood group antigen and human milk oligosaccharides: Genetic polymorphism and risk of infectious diseases. Adv. Exp. Med. Biol. 2004, 554, 135–143. [Google Scholar] [PubMed]
- Watkins, W.M. Biochemistry and genetics of the ABO, Lewis and P blood group systems. Adv. Hum. Genet. 1980, 10, 1–136. [Google Scholar] [PubMed]
- Stanley, P.; Cummings, R.D. Structures Common to Different Glycans. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 161–178. [Google Scholar]
- Henry, S.; Oriol, R.; Samuelsson, B. Lewis histo-blood group system and associated secretory phenotypes. Vox Sang. 1995, 69, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Donald, A.S.; Soh, C.P.; Yates, A.D.; Feeney, J.; Morgan, W.T.; Watkins, W.M. Structure, biosynthesis and genetics of the Sda antigen. Biochem. Soc. Trans. 1987, 15, 606–608. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.I. Sda blood group carbohydrate antigen, exclusively expressed in normal gastrointestinal mucosa but not in cancer tissues. Seikagaku 2008, 80, 425–429. [Google Scholar] [PubMed]
- Childs, R.A.; Kapadia, A.; Feizi, T. Expression of blood group I and I active carbohydrate sequences on cultured human and animal cell lines assessed by radioimmunoassays with monoclonal cold agglutinins. Eur. J. Immunol. 1980, 10, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Landsteiner, K. On agglutination of normal human blood. Transfusion 1961, 1, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Kabat, E.A. Philip Levine Award Lecture. Contributions of quantitative immunochemistry to knowledge of blood group A, B, H, Le, I and I antigens. Am. J. Clin. Pathol. 1982, 78, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, F.; Cid, E.; Yamamoto, M.; Saitou, N.; Bertranpetit, J.; Blancher, A. An integrative evolution theory of histo-blood group ABO and related genes. Sci. Rep. 2014, 4, 6601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooling, L. Blood Groups in Infection and Host Susceptibility. Clin. Microbiol. Rev. 2015, 28, 801–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dotz, V.; Wuhrer, M. Histo-blood group glycans in the context of personalized medicine. Biochim. Biophys. Acta 2016, 1860, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.L.; McGuckin, M.A.; Wesselingh, S.; Rogers, G.B. Infection’s Sweet Tooth: How Glycans Mediate Infection and Disease Susceptibility. Trends Microbiol. 2018, 26, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Heggelund, J.E.; Varrot, A.; Imberty, A.; Krengel, U. Histo-blood group antigens as mediators of infections. Curr. Opin. Struct. Biol. 2017, 44, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; Hurtado-Ziola, N.; Varki, A. ABO blood group glycans modulate sialic acid recognition on erythrocytes. Blood 2009, 114, 3668–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomsson, K.A.; Schulz, B.L.; Packer, N.H.; Karlsson, N.G. MUC5B Glycosylation in Human Saliva Reflects Blood Group and Secretor Status. Glycobiology 2005, 15, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Malagolini, N.; Dall’Olio, F.; Di Stefano, G.; Minni, F.; Marrano, D.; Serafini-Cessi, F. Expression of UDP-GalNAc:NeuAc alpha 2,3Gal beta-R beta 1,4(GalNAc to Gal) N-acetylgalactosaminyltransferase involved in the synthesis of Sda antigen in human large intestine and colorectal carcinomas. Cancer Res. 1989, 49, 6466–6470. [Google Scholar] [PubMed]
- Malagolini, N.; Santini, D.; Chiricolo, M.; Dall’Olio, F. Biosynthesis and expression of the Sda and sialyl Lewis x antigens in normal and cancer colon. Glycobiology 2007, 17, 688–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Culling, C.F.; Reid, P.E.; Dunn, W.L. A histochemical comparison of the O-acylated sialic acids of the epithelial mucins in ulcerative colitis, Crohn’s disease, and normal controls. J. Clin. Pathol. 1979, 32, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Reid, P.E.; Culling, C.F.A.; Dunn, W.L.; Ramey, C.W.; Clay, M.G. Clinical and histochemical studies of normal and diseased human gastrointestinal tract. I. A comparison between histologically normal colon, colon tumors, ulcerative colitis and diverticular disease. Histochem. J. 1984, 16, 235–251. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Myerscough, N.; Warren, B.F.; Durdey, P.; Paraskeva, C.; Schauer, R. Reduction of sialic acid O-acetylation in human colonic mucins in the adenoma-carcinoma sequence. Glycoconj. J. 1999, 16, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F. Host responses to the human microbiome. Nutr. Rev. 2012, 70 (Suppl. 1), S14–S17. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J. The impact of nutrition on the human microbiome. Nutr. Rev. 2012, 70 (Suppl. 1), S10–S13. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Sonnenburg, J.L.; Manchester, J.K.; Hansen, E.E.; Chiang, H.C.; Gordon, J.I. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc. Natl. Acad. Sci. USA 2006, 103, 8834–8839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravcheev, D.A.; Thiele, I. Comparative Genomic Analysis of the Human Gut Microbiome Reveals a Broad Distribution of Metabolic Pathways for the Degradation of Host-Synthetized Mucin Glycans and Utilization of Mucin-Derived Monosaccharides. Front. Genet. 2017, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Colomb, F.; Robbe-Masselot, C.; Groux-Degroote, S.; Bouckaert, J.; Delannoy, P.; Michalski, J.-C. Epithelial mucins and bacterial adhesion. Carbohydr. Chem. 2014, 40, 596–623. [Google Scholar]
- Argueso, P.; Guzman-Aranguez, A.; Mantelli, F.; Cao, Z.; Ricciuto, J.; Panjwani, N. Association of cell surface mucins with galectin-3 contributes to the ocular surface epithelial barrier. J. Biol. Chem. 2009, 284, 23037–23045. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.; Ellingham, R.B.; Corfield, A.P. Membrane-associated mucins in normal human conjunctiva. Investig. Ophthalmol. Vis. Sci. 2000, 41, 398–403. [Google Scholar]
- Jonckheere, N.; Van Seuningen, I. The membrane-bound mucins: How large O-glycoproteins play key roles in epithelial cancers and hold promise as biological tools for gene-based and immunotherapies. Crit. Rev. Oncog. 2008, 14, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, B.; Gipson, I.K. Membrane-tethered mucins have multiple functions on the ocular surface. Exp. Eye Res. 2010, 90, 655–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki-Kinoshita, K.F. Using databases and web resources for glycomics research. Mol. Cell. Proteom. 2013, 12, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Imperiali, B. The chemistry-glycobiology frontier. J. Am. Chem. Soc. 2012, 134, 17835–17839. [Google Scholar] [CrossRef] [PubMed]
- Cummings, R.D.; Schnaar, R.L.; Esko, J.D.; Drickamer, K.; Taylor, M.E. Principles of Glycan Recognition. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 373–385. [Google Scholar]
- Dakour, J.; Hansson, G.C.; Lundblad, A.; Zopf, D. Separation and partial sequence analysis of blood group A-active oligosaccharides by affinity chromatography using monoclonal antibodies. Arch. Biochem. Biophys. 1986, 248, 677–683. [Google Scholar] [CrossRef]
- Harada, H.; Kondo, M.; Yanagisawa, M.; Sunada, S. Mucin-specific bark lectin from elderberry Sambucus sieboldiana and its applications to the affinity chromatography of mucin. Anal. Biochem. 1990, 189, 262–266. [Google Scholar] [CrossRef]
- Sato, C.; Yamakawa, N.; Kitajima, K. Measurement of glycan-based interactions by frontal affinity chromatography and surface plasmon resonance. Methods Enzymol. 2010, 478, 219–232. [Google Scholar] [PubMed]
- Sakka, K.; Nakanishi, M.; Sogabe, M.; Arai, T.; Ohara, H.; Tanaka, A.; Kimura, T.; Ohmiya, K. Isothermal titration calorimetric studies on the binding of a family 6 carbohydrate-binding module of Clostridium thermocellum xynA with xlylooligosaccharides. Biosci. Biotechnol. Biochem. 2003, 67, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Valerio-Lepiniec, M.; Aumont-Nicaise, M.; Roux, C.; Raynal, B.; England, P.; Badet, B.; Badet-Denisot, M.A.; Desmadril, M. Analysis of the Escherichia coli glucosamine-6-phosphate synthase activity by isothermal titration calorimetry and differential scanning calorimetry. Arch. Biochem. Biophys. 2010, 498, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Bellapadrona, G.A.; Tesler, B.; Grunstein, D.; Hossain, L.H.; Kikkeri, R.; Seeberger, P.H.; Vaskevich, A.; Rubinstein, I. Optimization of localized surface plasmon resonance transducers for studying carbohydrate-protein interactions. Anal. Chem. 2012, 84, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Houngkamhang, N.; Vongsakulyanon, A.; Peungthum, P.; Sudprasert, K.; Kitpoka, P.; Kunakorn, M.; Sutapun, B.; Amarit, R.; Somboonkaew, A.; Srikhirin, T. ABO blood-typing using an antibody array technique based on surface plasmon resonance imaging. Sensors (Basel) 2013, 13, 11913–11922. [Google Scholar] [CrossRef] [PubMed]
- Dell, A.; Morris, H.R. Glycoprotein structure determination by mass spectrometry. Science 2001, 291, 2351–2356. [Google Scholar] [CrossRef] [PubMed]
- Robbe, C.; Michalski, J.C.; Capon, C. Structural determination of O-glycans by tandem mass spectrometry. Methods Mol. Biol. 2006, 347, 109–123. [Google Scholar] [PubMed]
- Cooper, C.A.; Gasteiger, E.; Packer, N.H. GlycoMod–a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 2001, 1, 340–349. [Google Scholar] [CrossRef]
- Kronewitter, S.R.; De Leoz, M.L.; Strum, J.S.; An, H.J.; Dimapasoc, L.M.; Guerrero, A.; Miyamoto, S.; Lebrilla, C.B.; Leiserowitz, G.S. The glycolyzer: Automated glycan annotation software for high performance mass spectrometry and its application to ovarian cancer glycan biomarker discovery. Proteomics 2012, 12, 2523–2538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bones, J.; Byrne, J.C.; O’Donoghue, N.; McManus, C.; Scaife, C.; Boissin, H.; Nastase, A.; Rudd, P.M. Glycomic and glycoproteomic analysis of serum from patients with stomach cancer reveals potential markers arising from host defense response mechanisms. J. Proteome Res. 2011, 10, 1246–1265. [Google Scholar] [CrossRef] [PubMed]
- Bones, J.; Mittermayr, S.; O’Donoghue, N.; Guttman, A.; Rudd, P.M. Ultra performance liquid chromatographic profiling of serum N-glycans for fast and efficient identification of cancer associated alterations in glycosylation. Anal. Chem. 2010, 82, 10208–10215. [Google Scholar] [CrossRef] [PubMed]
- Andre, S.; Kaltner, H.; Manning, J.C.; Murphy, P.V.; Gabius, H.J. Lectins: Getting familiar with translators of the sugar code. Molecules 2015, 20, 1788–1823. [Google Scholar] [CrossRef] [PubMed]
- Manning, J.C.; Romero, A.; Habermann, F.A.; Garcia Caballero, G.; Kaltner, H.; Gabius, H.J. Lectins: A primer for histochemists and cell biologists. Histochem. Cell Biol. 2017, 147, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Lis, H.; Sharon, N. Lectins: Carbohydrate-Specific Proteins That Mediate Cellular Recognition. Chem. Rev. 1998, 98, 637–674. [Google Scholar] [CrossRef] [PubMed]
- De Bentzmann, S.; Varrot, A.; Imberty, A. Monitoring lectin interactions with carbohydrates. Methods Mol. Biol. 2014, 1149, 403–414. [Google Scholar] [PubMed]
- Varki, A. Selectins and other mammalian sialic acid-binding lectins. Curr. Opin. Cell Biol. 1992, 4, 257–266. [Google Scholar] [CrossRef]
- Crocker, P.R.; Varki, A. Siglecs, sialic acids and innate immunity. Trends Immunol. 2001, 22, 337–342. [Google Scholar] [CrossRef]
- Juge, N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012, 20, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Ringot-Destrez, B.; Kalach, N.; Mihalache, A.; Gosset, P.; Michalski, J.C.; Leonard, R.; Robbe-Masselot, C. How do they stick together? Bacterial adhesins implicated in the binding of bacteria to the human gastrointestinal mucins. Biochem. Soc. Trans. 2017, 45, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Stanley, P.; Hart, G.W.; Aebi, M.; Darvill, A.G.; Kinoshita, T.; Packer, N.H.; Prestegard, J.H.; et al. Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; p. 823. [Google Scholar]
- Cummings, R.D.; Schnaar, R.L. R-Type Lectins. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 401–412. [Google Scholar]
- Cummings, R.D.; Etzler, M.E.; Surolia, A. L-Type Lectins. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 413–422. [Google Scholar]
- Varki, A.; Kornfeld, S. P-Type Lectins. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 423–433. [Google Scholar]
- Crocker, P.R. Siglecs in innate immunity. Curr. Opin. Pharmacol. 2005, 5, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Crocker, P.R.; Redelinghuys, P. Siglecs as positive and negative regulators of the immune system. Biochem. Soc. Trans. 2008, 36 Pt 6, 1467–1471. [Google Scholar] [CrossRef] [PubMed]
- Cummings, R.D.; Liu, F.-T.; Vasta, G.R. Galectins. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 469–480. [Google Scholar]
- Kaltner, H.; Toegel, S.; Caballero, G.G.; Manning, J.C.; Ledeen, R.W.; Gabius, H.J. Galectins: Their network and roles in immunity/tumor growth control. Histochem. Cell Biol. 2017, 147, 239–256. [Google Scholar] [CrossRef] [PubMed]
- Hellebo, A.; Vilas, U.; Falk, K.; Vlasak, R. Infectious salmon anemia virus specifically binds to and hydrolyzes 4-O-acetylated sialic acids. J. Virol. 2004, 78, 3055–3062. [Google Scholar] [CrossRef] [PubMed]
- Strasser, P.; Unger, U.; Strobl, B.; Vilas, U.; Vlasak, R. Recombinant viral sialate-O-acetylesterases. Glycoconj. J. 2004, 20, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Vlasak, R.; Luytjes, W.; Spaan, W.; Palese, P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. USA 1988, 85, 4526–4529. [Google Scholar] [CrossRef] [PubMed]
- Vlasak, R.; Krystal, M.; Nacht, M.; Palese, P. The influenza C virus glycoprotein (HE) exhibits receptor-binding (hemagglutinin) and receptor-destroying (esterase) activities. Virology 1987, 160, 419–425. [Google Scholar] [CrossRef]
- Langereis, M.A.; Bakkers, M.J.; Deng, L.; Padler-Karavani, V.; Vervoort, S.J.; Hulswit, R.J.; van Vliet, A.L.; Gerwig, G.J.; de Poot, S.A.; Boot, W.; et al. Complexity and Diversity of the Mammalian Sialome Revealed by Nidovirus Virolectins. Cell Rep. 2015, 11, 1966–1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackenzie, D.A.; Jeffers, F.; Parker, M.L.; Vibert-Vallet, A.; Bongaerts, R.J.; Roos, S.; Walter, J.; Juge, N. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology 2010, 156 Pt 11, 3368–3378. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382 Pt 3, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Ficko-Blean, E.; Boraston, A.B. Insights into the recognition of the human glycome by microbial carbohydrate-binding modules. Curr. Opin. Struct. Biol. 2012, 22, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.C.; Phan, N.N.; Chen, Y.; Reilly, P.J. Carbohydrate-binding module tribes. Biopolymers 2015, 103, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Blixt, O.; Head, S.; Mondala, T.; Scanlan, C.; Huflejt, M.E.; Alvarez, R.; Bryan, M.C.; Fazio, F.; Calarese, D.; Stevens, J.; et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17033–17038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briard, J.G.; Jiang, H.; Moremen, K.W.; Macauley, M.S.; Wu, P. Cell-based glycan arrays for probing glycan-glycan binding protein interactions. Nat. Commun. 2018, 9, 880. [Google Scholar] [CrossRef] [PubMed]
- Campanero-Rhodes, M.A.; Childs, R.A.; Kiso, M.; Komba, S.; Le Narvor, C.; Warren, J.; Otto, D.; Crocker, P.R.; Feizi, T. Carbohydrate microarrays reveal sulphation as a modulator of siglec binding. Biochem. Biophys. Res. Commun. 2006, 344, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- de Boer, A.R.; Hokke, C.H.; Deelder, A.M.; Wuhrer, M. General microarray technique for immobilization and screening of natural glycans. Anal. Chem. 2007, 79, 8107–8113. [Google Scholar] [CrossRef] [PubMed]
- de Paz, J.L.; Seeberger, P.H. Recent advances and future challenges in glycan microarray technology. Methods Mol. Biol. 2012, 808, 1–12. [Google Scholar] [PubMed]
- Dyukova, V.I.; Shilova, N.V.; Galanina, O.E.; Rubina, A.Y.; Bovin, N.V. Design of carbohydrate multiarrays. Biochim. Biophys. Acta 2006, 1760, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Hoang, A.; Laigre, E.; Goyard, D.; Defrancq, E.; Vinet, F.; Dumy, P.; Renaudet, O. An oxime-based glycocluster microarray. Org. Biomol. Chem. 2017, 15, 5135–5139. [Google Scholar] [CrossRef] [PubMed]
- Padler-Karavani, V.; Song, X.; Yu, H.; Hurtado-Ziola, N.; Huang, S.; Muthana, S.; Chokhawala, H.A.; Cheng, J.; Verhagen, A.; Langereis, M.A.; et al. Cross-comparison of protein recognition of sialic acid diversity on two novel sialoglycan microarrays. J. Biol. Chem. 2012, 287, 22593–22608. [Google Scholar] [CrossRef] [PubMed]
- Rillahan, C.D.; Paulson, J.C. Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 2011, 80, 797–823. [Google Scholar] [CrossRef] [PubMed]
- Comelli, E.M.; Head, S.R.; Gilmartin, T.; Whisenant, T.; Haslam, S.M.; North, S.J.; Wong, N.K.; Kudo, T.; Narimatsu, H.; Esko, J.D.; et al. A focused microarray approach to functional glycomics: Transcriptional regulation of the glycome. Glycobiology 2006, 16, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Disney, M.D.; Seeberger, P.H. The use of carbohydrate microarrays to study carbohydrate-cell interactions and to detect pathogens. Chem. Biol. 2004, 11, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Geissner, A.; Seeberger, P.H. Glycan Arrays: From Basic Biochemical Research to Bioanalytical and Biomedical Applications. Annu. Rev. Anal. Chem. (Palo Alto CA) 2016, 9, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Godula, K.; Rabuka, D.; Nam, K.T.; Bertozzi, C.R. Synthesis and microcontact printing of dual end-functionalized mucin-like glycopolymers for microarray applications. Angew. Chem. Int. Ed. Engl. 2009, 48, 4973–4976. [Google Scholar] [CrossRef] [PubMed]
- Stowell, S.R.; Arthur, C.M.; McBride, R.; Berger, O.; Razi, N.; Heimburg-Molinaro, J.; Rodrigues, L.C.; Gourdine, J.P.; Noll, A.J.; von Gunten, S.; et al. Microbial glycan microarrays define key features of host-microbial interactions. Nat. Chem. Biol. 2014, 10, 470–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flannery, A.; Gerlach, J.Q.; Joshi, L.; Kilcoyne, M. Assessing Bacterial Interactions Using Carbohydrate-Based Microarrays. Microarrays (Basel) 2015, 4, 690–713. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, S.; Trummer, B.J.; Deng, C.; Wang, A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat. Biotechnol. 2002, 20, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Feizi, T.; Fazio, F.; Chai, W.; Wong, C.H. Carbohydrate microarrays—A new set of technologies at the frontiers of glycomics. Curr. Opin. Struct. Biol. 2003, 13, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Brun, M.A.; Disney, M.D.; Seeberger, P.H. Miniaturization of microwave-assisted carbohydrate functionalization to create oligosaccharide microarrays. Chembiochem 2006, 7, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Lonardi, E.; Deelder, A.M.; Wuhrer, M.; Balog, C.I. Microarray technology using glycans extracted from natural sources for serum antibody fluorescent detection. Methods Mol. Biol. 2012, 808, 285–302. [Google Scholar] [PubMed]
- Serna, S.; Etxebarria, J.; Ruiz, N.; Martin-Lomas, M.; Reichardt, N.C. Construction of N-glycan microarrays by using modular synthesis and on-chip nanoscale enzymatic glycosylation. Chemistry 2010, 16, 13163–13175. [Google Scholar] [CrossRef] [PubMed]
- Tseng, S.Y.; Wang, C.C.; Lin, C.W.; Chen, C.L.; Yu, W.Y.; Chen, C.H.; Wu, C.Y.; Wong, C.H. Glycan arrays on aluminum-coated glass slides. Chem. Asian J. 2008, 3, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Zhi, Z.L.; Laurent, N.; Powell, A.K.; Karamanska, R.; Fais, M.; Voglmeir, J.; Wright, A.; Blackburn, J.M.; Crocker, P.R.; Russell, D.A.; et al. A versatile gold surface approach for fabrication and interrogation of glycoarrays. Chembiochem 2008, 9, 1568–1575. [Google Scholar] [CrossRef] [PubMed]
- Laurent, N.; Voglmeir, J.; Flitsch, S.L. Glycoarrays–tools for determining protein-carbohydrate interactions and glycoenzyme specificity. Chem. Commun. (Camb.) 2008, 4400–4412. [Google Scholar] [CrossRef] [PubMed]
- Laurent, N.; Voglmeir, J.; Wright, A.; Blackburn, J.; Pham, N.T.; Wong, S.C.; Gaskell, S.J.; Flitsch, S.L. Enzymatic glycosylation of peptide arrays on gold surfaces. Chembiochem 2008, 9, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Reis, C.A.; Campos, D.; Osorio, H.; Santos, L.L. Glycopeptide microarray for autoantibody detection in cancer. Expert Rev. Proteom. 2011, 8, 435–437. [Google Scholar] [CrossRef] [PubMed]
- Brewer, C.F.; Miceli, M.C.; Baum, L.G. Clusters, bundles, arrays and lattices: Novel mechanisms for lectin-saccharide-mediated cellular interactions. Curr. Opin. Struct. Biol. 2002, 12, 616–623. [Google Scholar] [CrossRef]
- Godula, K.; Bertozzi, C.R. Density variant glycan microarray for evaluating cross-linking of mucin-like glycoconjugates by lectins. J. Am. Chem. Soc. 2012, 134, 15732–15742. [Google Scholar] [CrossRef] [PubMed]
- Clyne, M.; Duggan, G.; Naughton, J.; Bourke, B. Methods to Assess the Direct Interaction of C. jejuni with Mucins. Methods Mol. Biol. 2017, 1512, 107–115. [Google Scholar] [PubMed]
- Naughton, J.A.; Marino, K.; Dolan, B.; Reid, C.; Gough, R.; Gallagher, M.E.; Kilcoyne, M.; Gerlach, J.Q.; Joshi, L.; Rudd, P.; et al. Divergent mechanisms of interaction of Helicobacter pylori and Campylobacter jejuni with mucus and mucins. Infect. Immun. 2013, 81, 2838–2850. [Google Scholar] [CrossRef] [PubMed]
- Deacon, M.P.; McGurk, S.; Roberts, C.J.; Williams, P.M.; Tendler, S.J.; Davies, M.C.; Davis, S.S.; Harding, S.E. Atomic force microscopy of gastric mucin and chitosan mucoadhesive systems. Biochem. J. 2000, 348 Pt 3, 557–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, Z.; Chasan, B.; Bansil, R.; Turner, B.S.; Bhaskar, K.R.; Afdhal, N.H. Atomic force microscopy reveals aggregation of gastric mucin at low pH. Biomacromolecules 2005, 6, 3458–3466. [Google Scholar] [CrossRef] [PubMed]
- McMaster, T.J.; Berry, M.; Corfield, A.P.; Miles, M.J. Atomic force microscopy of the submolecular architecture of hydrated ocular mucins. Biophys. J. 1999, 77, 533–541. [Google Scholar] [CrossRef]
- Parreira, P.; Shi, Q.; Magalhaes, A.; Reis, C.A.; Bugaytsova, J.; Boren, T.; Leckband, D.; Martins, M.C. Atomic force microscopy measurements reveal multiple bonds between Helicobacter pylori blood group antigen binding adhesin and Lewis b ligand. J. R. Soc. Interface R. Soc. 2014, 11, 20141040. [Google Scholar] [CrossRef] [PubMed]
- Round, A.N.; Berry, M.; McMaster, T.J.; Corfield, A.P.; Miles, M.J. Glycopolymer charge density determines conformation in human ocular mucin gene products: An atomic force microscope study. J. Struct. Biol. 2004, 145, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Round, A.N.; Berry, M.; McMaster, T.J.; Stoll, S.; Gowers, D.; Corfield, A.P.; Miles, M.J. Heterogeneity and persistence length in human ocular mucins. Biophys. J. 2002, 83, 1661–1670. [Google Scholar] [CrossRef] [Green Version]
- Round, A.N.; Rigby, N.M.; Garcia de la Torre, A.; Macierzanka, A.; Mills, E.N.; Mackie, A.R. Lamellar structures of MUC2-rich mucin: A potential role in governing the barrier and lubricating functions of intestinal mucus. Biomacromolecules 2012, 13, 3253–3261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sicard, D.; Chevolot, Y.; Souteyrand, E.; Imberty, A.; Vidal, S.; Phaner-Goutorbe, M. Molecular arrangement between multivalent glycocluster and Pseudomonas aeruginosa LecA (PA-IL) by atomic force microscopy: Influence of the glycocluster concentration. J. Mol. Recognit. 2013, 26, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.; Karlsson, N.G.; Khoo, K.-H.; Packer, N.H. Glycomics and Glycoproteomics. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 653–666. [Google Scholar]
- Fellmann, C.; Gowen, B.G.; Lin, P.C.; Doudna, J.A.; Corn, J.E. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 2017, 16, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Rouet, R.; Thuma, B.A.; Roy, M.D.; Lintner, N.G.; Rubitski, D.M.; Finley, J.E.; Wisniewska, H.M.; Mendonsa, R.; Hirsh, A.; de Onate, L.; et al. Receptor-Mediated Delivery of CRISPR-Cas9 Endonuclease for Cell-Type-Specific Gene Editing. J. Am. Chem. Soc. 2018, 140, 6596–6603. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Nowicki, M.; Fisher, J.P.; Zhang, L.G. 3D Bioprinting for Organ Regeneration. Adv. Healthc. Mater. 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Gungor-Ozkerim, P.S.; Inci, I.; Zhang, Y.S.; Khademhosseini, A.; Dokmeci, M.R. Bioinks for 3D bioprinting: An overview. Biomater. Sci. 2018, 6, 915–946. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yang, F.; Zhao, H.; Gao, Q.; Xia, B.; Fu, J. Research on the printability of hydrogels in 3D bioprinting. Sci. Rep. 2016, 6, 29977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, N.; Yang, G.H.; Lee, J.; Kim, G. 3D bioprinting and its in vivo applications. J. Biomed. Mater. Res. B Appl. Biomater. 2018, 106, 444–459. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.S.; Yesantharao, P.; Huang, C.Y.; Mattson, G.; Boktor, J.; Fukunishi, T.; Zhang, H.; Hibino, N. 3D bioprinting using stem cells. Pediatr. Res. 2018, 83, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Datta, P.; Ayan, B.; Ozbolat, V.; Sosnoski, D.; Ozbolat, I.T. 3D bioprinting for drug discovery and development in pharmaceutics. Acta Biomater. 2017, 57, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Wlodarczyk-Biegun, M.K.; Del Campo, A. 3D bioprinting of structural proteins. Biomaterials 2017, 134, 180–201. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.P.; Aoki-Kinoshita, K.F.; Lisacek, F.; York, W.S.; Packer, N.H. Glycoinformatics. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 667–679. [Google Scholar]
- Adibekian, A.; Stallforth, P.; Hecht, M.L.; Werz, D.B.; Gagneux, P.; Seeberger, P.H. Comparative bioinformatics analysis of the mammalian and bacterial glycomes. Chem. Sci. 2011, 2, 337–344. [Google Scholar] [CrossRef]
- Ceroni, A.; Dell, A.; Haslam, S.M. The GlycanBuilder: A fast, intuitive and flexible software tool for building and displaying glycan structures. Source Code Biol. Med. 2007, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Brockhausen, I. O-linked chain glycosyltransferases. Methods Mol. Biol. 2000, 125, 273–293. [Google Scholar] [PubMed]
- Brockhausen, I.; Stanley, P. O-GalNAc Glycans. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 113–123. [Google Scholar]
- Freeze, H.H.; Hart, G.W.; Schnaar, R.L. Glycosylation Precursors. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 51–63. [Google Scholar]
- Hinderlich, S.; Stasche, R.; Zeitler, R.; Reutter, W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J. Biol. Chem. 1997, 272, 24313–24318. [Google Scholar] [CrossRef] [PubMed]
- Keppler, O.T.; Hinderlich, S.; Langner, J.; Schwartz-Albiez, R.; Reutter, W.; Pawlita, M. UDP-GlcNAc 2-epimerase: A regulator of cell surface sialic acid. Science 1999, 284, 1372–1376. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.K.; Hinderlich, S.; Reutter, W.; Tanner, M.E. Sialic acid biosynthesis: Stereochemistry and mechanism of the reaction catalyzed by the mammalian UDP-N-acetylglucosamine 2-epimerase. J. Am. Chem. Soc. 2003, 125, 2455–2461. [Google Scholar] [CrossRef] [PubMed]
- Hinderlich, S.; Nohring, S.; Weise, C.; Franke, P.; Stasche, R.; Reutter, W. Purification and characterization of N-acetylglucosamine kinase from rat live–comparison with UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Eur. J. Biochem. 1998, 252, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Vimr, E.R. Unified theory of bacterial sialometabolism: How and why bacteria metabolize host sialic acids. ISRN Microbiol. 2013, 2013, 816713. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R. Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struct. Biol. 2009, 19, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R.; Sommer, U.; Kruger, D.; van Unen, H.; Traving, C. The terminal enzymes of sialic acid metabolism: Acylneuraminate pyruvate-lyases. Biosci. Rep. 1999, 19, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T.A.; Revoredo, L.; Thome, J.J.; Tabak, L.A.; Vester-Christensen, M.B.; Clausen, H.; Gahlay, G.K.; Jarvis, D.L.; Johnson, R.W.; Moniz, H.A.; et al. The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J. Biol. Chem. 2013, 288, 19900–19914. [Google Scholar] [CrossRef] [PubMed]
- Revoredo, L.; Wang, S.; Bennett, E.P.; Clausen, H.; Moremen, K.W.; Jarvis, D.L.; Ten Hagen, K.G.; Tabak, L.A.; Gerken, T.A. Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. Glycobiology 2016, 26, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Carrington, S.D.; Irwin, J.A.; Liu, L.; Rudd, P.M.; Matthews, E.; Corfield, A.P. Analysing mucin degradation. Methods Mol. Biol. 2012, 842, 191–215. [Google Scholar] [PubMed]
- Crost, E.H.; Tailford, L.E.; Monestier, M.; Swarbreck, D.; Henrissat, B.; Crossman, L.C.; Juge, N. The mucin-degradation strategy of Ruminococcus gnavus: The importance of intramolecular trans-sialidases. Gut Microbes 2016, 7, 302–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef] [PubMed]
- Rho, J.H.; Wright, D.P.; Christie, D.L.; Clinch, K.; Furneaux, R.H.; Roberton, A.M. A novel mechanism for desulfation of mucin: Identification and cloning of a mucin-desulfating glycosidase (sulfoglycosidase) from Prevotella strain RS2. J. Bacteriol. 2005, 187, 1543–1551. [Google Scholar] [CrossRef] [PubMed]
- Roberton, A.M.; McKenzie, C.G.; Sharfe, N.; Stubbs, L.B. A glycosulphatase that removes sulphate from mucus glycoprotein. Biochem. J. 1993, 293 Pt 3, 683–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberton, A.M.; Wiggins, R.; Horner, P.J.; Greenwood, R.; Crowley, T.; Fernandes, A.; Berry, M.; Corfield, A.P. A novel bacterial mucinase, glycosulfatase, is associated with bacterial vaginosis. J. Clin. Microbiol. 2005, 43, 5504–5508. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, R.K.; Roberton, A.M. A novel glycosulphatase isolated from a mucus glycopeptide-degrading Bacteroides sp. FEMS Microbiol. Lett. 1988, 50, 195–199. [Google Scholar] [CrossRef]
- Jansen, H.J.; Hart, C.A.; Rhodes, J.M.; Saunders, J.R.; Smalley, J.W. A novel mucin-sulphatase activity found in Burkholderia cepacia and Pseudomonas aeruginosa. J. Med. Microbiol. 1999, 48, 551–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smalley, J.W.; Dwarakanath, D.; Rhodes, J.M.; Hart, C.A. Mucin-sulphatase activity of some oral streptococci. Caries Res. 1994, 28, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.H.; Dwarakanath, A.D.; Hart, C.A.; Milton, J.D.; Rhodes, J.M. Increased faecal mucin sulphatase activity in ulcerative colitis: A potential target for treatment. Gut 1995, 36, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.H.; Sunderland, D.; Gibson, G.R.; Hart, C.A.; Rhodes, J.M. A novel mucin sulphatase from human faeces: Its identification, purification and characterization. Clin. Sci. 1992, 82, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.; Jiang, H.; O’Connell Motherway, M.; Oscarson, S.; van Sinderen, D. Glycosulfatase-Encoding Gene Cluster in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 2016, 82, 6611–6623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Ravcheev, D.A.; Hu, D.; Zhang, F.; Gong, X.; Hao, L.; Cao, M.; Rodionov, D.A.; Wang, C.; Feng, Y. Two novel regulators of N-acetyl-galactosamine utilization pathway and distinct roles in bacterial infections. Microbiologyopen 2015, 4, 983–1000. [Google Scholar] [CrossRef] [PubMed]
- Muilerman, H.G.; Lasthuis, A.M.; Hooghwinkel, G.J.; Van Dijk, W. On the presence of two non-specific nucleotide-sugar-hydrolysing enzymes in rat liver. Biochem. J. 1984, 220, 95–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varki, A.; Kornfeld, S. Purification and characterization of rat liver alpha-N-acetylglucosaminyl phosphodiesterase. J. Biol. Chem. 1981, 256, 9937–9943. [Google Scholar] [PubMed]
- Kean, E.L.; Bighouse, K.J. Cytidine 5′-monophosphosialic acid hydrolase. Subcellular location and properties. J. Biol. Chem. 1974, 249, 7813–7823. [Google Scholar] [PubMed]
- van Dijk, W.; Maier, H.; van den Eijnden, D.H. Properties and subcellular localization of CMP-N-acetylneuraminic acid hydrolase of calf kidney. Biochim. Biophys. Acta 1976, 444, 816–834. [Google Scholar] [CrossRef]
- Van Dijk, W.; Maier, H.; Van den Eijnden, D.H. CMP-N-acetylneuraminic acid hydrolase, an ectoenzyme distributed unevenly over the hepatocyte surface. Biochim. Biophys. Acta 1977, 466, 187–197. [Google Scholar] [CrossRef]
- Ladeveze, S.; Tarquis, L.; Cecchini, D.A.; Bercovici, J.; Andre, I.; Topham, C.M.; Morel, S.; Laville, E.; Monsan, P.; Lombard, V.; et al. Role of glycoside phosphorylases in mannose foraging by human gut bacteria. J. Biol. Chem. 2013, 288, 32370–32383. [Google Scholar] [CrossRef] [PubMed]
- Lewis, W.G.; Robinson, L.S.; Gilbert, N.M.; Perry, J.C.; Lewis, A.L. Degradation, foraging, and depletion of mucus sialoglycans by the vagina-adapted Actinobacterium Gardnerella vaginalis. J. Biol. Chem. 2013, 288, 12067–12079. [Google Scholar] [CrossRef] [PubMed]
- Martens, E.C.; Chiang, H.C.; Gordon, J.I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell. Host Microbe 2008, 4, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Robinson, L.S.; Lewis, W.G.; Lewis, A.L. The sialate O-acetylesterase EstA from gut Bacteroidetes species enables sialidase-mediated cross-species foraging of 9-O-acetylated sialoglycans. J. Biol. Chem. 2017, 292, 11861–11872. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, J.L.; Xu, J.; Leip, D.D.; Chen, C.H.; Westover, B.P.; Weatherford, J.; Buhler, J.D.; Gordon, J.I. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 2005, 307, 1955–1959. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, J.L.; Angenent, L.T.; Gordon, J.I. Getting a grip on things: How do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 2004, 5, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.I.; Hooper, L.V.; McNevin, M.S.; Wong, M.; Bry, L. Epithelial cell growth and differentiation. III. Promoting diversity in the intestine: Conversations between the microflora, epithelium, and diffuse GALT. Am. J. Physiol. 1997, 273 Pt 1, G565–G570. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, B.P.; Klopfleisch, R.; Loh, G.; Blaut, M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS ONE 2013, 8, E74963. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353. [Google Scholar] [CrossRef] [PubMed]
- Arike, L.; Holmen-Larsson, J.; Hansson, G.C. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology 2017, 27, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Mackenzie, D.A.; Peterson, D.A.; Schmaltz, R.; Fangman, T.; Zhou, Y.; Zhang, C.; Benson, A.K.; Cody, L.A.; Mulholland, F.; et al. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet. 2013, 9, E1004057. [Google Scholar] [CrossRef] [PubMed]
- Macauley, M.S.; Kawasaki, N.; Peng, W.; Wang, S.H.; He, Y.; Arlian, B.M.; McBride, R.; Kannagi, R.; Khoo, K.H.; Paulson, J.C. Unmasking of CD22 Co-receptor on Germinal Center B-cells Occurs by Alternative Mechanisms in Mouse and Man. J. Biol. Chem. 2015, 290, 30066–30077. [Google Scholar] [CrossRef] [PubMed]
- Beura, L.K.; Hamilton, S.E.; Bi, K.; Schenkel, J.M.; Odumade, O.A.; Casey, K.A.; Thompson, E.A.; Fraser, K.A.; Rosato, P.C.; Filali-Mouhim, A.; et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 2016, 532, 512–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mege, D.; Bege, T.; Beyer-Berjot, L.; Loundou, A.; Grimaud, J.C.; Brunet, C.; Berdah, S. Does faecal diversion prevent morbidity after ileocecal resection for Crohn’s disease? Retrospective series of 80 cases. ANZ J. Surg. 2017, 87, E74–E79. [Google Scholar] [CrossRef] [PubMed]
- Nurkin, S.; Kakarla, V.R.; Ruiz, D.E.; Cance, W.G.; Tiszenkel, H.I. The role of faecal diversion in low rectal cancer: A review of 1791 patients having rectal resection with anastomosis for cancer, with and without a proximal stoma. Colorectal Dis. 2013, 15, e309–e316. [Google Scholar] [CrossRef] [PubMed]
- Mennigen, R.; Heptner, B.; Senninger, N.; Rijcken, E. Temporary fecal diversion in the management of colorectal and perianal Crohn’s disease. Gastroenterol. Res. Pract. 2015, 2015, 286315. [Google Scholar] [CrossRef] [PubMed]
- Longman, R.J.; Poulsom, R.; Corfield, A.P.; Warren, B.F.; Wright, N.A.; Thomas, M.G. Alterations in the Composition of the Supramucosal Defense Barrier in Relation to Disease Severity of Ulcerative Colitis. J. Histochem. Cytochem. 2006, 54, 1335–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swallow, D.M.; Griffiths, B.; Bramwell, M.; Wiseman, G.; Burchell, J. Detection of the urinary ‘PUM’ polymorphism by the tumour-binding monoclonal antibodies Ca1, Ca2, Ca3, HMFG1, and HMFG2. Dis. Mark. 1986, 4, 247–254. [Google Scholar]
- Hovenberg, H.W.; Davies, J.R.; Herrmann, A.; Linden, C.-J.; Carlstedt, I. MUC5AC, but not MUC2, is a prominent mucin in respiratory secretions. Glycoconj. J. 1996, 13, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Xing, P.X.; Prenzoska, J.; Apostolopoulos, V.; Karkaloutsos, J.; McKenzie, I.F.C. Monoclonal antibodies to a MUC4 peptide react with lung cancer. Int. J. Oncol. 1997, 11, 289–295. [Google Scholar] [PubMed]
- Ho, J.J.; Crawley, S.; Pan, P.L.; Farrelly, E.R.; Kim, Y.S. Secretion of MUC5AC mucin from pancreatic cancer cells in response to forskolin and VIP. Biochem. Biophys. Res. Commun. 2002, 294, 680–686. [Google Scholar] [CrossRef]
- Wickström, C.; Christersson, C.; Davies, J.R.; Carlstedt, I. Macromolecular organization of saliva: Identification of ‘insoluble’ MUC5B assemblies and non-mucin proteins in the gel phase [In Process Citation]. Biochem. J. 2000, 351, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.J.; McGuckin, M.A.; Gotley, D.C.; Eyre, H.J.; Sutherland, G.R.; Antalis, T.M. Two novel mucin genes down-regulated in colorectal cancer identified by differential display. Cancer Res. 1999, 59, 4083–4089. [Google Scholar] [PubMed]
- Walsh, M.D.; Young, J.P.; Leggett, B.A.; Williams, S.H.; Jass, J.R.; McGuckin, M.A. The MUC13 cell surface mucin is highly expressed by human colorectal carcinomas. Hum. Pathol. 2007, 38, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.K.; Metoki, R.; Hakomori, S.-I. Immunoglobulin G3 monoclonal antibody directed to Tn antigen (Tumor-associated a-N-acetylgalactosaminyl epitope) that does not cross-react with blood group A antigen. Cancer Res. 1988, 48, 4361–4367. [Google Scholar] [PubMed]
- Kjeldsen, T.; Clausen, H.; Hirohashi, S.; Ogawa, T.; Iijima, H.; Hakomori, S. Preparation and characterization of monoclonal antibodies directed to the tumor-associated O-linked sialosyl-2–6 alpha-N-acetylgalactosaminyl (sialosyl-Tn) epitope. Cancer Res. 1988, 48, 2214–2220. [Google Scholar] [PubMed]
- Yamanaka, T.; Kitagawa, Y.; Seki, H.; Kimura, K.; Sakurabayashi, I.; Kawai, T. A new carbohydrate antigen CA19-9 associated with malignancies of digestive system. II. Serum levels of CA19-9 in various digestive diseases and their clinical significance. Rinsho Byori 1984, 32, 786–792. [Google Scholar] [PubMed]
- Hanski, C.; Bornhoeft, G.; Topf, N.; Hermann, U.; Stein, H.; Riecken, O.E. Detection of a mucin marker for the adenoma-carcinoma sequence in human colonic mucosa by monoclonal antibody AM-3. J. Clin. Pathol. 1990, 43, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Irimura, T.; Wynn, D.M.; Hager, L.G.; Cleary, K.R.; Ota, D.M. Human colonic sulfomucin identified by a specific monoclonal antibody. Cancer Res. 1991, 51, 5728–5735. [Google Scholar] [PubMed]
- Veerman, E.C.I.; Bolscher, E.; Appelmelk, B.J.; Bloemena, E.; van den Berg, T.K.; Nieuw Amerongen, A.V. A monoclonal antibody directed against high Mr salivary mucins recognises the SO3-3Galb1-3GlcNAc moiety of sulpho-Lewisa: A histochemical survey of human and rat tissue. Glycobiology 1997, 7, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Richman, P.I.; Bodmer, W.F. Monoclonal antibodies to human colorectal epithelium: Markers for differentation and tumour characterization. Int. J. Cancer 1987, 39, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Smithson, J.E.; Campbell, A.; Andrews, J.M.; Milton, J.D.; Pigott, R.; Jewell, D.P. Altered expression of mucins through the colon in ulcerative colitis. Gut 1997, 40, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.C.; Cummings, R.D. The immobilized leukoagglutinin from the seeds of Maackia amurensis binds with high affinity to complex-type Asn-linked oligosaccharides containing terminal sialic acid-linked alpha-2,3 to penultimate galactose residues. J. Biol. Chem. 1988, 263, 4576–4585. [Google Scholar] [PubMed]
- Shibuya, N.; Goldstein, I.J.; Broekaert, W.F.; Nsimba-Lubaki, M.; Peeters, B.; Peumans, W.J. The elderberry (Sambucus nigra L.) bark lectin recognises the Neu5Ac(a2,6)Gal/GalNAc sequence. J. Biol. Chem. 1987, 262, 1596–1601. [Google Scholar] [PubMed]
- Duk, M.; Wu, A.M.; Lisowska, E. Vicia villosa B4 lectin is the second anti-Tn lectin shown to react better with blood group N than M antigen. Glycoconj. J. 1994, 11, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Baldus, S.E.; Thiele, J.; Park, Y.O.; Hanisch, F.G.; Bara, J.; Fischer, R. Characterization of the binding specificity of Anguilla anguilla agglutinin (AAA) in comparison to Ulex europaeus agglutinin I (UEA-I). Glycoconj. J. 1996, 13, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Lotan, R.; Skutelsky, E.; Danon, D.; Sharon, N. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea). J. Biol. Chem. 1975, 250, 8518–8523. [Google Scholar] [PubMed]
- Geisler, C.; Jarvis, D.L. Effective glycoanalysis with Maackia amurensis lectins requires a clear understanding of their binding specificities. Glycobiology 2011, 21, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Casset, F.; Peters, T.; Etzler, M.; Korchagina, E.; Nifant’ev, N.; Perez, S.; Imberty, A. Conformational analysis of blood group A trisaccharide in solution and in the binding site of Dolichos biflorus lectin using transient and transferred nuclear Overhauser enhancement (NOE) and rotating-frame NOE experiments. Eur. J. Biochem. 1996, 239, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Klisch, K.; Contreras, D.A.; Sun, X.; Brehm, R.; Bergmann, M.; Alberio, R. The Sda/GM2-glycan is a carbohydrate marker of porcine primordial germ cells and of a subpopulation of spermatogonia in cattle, pigs, horses and llama. Reproduction 2011, 142, 667–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corfield, A.P.; Myerscough, N.; Van Klinken, B.J.; Einerhand, A.W.; Dekker, J. Metabolic labeling methods for the preparation and biosynthetic study of mucin. Methods Mol. Biol. 2000, 125, 227–237. [Google Scholar] [PubMed]
- Corfield, A.P.; do Amaral Corfield, C.; Wagner, S.A.; Warren, B.F.; Mountford, R.A.; Bartolo, D.C.; Clamp, J.R. Loss of sulphate in human colonic mucins during ulcerative colitis. Biochem. Soc. Trans. 1992, 20, 95S. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Myerscough, N.; Bradfield, N.; Corfield Cdo, A.; Gough, M.; Clamp, J.R.; Durdey, P.; Warren, B.F.; Bartolo, D.C.; King, K.R.; et al. Colonic mucins in ulcerative colitis: Evidence for loss of sulfation. Glycoconj. J. 1996, 13, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Landemarre, L.; Cancellieri, P.; Duverger, E. Cell surface lectin array: Parameters affecting cell glycan signature. Glycoconj. J. 2013, 30, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, H.; Zhang, W.; Wei, L.; Yu, H.; Yang, P. Multiplex profiling of glycoproteins using a novel bead-based lectin array. Proteomics 2014, 14, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wang, D.; Yamada, M.; Wang, L.X. Chemoenzymatic synthesis and lectin array characterization of a class of N-glycan clusters. J. Am. Chem. Soc. 2009, 131, 17963–17971. [Google Scholar] [CrossRef] [PubMed]
- Borrebaeck, C.A.; Wingren, C. Antibody array generation and use. Methods Mol. Biol. 2014, 1131, 563–571. [Google Scholar] [PubMed]
- Haab, B.B.; Partyka, K.; Cao, Z. Using antibody arrays to measure protein abundance and glycosylation: Considerations for optimal performance. Curr. Protocols Protein Sci. 2013, 73, 27.6.1–27.6.16. [Google Scholar]
- Huang, W.; Whittaker, K.; Zhang, H.; Wu, J.; Zhu, S.W.; Huang, R.P. Integration of Antibody Array Technology into Drug Discovery and Development. Assay Drug Dev. Technol. 2018, 16, 74–95. [Google Scholar] [CrossRef] [PubMed]
- Geissner, A.; Anish, C.; Seeberger, P.H. Glycan arrays as tools for infectious disease research. Curr. Opin. Chem. Biol. 2014, 18, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Ratner, D.M.; Seeberger, P.H. Carbohydrate microarrays as tools in HIV glycobiology. Curr. Pharm. Des. 2007, 13, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Yue, T.; Haab, B.B. Microarrays in glycoproteomics research. Clin. Lab. Med. 2009, 29, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Petersson, J.; Schreiber, O.; Hansson, G.C.; Gendler, S.J.; Velcich, A.; Lundberg, J.O.; Roos, S.; Holm, L.; Phillipson, M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G327–G333. [Google Scholar] [CrossRef] [PubMed]
- Van der Sluis, M.; De Koning, B.A.; De Bruijn, A.C.; Velcich, A.; Meijerink, J.P.; Van Goudoever, J.B.; Buller, H.A.; Dekker, J.; Van Seuningen, I.; Renes, I.B.; et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 2006, 131, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Velcich, A.; Yang, W.; Heyer, J.; Fragale, A.; Nicholas, C.; Viani, S.; Kucherlapati, R.; Lipkin, M.; Yang, K.; Augenlicht, L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 2002, 295, 1726–1729. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, L.L.; Gavin, J.B.; Tasman-Jones, C. Relation of Helicobacter pylori to the human gastric mucosa in chronic gastritis of the antrum. Gut 1990, 31, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- Van de Bovenkamp, J.H.; Mahdavi, J.; Korteland-Van Male, A.M.; Buller, H.A.; Einerhand, A.W.; Boren, T.; Dekker, J. The MUC5AC glycoprotein is the primary receptor for Helicobacter pylori in the human stomach. Helicobacter 2003, 8, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, J.; Sonden, B.; Hurtig, M.; Olfat, F.O.; Forsberg, L.; Roche, N.; Angstrom, J.; Larsson, T.; Teneberg, S.; Karlsson, K.A.; et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 2002, 297, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Rossez, Y.; Gosset, P.; Boneca, I.G.; Magalhaes, A.; Ecobichon, C.; Reis, C.A.; Cieniewski-Bernard, C.; Joncquel Chevalier Curt, M.; Leonard, R.; Maes, E.; et al. The lacdiNAc-specific adhesin LabA mediates adhesion of Helicobacter pylori to human gastric mucosa. J. Infect. Dis. 2014, 210, 1286–1295. [Google Scholar] [CrossRef] [PubMed]
- Naughton, J.; Duggan, G.; Bourke, B.; Clyne, M. Interaction of microbes with mucus and mucins: Recent developments. Gut Microbes 2014, 5, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Cody, A.J.; McCarthy, N.D.; Jansen van Rensburg, M.; Isinkaye, T.; Bentley, S.D.; Parkhill, J.; Dingle, K.E.; Bowler, I.C.; Jolley, K.A.; Maiden, M.C. Real-time genomic epidemiological evaluation of human Campylobacter isolates by use of whole-genome multilocus sequence typing. J. Clin. Microbiol. 2013, 51, 2526–2534. [Google Scholar] [CrossRef] [PubMed]
- Parkhill, J.; Wren, B.W.; Mungall, K.; Ketley, J.M.; Churcher, C.; Basham, D.; Chillingworth, T.; Davies, R.M.; Feltwell, T.; Holroyd, S.; et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000, 403, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Byrne, C.M.; Clyne, M.; Bourke, B. Campylobacter jejuni adhere to and invade chicken intestinal epithelial cells in vitro. Microbiology 2007, 153 Pt 2, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Alemka, A.; Whelan, S.; Gough, R.; Clyne, M.; Gallagher, M.E.; Carrington, S.D.; Bourke, B. Purified chicken intestinal mucin attenuates Campylobacter jejuni pathogenicity in vitro. J. Med. Microbiol. 2010, 59 Pt 8, 898–903. [Google Scholar] [CrossRef] [PubMed]
- Pryor, W.M.; Freiman, J.S.; Gillies, M.A.; Tuck, R.R. Guillain-Barre syndrome associated with Campylobacter infection. Aust. N. Z. J. Med. 1984, 14, 687–688. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.; Lizak, C.; Michaud, G.; Bucher, M.; Darbre, T.; Aebi, M.; Reymond, J.L.; Locher, K.P. Mechanism of bacterial oligosaccharyltransferase: In vitro quantification of sequon binding and catalysis. J. Biol. Chem. 2013, 288, 8849–8861. [Google Scholar] [CrossRef] [PubMed]
- Alemka, A.; Nothaft, H.; Zheng, J.; Szymanski, C.M. N-glycosylation of Campylobacter jejuni surface proteins promotes bacterial fitness. Infect. Immun. 2013, 81, 1674–1682. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.F.; Wacker, M.; Hernandez, M.; Hitchen, P.G.; Marolda, C.L.; Kowarik, M.; Morris, H.R.; Dell, A.; Valvano, M.A.; Aebi, M. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. USA 2005, 102, 3016–3021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glover, K.J.; Weerapana, E.; Chen, M.M.; Imperiali, B. Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 2006, 45, 5343–5350. [Google Scholar] [CrossRef] [PubMed]
- Glover, K.J.; Weerapana, E.; Imperiali, B. In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. USA 2005, 102, 14255–14259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heikema, A.P.; Bergman, M.P.; Richards, H.; Crocker, P.R.; Gilbert, M.; Samsom, J.N.; van Wamel, W.J.; Endtz, H.P.; van Belkum, A. Characterization of the specific interaction between sialoadhesin and sialylated Campylobacter jejuni lipooligosaccharides. Infect. Immun. 2010, 78, 3237–3246. [Google Scholar] [CrossRef] [PubMed]
- Avril, T.; Wagner, E.R.; Willison, H.J.; Crocker, P.R. Sialic acid-binding immunoglobulin-like lectin 7 mediates selective recognition of sialylated glycans expressed on Campylobacter jejuni lipooligosaccharides. Infect. Immun. 2006, 74, 4133–4141. [Google Scholar] [CrossRef] [PubMed]
- Linton, D.; Karlyshev, A.V.; Hitchen, P.G.; Morris, H.R.; Dell, A.; Gregson, N.A.; Wren, B.W. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: Identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol. Microbiol. 2000, 35, 1120–1134. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yu, H.; Lau, K.; Huang, S.; Chokhawala, H.A.; Li, Y.; Tiwari, V.K.; Chen, X. Multifunctionality of Campylobacter jejuni sialyltransferase CstII: Characterization of GD3/GT3 oligosaccharide synthase, GD3 oligosaccharide sialidase, and trans-sialidase activities. Glycobiology 2008, 18, 686–697. [Google Scholar] [CrossRef] [PubMed]
- Houliston, R.S.; Endtz, H.P.; Yuki, N.; Li, J.; Jarrell, H.C.; Koga, M.; van Belkum, A.; Karwaski, M.F.; Wakarchuk, W.W.; Gilbert, M. Identification of a sialate O-acetyltransferase from Campylobacter jejuni: Demonstration of direct transfer to the C-9 position of terminalalpha-2, 8-linked sialic acid. J. Biol. Chem. 2006, 281, 11480–11486. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.L.; Hensler, M.E.; Varki, A.; Nizet, V. The group B streptococcal sialic acid O-acetyltransferase is encoded by neuD, a conserved component of bacterial sialic acid biosynthetic gene clusters. J. Biol. Chem. 2006, 281, 11186–11192. [Google Scholar] [CrossRef] [PubMed]
- Hitchen, P.; Brzostek, J.; Panico, M.; Butler, J.A.; Morris, H.R.; Dell, A.; Linton, D. Modification of the Campylobacter jejuni flagellin glycan by the product of the Cj1295 homopolymeric-tract-containing gene. Microbiology 2010, 156 Pt 7, 1953–1962. [Google Scholar] [CrossRef] [PubMed]
- McNally, D.J.; Schoenhofen, I.C.; Houliston, R.S.; Khieu, N.H.; Whitfield, D.M.; Logan, S.M.; Jarrell, H.C.; Brisson, J.R. CMP-pseudaminic acid is a natural potent inhibitor of PseB, the first enzyme of the pseudaminic acid pathway in Campylobacter jejuni and Helicobacter pylori. ChemMedChem 2008, 3, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Bode, L.; Jantscher-Krenn, E. Structure-function relationships of human milk oligosaccharides. Adv. Nutr. (Bethesda Md.) 2012, 3, 383S–391S. [Google Scholar] [CrossRef] [PubMed]
- Tu, Q.V.; McGuckin, M.A.; Mendz, G.L. Campylobacter jejuni response to human mucin MUC2: Modulation of colonization and pathogenicity determinants. J. Med. Microbiol. 2008, 57 Pt 7, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Xue, M.; Gu, G.; Liu, X.W.; Wang, P.G. A chemoenzymatic route to synthesize unnatural sugar nucleotides using a novel N-acetylglucosamine-1-phosphate pyrophosphorylase from Camphylobacter jejuni NCTC 11168. Bioorg. Med. Chem. Lett. 2013, 23, 4303–4307. [Google Scholar] [CrossRef] [PubMed]
- Hug, I.; Zheng, B.; Reiz, B.; Whittal, R.M.; Fentabil, M.A.; Klassen, J.S.; Feldman, M.F. Exploiting bacterial glycosylation machineries for the synthesis of a Lewis antigen-containing glycoprotein. J. Biol. Chem. 2011, 286, 37887–37894. [Google Scholar] [CrossRef] [PubMed]
- Iwashkiw, J.A.; Fentabil, M.A.; Faridmoayer, A.; Mills, D.C.; Peppler, M.; Czibener, C.; Ciocchini, A.E.; Comerci, D.J.; Ugalde, J.E.; Feldman, M.F. Exploiting the Campylobacter jejuni protein glycosylation system for glycoengineering vaccines and diagnostic tools directed against brucellosis. Microb. Cell Fact. 2012, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Lindhout, T.; Iqbal, U.; Willis, L.M.; Reid, A.N.; Li, J.; Liu, X.; Moreno, M.; Wakarchuk, W.W. Site-specific enzymatic polysialylation of therapeutic proteins using bacterial enzymes. Proc. Natl. Acad. Sci. USA 2011, 108, 7397–7402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandhal, J.; Wright, P.C. N-Linked glycoengineering for human therapeutic proteins in bacteria. Biotechnol. Lett. 2010, 32, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Sokurenko, E.V.; Chesnokova, V.; Dykhuizen, D.E.; Ofek, I.; Wu, X.R.; Krogfelt, K.A.; Struve, C.; Schembri, M.A.; Hasty, D.L. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. USA 1998, 95, 8922–8926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aprikian, P.; Tchesnokova, V.; Kidd, B.; Yakovenko, O.; Yarov-Yarovoy, V.; Trinchina, E.; Vogel, V.; Thomas, W.; Sokurenko, E. Interdomain interaction in the FimH adhesin of Escherichia coli regulates the affinity to mannose. J. Biol. Chem. 2007, 282, 23437–23446. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, J.; Mackenzie, J.; de Paz, J.L.; Chipwaza, B.; Choudhury, D.; Zavialov, A.; Mannerstedt, K.; Anderson, J.; Pierard, D.; Wyns, L.; et al. The affinity of the FimH fimbrial adhesin is receptor-driven and quasi-independent of Escherichia coli pathotypes. Mol. Microbiol. 2006, 61, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Chalopin, T.; Brissonnet, Y.; Sivignon, A.; Deniaud, D.; Cremet, L.; Barnich, N.; Bouckaert, J.; Gouin, S.G. Inhibition profiles of mono- and polyvalent FimH antagonists against 10 different Escherichia coli strains. Org. Biomol. Chem. 2015, 13, 11369–11375. [Google Scholar] [CrossRef] [PubMed]
- de Ruyck, J.; Lensink, M.F.; Bouckaert, J. Structures of C-mannosylated anti-adhesives bound to the type 1 fimbrial FimH adhesin. IUCrJ 2016, 3 Pt 3, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Swanson, J.L.; Barela, T.J.; Brown, J.J. Receptor specificities of variant Gal(alpha1-4)Gal-binding PapG adhesins of uropathogenic Escherichia coli as assessed by hemagglutination phenotypes. J. Infect. Dis. 1997, 175, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Spaulding, C.N.; Klein, R.D.; Ruer, S.; Kau, A.L.; Schreiber, H.L.; Cusumano, Z.T.; Dodson, K.W.; Pinkner, J.S.; Fremont, D.H.; Janetka, J.W.; et al. Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist. Nature 2017, 546, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, J.A.; Wu, K.J.; Hryckowian, A.J.; Bouley, D.M.; Weimer, B.C.; Sonnenburg, J.L. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 2014, 16, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Broecker, F.; Hanske, J.; Martin, C.E.; Baek, J.Y.; Wahlbrink, A.; Wojcik, F.; Hartmann, L.; Rademacher, C.; Anish, C.; Seeberger, P.H. Multivalent display of minimal Clostridium difficile glycan epitopes mimics antigenic properties of larger glycans. Nat. Commun. 2016, 7, 11224. [Google Scholar] [CrossRef] [PubMed]
- Crost, E.H.; Tailford, L.E.; Le Gall, G.; Fons, M.; Henrissat, B.; Juge, N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS ONE 2013, 8, e76341. [Google Scholar] [CrossRef] [PubMed]
- Hata, D.J.; Smith, D.S. Blood group B degrading activity of Ruminococcus gnavus alpha-galactosidase. Artif. Cells Blood Substit. Immobil. Biotechnol. 2004, 32, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Cervera-Tison, M.; Tailford, L.E.; Fuell, C.; Bruel, L.; Sulzenbacher, G.; Henrissat, B.; Berrin, J.G.; Fons, M.; Giardina, T.; Juge, N. Functional analysis of family GH36 alpha-galactosidases from Ruminococcus gnavus E1: Insights into the metabolism of a plant oligosaccharide by a human gut symbiont. Appl. Environ. Microbiol. 2012, 78, 7720–7732. [Google Scholar] [CrossRef] [PubMed]
- Png, C.W.; Linden, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R.; et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Chua, H.H.; Chou, H.C.; Tung, Y.L.; Chiang, B.L.; Liao, C.C.; Liu, H.H.; Ni, Y.H. Intestinal Dysbiosis Featuring Abundance of Ruminococcus gnavus Associates With Allergic Diseases in Infants. Gastroenterology 2018, 154, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Graziani, F.; Pujol, A.; Nicoletti, C.; Dou, S.; Maresca, M.; Giardina, T.; Fons, M.; Perrier, J. Ruminococcus gnavus E1 modulates mucin expression and intestinal glycosylation. J. Appl. Microbiol. 2016, 120, 1403–1417. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, W.; Caplan, M.S.; Qu, X.-W.; Tan, X.-D.; de Plaen, I.G.; Gonzalez-Crussi, F. Neonatal Necrotising Enterocolitis: Clinical Considerations and Pathogenic Concepts. Pediatr. Dev. Pathol. 2002, 6, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Morowitz, M.J.; Denef, V.J.; Costello, E.K.; Thomas, B.C.; Poroyko, V.; Relman, D.A.; Banfield, J.F. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc. Natl. Acad. Sci. USA 2011, 108, 1128–1133. [Google Scholar] [CrossRef] [PubMed]
- Vieten, D.; Corfield, A.; Carroll, D.; Ramani, P.; Spicer, R. Impaired mucosal regeneration in neonatal necrotising enterocolitis. Pediatr. Surg. Int. 2005, 21, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Vieten, D.; Corfield, A.; Ramani, P.; Spicer, R. Proliferative response in necrotising enterocolitis is insufficient to prevent disease progression. Pediatr. Surg. Int. 2006, 22, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Schaart, M.W.; de Bruijn, A.C.; Bouwman, D.M.; de Krijger, R.R.; van Goudoever, J.B.; Tibboel, D.; Renes, I.B. Epithelial functions of the residual bowel after surgery for necrotising enterocolitis in human infants. J. Pediatr. Gastroenterol. Nutr. 2009, 49, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Itani, T.; Ayoub Moubareck, C.; Melki, I.; Rousseau, C.; Mangin, I.; Butel, M.J.; Karam-Sarkis, D. Preterm infants with necrotising enterocolitis demonstrate an unbalanced gut microbiota. Acta Paediatr. 2018, 107, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Abrahamsson, T.R. Using probiotics to prevent necrotising enterocolitis. Acta Paediatr. 2017, 106, 1718–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Embleton, N.; Berrington, J.E. Probiotics reduce the risk of necrotising enterocolitis (NEC) in preterm infants. Evid. Based Med. 2013, 18, 219–220. [Google Scholar] [CrossRef] [PubMed]
- Uberos, J.; Aguilera-Rodriguez, E.; Jerez-Calero, A.; Molina-Oya, M.; Molina-Carballo, A.; Narbona-Lopez, E. Probiotics to prevent necrotising enterocolitis and nosocomial infection in very low birth weight preterm infants. Br. J. Nutr. 2017, 117, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Autran, C.A.; Kellman, B.P.; Kim, J.H.; Asztalos, E.; Blood, A.B.; Spence, E.C.; Patel, A.L.; Hou, J.; Lewis, N.E.; Bode, L. Human milk oligosaccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut 2018, 67, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Jantscher-Krenn, E.; Zherebtsov, M.; Nissan, C.; Goth, K.; Guner, Y.S.; Naidu, N.; Choudhury, B.; Grishin, A.V.; Ford, H.R.; Bode, L. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut 2012, 61, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Butel, M.J.; Waligora-Dupriet, A.J.; Szylit, O. Oligofructose and experimental model of neonatal necrotising enterocolitis. Br. J. Nutr. 2002, 87 (Suppl. 2), S213–S219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Autran, C.A.; Schoterman, M.H.; Jantscher-Krenn, E.; Kamerling, J.P.; Bode, L. Sialylated galacto-oligosaccharides and 2′-fucosyllactose reduce necrotising enterocolitis in neonatal rats. Br. J. Nutr. 2016, 116, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Osborn, D.A.; Lui, K.; Pussell, P.; Jana, A.K.; Desai, A.S.; Cole, M. T and Tk antigen activation in necrotising enterocolitis: Manifestations, severity of illness, and effectiveness of testing. Arch. Dis. Child. Fetal Neonatal. Ed. 1999, 80, F192–F197. [Google Scholar] [CrossRef] [PubMed]
- McElroy, S.J.; Underwood, M.A.; Sherman, M.P. Paneth cells and necrotizing enterocolitis: A novel hypothesis for disease pathogenesis. Neonatology 2013, 103, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Xavier, R.J.; Podolsky, D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007, 448, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011, 474, 307–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellinghaus, D.; Bethune, J.; Petersen, B.S.; Franke, A. The genetics of Crohn’s disease and ulcerative colitis--status quo and beyond. Scand. J. Gastroenterol. 2015, 50, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Gustafsson, J.K.; Holmen-Larsson, J.; Jabbar, K.S.; Xia, L.; Xu, H.; Ghishan, F.K.; Carvalho, F.A.; Gewirtz, A.T.; Sjovall, H.; et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 2014, 63, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Larsson, J.M.; Karlsson, H.; Crespo, J.G.; Johansson, M.E.; Eklund, L.; Sjovall, H.; Hansson, G.C. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm. Bowel Dis. 2011, 17, 2299–2307. [Google Scholar] [CrossRef] [PubMed]
- Renes, I.; Van Seuningen, I. Mucins in intestinal inflammatory diseases: Their expression patterns, regulation and roles. In The Epithelial Mucins: Structure/Function Roles in Cancer and Inflammatory Diseases; Van Seuningen, I., Ed.; Research Signpost: Kerala, India, 2008; pp. 211–232. [Google Scholar]
- Buisine, M.P.; Desreumaux, P.; Debailleul, V.; Gambiez, L.; Geboes, K.; Ectors, N.; Delescaut, M.P.; Degand, P.; Aubert, J.P.; Colombel, J.F.; et al. Abnormalities in mucin gene expression in Crohn’s disease. Inflamm. Bowel Dis. 1999, 5, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Buisine, M.P.; Desreumaux, P.; Leteurtre, E.; Copin, M.C.; Colombel, J.F.; Porchet, N.; Aubert, J.P. Mucin gene expression in intestinal epithelial cells in Crohn’s disease. Gut 2001, 49, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.A.; Babyatsky, M.W.; Ogata, S.; Chen, A.; Itzkowitz, S.H. Expression of MUC2 and MUC3 mRNA in human normal, malignant, and inflammatory intestinal tissues. J. Histochem. Cytochem. 1996, 44, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Senapati, S.; Ho, S.B.; Sharma, P.; Das, S.; Chakraborty, S.; Kaur, S.; Niehans, G.; Batra, S.K. Expression of intestinal MUC17 membrane-bound mucin in inflammatory and neoplastic diseases of the colon. J. Clin. Pathol. 2010, 63, 702–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tytgat, K.M.A.J.; Van der Wal, T.-W.; Einerhand, A.W.C.; Buller, H.A.; Dekker, J. Quantitative analysis of MUC2 synthesis in ulcerative colitis. Biochem. Biophys. Res. Commun. 1996, 224, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Forgue-Lafitte, M.E.; Fabiani, B.; Levy, P.P.; Maurin, N.; Flejou, J.F.; Bara, J. Abnormal expression of M1/MUC5AC mucin in distal colon of patients with diverticulitis, ulcerative colitis and cancer. Int. J. Cancer 2007, 121, 1543–1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaoul, R.; Okada, Y.; Cutz, E.; Marcon, M.A. Colonic expression of MUC2, MUC5AC, and TFF1 in inflammatory bowel disease in children. J. Pediatr. Gastroenterol. Nutr. 2004, 38, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Van Klinken, B.J.; Dekker, J.; van Gool, S.A.; van Marle, J.; Buller, H.A.; Einerhand, A.W. MUC5B is the prominent mucin in human gallbladder and is also expressed in a subset of colonic goblet cells. Am. J. Physiol. 1998, 274 Pt 1, G871–G878. [Google Scholar] [CrossRef] [PubMed]
- McMahon, R.F.; Warren, B.F.; Jones, C.J.; Mayberry, J.F.; Probert, C.S.; Corfield, A.P.; Stoddart, R.W. South Asians with ulcerative colitis exhibit altered lectin binding compared with matched European cases. Histochem. J. 1997, 29, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Probert, C.S.; Warren, B.F.; Perry, T.; Mackay, E.H.; Mayberry, J.F.; Corfield, A.P. South Asian and European colitics show characteristic differences in colonic mucus glycoprotein type and turnover. Gut 1995, 36, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Ghazi, L.J.; Lydecker, A.D.; Patil, S.A.; Rustgi, A.; Cross, R.K.; Flasar, M.H. Racial differences in disease activity and quality of life in patients with Crohn’s disease. Dig. Dis. Sci. 2014, 59, 2508–2513. [Google Scholar] [CrossRef] [PubMed]
- Marteau, P. Bacterial flora in inflammatory bowel disease. Dig. Dis. (Basel Switz.) 2009, 27 (Suppl. 1), 99–103. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Ladhoff, A.; Pernthaler, A.; Swidsinski, S.; Loening-Baucke, V.; Ortner, M.; Weber, J.; Hoffmann, U.; Schreiber, S.; Dietel, M.; et al. Mucosal flora in inflammatory bowel disease. Gastroenterology 2002, 122, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, F. The microbiota in inflammatory bowel disease: Friend, bystander, and sometime-villain. Nutr. Rev. 2012, 70 (Suppl. 1), S31–S37. [Google Scholar] [CrossRef] [PubMed]
- Theodoratou, E.; Campbell, H.; Ventham, N.T.; Kolarich, D.; Pucic-Bakovic, M.; Zoldos, V.; Fernandes, D.; Pemberton, I.K.; Rudan, I.; Kennedy, N.A.; et al. The role of glycosylation in IBD. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 588–600. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.J.; Yu, L.G.; Rhodes, J.M. Altered glycosylation in inflammatory bowel disease: A possible role in cancer development. Glycoconj. J. 2001, 18, 851–858. [Google Scholar] [CrossRef] [PubMed]
- An, G.; Wei, B.; Xia, B.; McDaniel, J.M.; Ju, T.; Cummings, R.D.; Braun, J.; Xia, L. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J. Exp. Med. 2007, 204, 1417–1429. [Google Scholar] [CrossRef] [PubMed]
- Iwai, T.; Kudo, T.; Kawamoto, R.; Kubota, T.; Togayachi, A.; Hiruma, T.; Okada, T.; Kawamoto, T.; Morozumi, K.; Narimatsu, H. Core 3 synthase is down-regulated in colon carcinoma and profoundly suppresses the metastatic potential of carcinoma cells. Proc. Natl. Acad. Sci. USA 2005, 102, 4572–4577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, L. Core 3-derived O-glycans are essential for intestinal mucus barrier function. Methods Enzymol. 2010, 479, 123–141. [Google Scholar] [PubMed]
- Bergstrom, K.; Fu, J.; Johansson, M.E.; Liu, X.; Gao, N.; Wu, Q.; Song, J.; McDaniel, J.M.; McGee, S.; Chen, W.; et al. Core 1- and 3-derived O-glycans collectively maintain the colonic mucus barrier and protect against spontaneous colitis in mice. Mucosal Immunol. 2017, 10, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wei, B.; Wen, T.; Johansson, M.E.; Liu, X.; Bradford, E.; Thomsson, K.A.; McGee, S.; Mansour, L.; Tong, M.; et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J. Clin. Investig. 2011, 121, 1657–1666. [Google Scholar] [CrossRef] [PubMed]
- Itzkowitz, S.H.; Bloom, E.J.; Kokal, W.A.; Modin, G.; Hakomori, S.-I.; Kim, Y.S. Sialosyl-Tn: A novel mucin antigen associated with prognosis in colorectal cancer patients. Cancer 1990, 66, 1960–1966. [Google Scholar] [CrossRef]
- Campbell, B.J.; Rowe, G.E.; Leiper, K.; Rhodes, J.M. Increasing the intra-Golgi pH of cultured LS174T goblet-differentiated cells mimics the decreased mucin sulfation and increased Thomsen- Friedenreich antigen (Galbeta1-3GalNacalpha-) expression seen in colon cancer. Glycobiology 2001, 11, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Raouf, A.H.; Tsai, H.H.; Parker, N.; Hoffman, J.; Walker, R.J.; Rhodes, J.M. Sulphation of colonic and rectal mucin in inflammatory bowel disease: Reduced sulphation of rectal mucus in ulcerative colitis. Clin. Sci. 1992, 83, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Williams, A.J.; Clamp, J.R.; Wagner, S.A.; Mountford, R.A. Degradation by bacterial enzymes of colonic mucus from normal subjects and patients with inflammatory bowel disease: The role of sialic acid metabolism and the detection of a novel O-acetylsialic acid esterase. Clin. Sci. (Lond.) 1988, 74, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, F. Probiotics in inflammatory bowel disease--therapeutic rationale and role. Adv. Drug Deliv. Rev. 2004, 56, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; de Vos, W.M. Next-Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Front. Microbiol. 2017, 8, 1765. [Google Scholar] [CrossRef] [PubMed]
- Ouwerkerk, J.P.; Aalvink, S.; Belzer, C.; De Vos, W.M. Preparation and preservation of viable Akkermansia muciniphila cells for therapeutic interventions. Benef. Microbes 2017, 8, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Le Bastard, Q.; Ward, T.; Sidiropoulos, D.; Hillmann, B.M.; Chun, C.L.; Sadowsky, M.J.; Knights, D.; Montassier, E. Fecal microbiota transplantation reverses antibiotic and chemotherapy-induced gut dysbiosis in mice. Sci. Rep. 2018, 8, 6219. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhou, Q.; Zhang, D.; Jiang, F.; Wu, J.; Zhou, J.Y.; Zheng, X.; Chen, Y.G. Effect of Faecal Microbiota Transplantation for Treatment of Clostridium difficile Infection in Patients with Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis of Cohort Studies. J. Crohn’s Colitis 2018, 12, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Vazquez-Baeza, Y.; Gonzalez, A.; Weiss, S.; Schmidt, B.; Muniz-Pedrogo, D.A.; Rainey, J.F., 3rd; Kammer, P.; Nelson, H.; Sadowsky, M.; et al. Changes in microbial ecology after fecal microbiota transplantation for recurrent C. difficile infection affected by underlying inflammatory bowel disease. Microbiome 2017, 5, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, K.M.; Rank, K.M.; Vaughn, B.P.; Khoruts, A. Treatment of recurrent Clostridium difficile infection using fecal microbiota transplantation in patients with inflammatory bowel disease. Gut Microbes 2017, 8, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.M.; Corfield, A.P.; Jewell, D.P.; Biddolph, S.; Warren, B.F. Mucin bound O-acetylated sialic acids and diversion colitis. Gut 2000, 46, A81. [Google Scholar]
- Kaser, A.; Blumberg, R.S. Endoplasmic reticulum stress in the intestinal epithelium and inflammatory bowel disease. Semin. Immunol. 2009, 21, 156–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz, T.; Niederreiter, L.; Adolph, T.; Blumberg, R.S.; Kaser, A. Crohn’s disease: NOD2, autophagy and ER stress converge. Gut 2011, 60, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.R.; Vogelstein, B. Genetic alterations in the adenoma—Carcinoma sequence. Cancer 1992, 70 (Suppl. 6), 1727–1731. [Google Scholar] [CrossRef]
- Riddell, R.H. The adenoma-carcinoma sequence. Prog. Clin. Biol. Res. 1988, 279, 23–33. [Google Scholar] [PubMed]
- Cao, Y.; Blohm, D.; Ghadimi, B.M.; Stosiek, P.; Xing, P.X. Mucins (MUC1 and MUC3) of gastrointestinal and breast epithelia reveal different and heterogeneous tumour-associated aberrations in glycosylation. J. Histochem. Cytochem. 1997, 45, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Buisine, M.-P.; Janin, A.; Mauroury, V.; Audie, J.-P.; Delescaut, M.-P.; Copin, M.-C.; Colombel, J.-F.; Degand, P.; Aubert, J.-P.; Porchet, N. Abberant expression of a human mucin gene (MUC5AC) in rectosigmoid villous adenoma. Gastroenterology 1996, 110, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Myerscough, N.; Sylvester, P.A.; Warren, B.F.; Biddolph, S.; Durdey, P.; Thomas, M.G.; Carlstedt, I.; Corfield, A.P. Abnormal subcellular distribution of mature MUC2 and de novo MUC5AC mucins in adenomas of the rectum: Immunohistochemical detection using non-VNTR antibodies to MUC2 and MUC5AC peptide. Glycoconj. J. 2001, 18, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Bara, J.; Forgue-Lafitte, M.E.; Maurin, N.; Flejou, J.F.; Zimber, A. Abnormal expression of gastric mucin in human and rat aberrant crypt foci during colon carcinogenesis. Tumour Biol. 2003, 24, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Wang, L.; Wei, P.; Chi, Y.; Li, D.; Wang, Q.; Ni, S.; Tan, C.; Sheng, W.; Sun, M.; et al. Role of MUC20 overexpression as a predictor of recurrence and poor outcome in colorectal cancer. J. Transl. Med. 2013, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.B.; Niehans, G.A.; Lyftogt, C.; Yan, P.S.; Cherwitz, D.L.; Gum, E.T.; Dahiya, R.; Kim, Y.S. Heterogeneity of mucin gene expression in normal and neoplastic tissues. Cancer Res. 1993, 53, 641–651. [Google Scholar] [PubMed]
- Mihalache, A.; Delplanque, J.F.; Ringot-Destrez, B.; Wavelet, C.; Gosset, P.; Nunes, B.; Groux-Degroote, S.; Leonard, R.; Robbe-Masselot, C. Structural Characterization of Mucin O-Glycosylation May Provide Important Information to Help Prevent Colorectal Tumor Recurrence. Front. Oncol. 2015, 5, 217. [Google Scholar] [CrossRef] [PubMed]
- Vavasseur, F.; Dole, K.; Yang, J.; Matta, K.L.; Myerscough, N.; Corfield, A.; Paraskeva, C.; Brockhausen, I. O-glycan biosynthesis in human colorectal adenoma cells during progression to cancer. Eur. J. Biochem. 1994, 222, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Chik, J.H.; Zhou, J.; Moh, E.S.; Christopherson, R.; Clarke, S.J.; Molloy, M.P.; Packer, N.H. Comprehensive glycomics comparison between colon cancer cell cultures and tumours: Implications for biomarker studies. J. Proteom. 2014, 108, 146–162. [Google Scholar] [CrossRef] [PubMed]
- Brockhausen, I. Pathways of O-glycan biosynthesis in cancer cells. Biochim. Biophys. Acta 1999, 1473, 67–95. [Google Scholar] [CrossRef]
- Brockhausen, I. Sulphotransferases acting on mucin-type oligosaccharides. Biochem. Soc. Trans. 2003, 31, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Myerscough, N.; Wagner, S.A.; Gough, M.; Clamp, J.R.; Paraskeva, C.; King, R.K.; Williams, J.M. Mucin sulphation in colorectal disease. Glycobiology 1992, 2, 499. [Google Scholar]
- Bodger, K.; Campbell, F.; Rhodes, J.M. Detection of sulfated glycoproteins in intestinal metaplasia: A comparison of traditional mucin staining with immunohistochemistry for the sulfo-Lewis(a) carbohydrate epitope. J. Clin. Pathol. 2003, 56, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, Y.; Yamamoto, N.; Shirahama, H.; Tanaka, S.; Yonezawa, S.; Yamori, T.; Irimura, T.; Sato, E. Expression of sulfomucins in normal mucosae, colorectal adenocarcinomas, and metastases. Jpn. J. Cancer Res. 1995, 86, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Jass, J.R.; Allison, L.J.; Edgar, S.G. Distribution of sialosyl-Tn and Tn antigens within normal and malignant colorectal epithelium. J. Pathol. 1995, 176, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Brockhausen, I.; Yang, J.; Dickinson, N.; Ogata, S.; Itzkowitz, S.H. Enzymatic basis for sialyl-Tn expression in human colon cancer cells. Glycoconj. J. 1998, 15, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Marcos, N.T.; Bennett, E.P.; Gomes, J.; Magalhaes, A.; Gomes, C.; David, L.; Dar, I.; Jeanneau, C.; DeFrees, S.; Krustrup, D.; et al. ST6GalNAc-I controls expression of sialyl-Tn antigen in gastrointestinal tissues. Front. Biosci. (Elite Ed.) 2011, 3, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- King, M.J.; Chan, A.; Roe, R.; Warren, B.F.; Dell, A.; Morris, H.R.; Bartolo, D.C.; Durdey, P.; Corfield, A.P. Two different glycosyltransferase defects that result in GalNAc alpha-O-peptide (Tn) expression. Glycobiology 1994, 4, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.J.; Finnie, I.A.; Hounsell, E.F.; Rhodes, J.M. Direct demonstration of increased expression of Thomsen-Friedenreich (TF) antigen in colonic adenocarcinoma and ulcerative colitis mucin and its concealment in normal mucin. J. Clin. Investig. 1995, 95, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Springer, G.F. Immunoreactive T and Tn epitopes in cancer diagnosis, prognosis, and immunotherapy. J. Mol. Med. 1997, 75, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Robbe-Masselot, C.; Herrmann, A.; Maes, E.; Carlstedt, I.; Michalski, J.C.; Capon, C. Expression of a core 3 disialyl-Le(x) hexasaccharide in human colorectal cancers: A potential marker of malignant transformation in colon. J. Proteome Res. 2009, 8, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, P.; Mann, B.; Mansmann, U.; Lovin, N.; Foss, H.D.; Berger, G.; Scherubl, H.; Riecken, E.O.; Buhr, H.J.; Hanski, C. Expression of SIALYL-Le(x) antigen defined by MAb AM-3 is an independent prognostic marker in colorectal carcinoma patients. Int. J. Cancer 2000, 88, 281–286. [Google Scholar] [CrossRef]
- Dall’Olio, F.; Malagolini, N.; Trinchera, M.; Chiricolo, M. Sialosignaling: Sialyltransferases as engines of self-fueling loops in cancer progression. Biochim. Biophys. Acta 2014, 1840, 2752–2764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldus, S.E.; Monig, S.P.; Hanisch, F.G.; Zirbes, T.K.; Flucke, U.; Oelert, S.; Zilkens, G.; Madejczik, B.; Thiele, J.; Schneider, P.M.; et al. Comparative evaluation of the prognostic value of MUC1, MUC2, sialyl- Lewis(a) and sialyl-Lewis(x) antigens in colorectal adenocarcinoma. Histopathology 2002, 40, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Wagner, S.A.; Paraskeva, C.; Clamp, J.R.; Durdey, P.; Reuter, G.; Schauer, R. Loss of sialic acid O-acetylation in human colorectal cancer cells. Biochem. Soc. Trans. 1992, 20, 94S. [Google Scholar] [CrossRef] [PubMed]
- Mann, B.; Klussmann, E.; Vandamme-Feldhaus, V.; Iwersen, M.; Hanski, M.L.; Riecken, E.O.; Buhr, H.J.; Schauer, R.; Kim, Y.S.; Hanski, C. Low O-acetylation of sialyl-Le(x) contributes to its overexpression in colon carcinoma metastases. Int. J. Cancer 1997, 72, 258–264. [Google Scholar] [CrossRef]
- Shen, Y.; Kohla, G.; Lrhorfi, A.L.; Sipos, B.; Kalthoff, H.; Gerwig, G.J.; Kamerling, J.P.; Schauer, R.; Tiralongo, J. O-acetylation and de-O-acetylation of sialic acids in human colorectal carcinoma. Eur. J. Biochem. 2004, 271, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Wagner, S.A.; O’Donnell, L.J.; Durdey, P.; Mountford, R.A.; Clamp, J.R. The roles of enteric bacterial sialidase, sialate O-acetyl esterase and glycosulfatase in the degradation of human colonic mucin. Glycoconj. J. 1993, 10, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Corfield, T. Bacterial sialidases–roles in pathogenicity and nutrition. Glycobiology 1992, 2, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P.; Wagner, S.A.; Clamp, J.R. Detection of a carbohydrate sulphatase in human faecal extracts. Biochem. Soc. Trans. 1987, 15, 1089. [Google Scholar] [CrossRef]
- Hart, G.W.; Varki, A. Future Directions in Glycosciences. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017; pp. 761–768. [Google Scholar]
- Agard, N.J.; Bertozzi, C.R. Chemical approaches to perturb, profile, and perceive glycans. Acc. Chem. Res. 2009, 42, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Bertozzi, C.R. Site-specific antibody-drug conjugates: The nexus of bioorthogonal chemistry, protein engineering, and drug development. Bioconjug. Chem. 2015, 26, 176–192. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Aebi, M.; Packer, N.H.; Seeberger, P.H.; Esko, J.D.; Stanley, P.; Hart, G.; Darvill, A.; Kinoshita, T.; et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 25, 1323–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
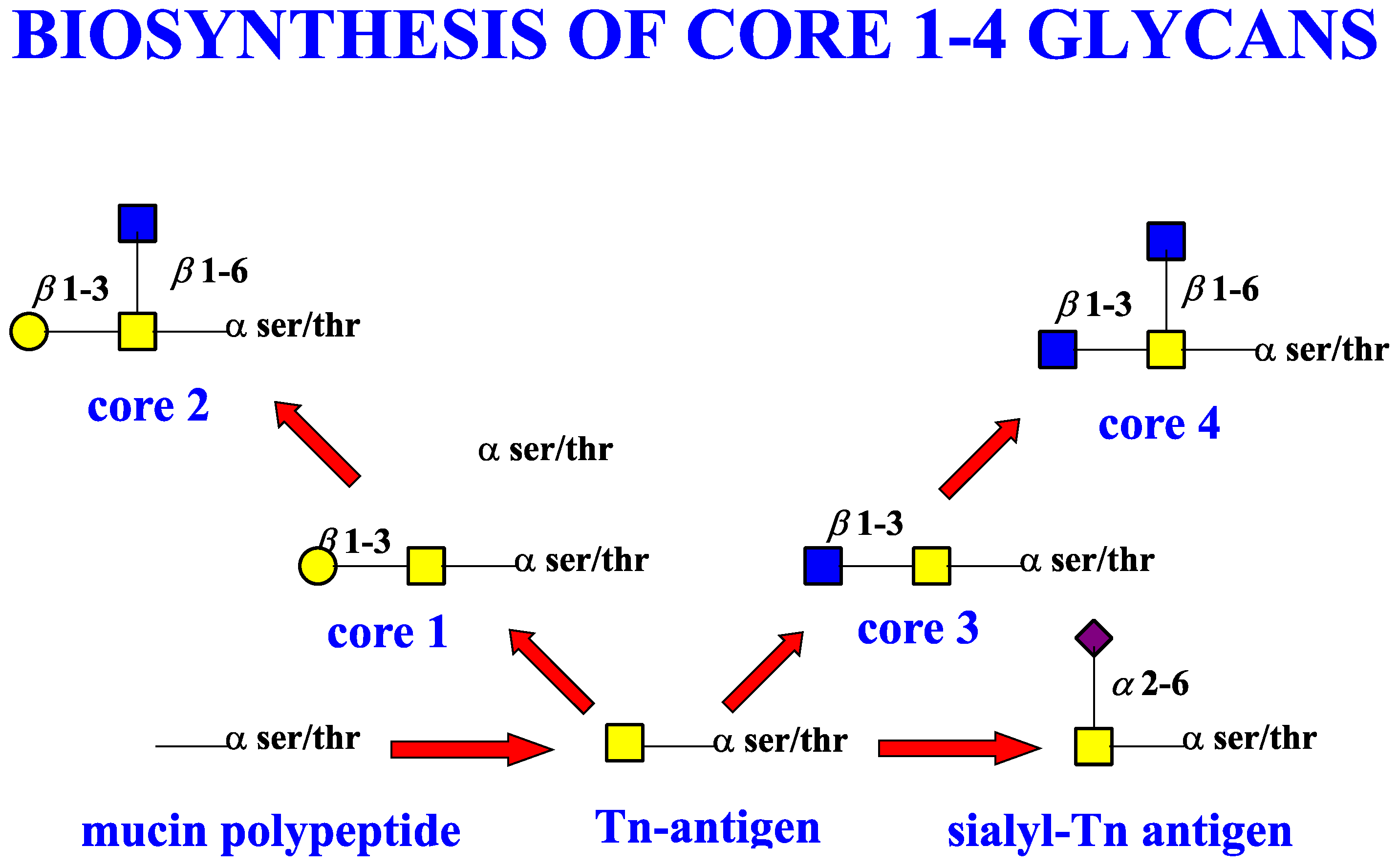
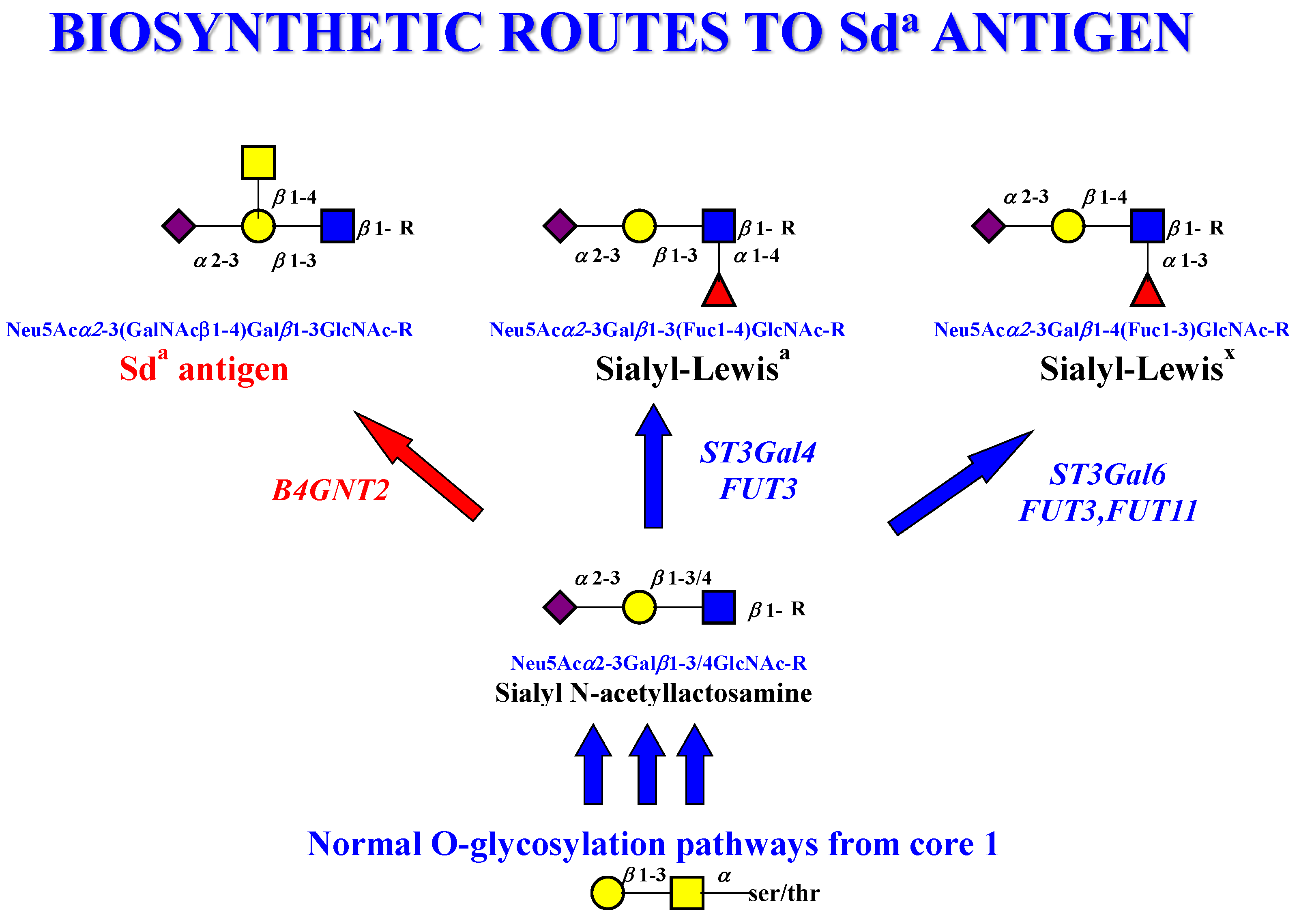
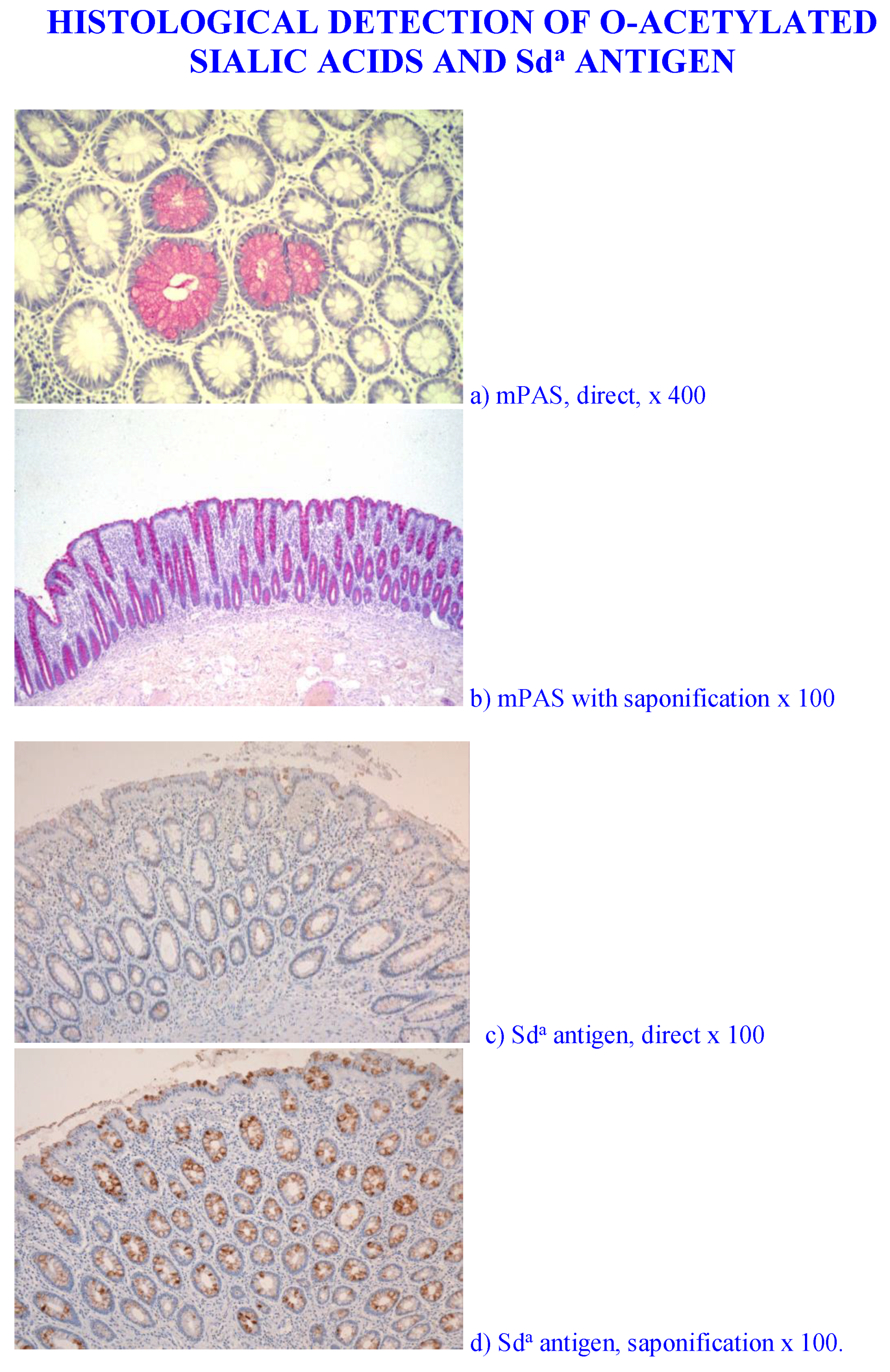
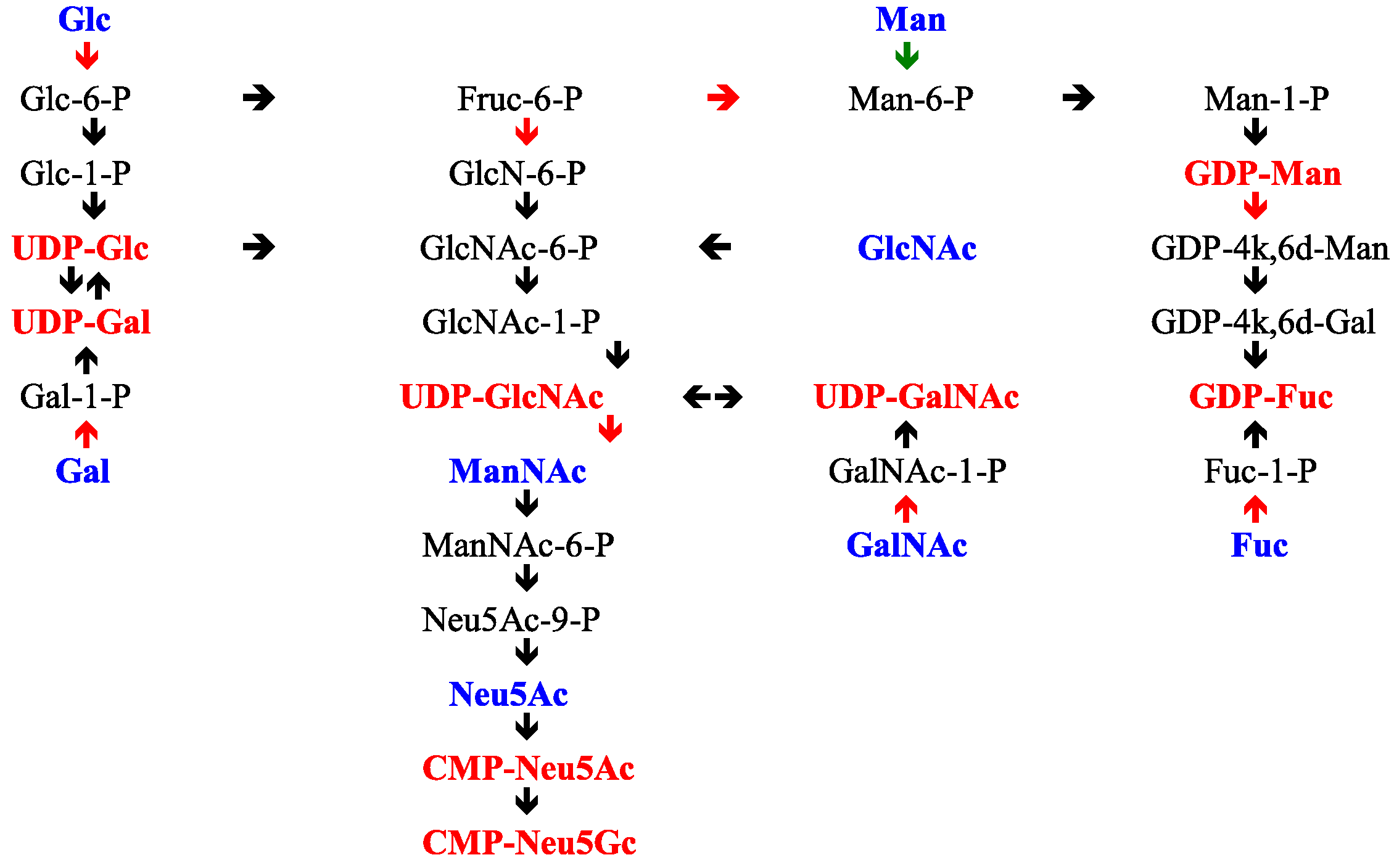

| MUC Gene | Chromosome | Tandem Repeat Size | N-Terminal Signal Sequence | Gastrointestinal Tract Location |
|---|---|---|---|---|
| Membrane Associated Mucins | ||||
| MUC1 | 1q21 | 20 | √ | Stomach, duodenum, ileum, colon |
| MUC3A/B | 7q22 | 17 | √ | Small intestine, colon |
| MUC4 | 3q29 | 16 | √ | Small intestine, colon |
| MUC12 | 7q22 | 28 | √ | Colon |
| MUC13 | 3q21.2 | 27 | √ | Small intestine, colon |
| MUC15 | 11p14.3 | none | √ | Small intestine, colon |
| MUC16 | 19p13.2 | 156 | √ | Not expressed |
| MUC17 | 7q22 | 59 | √ | Stomach, duodenum, colon |
| MUC20 | 3q29 | 18 | √ | Colon |
| MUC21 | 6p21 | 15 | √ | Colon |
| Secreted gel-forming mucins | ||||
| MUC2 | 11p15.5 | 23 | √ | Jejunum, ileum, colon |
| MUC5AC | 11p15.5 | 8 | √ | Stomach |
| MUC5B | 11p15.5 | 29 | √ | Salivary glands |
| MUC6 | 11p15.5 | 169 | √ | Stomach, ileum |
| MUC19 | 12q12 | 19 | √ | No reports for GI tract |
| Secreted non gel-forming mucins | ||||
| MUC7 | 4q13-q21 | 23 | √ | Salivary glands |
| MUC8 | 12q243 | 13/41 | √ | Not expressed |
| MUC9 | 1p13 | 15 | √ | Not expressed |
| Peptide Domain Type | Mucin | Mucin Type | Peptide Domain Function |
|---|---|---|---|
| Cysteine rich CYS domains | MUC2, MUC5AC, MUC5B, MUC19 | Secreted | Non-glycosylated multiple copy domains adjacent or interrupting tandem repeat domains. Important for various mucin–mucin interactions. |
| Cysteine Knot | MUC2, MUC5AC, MUC5B, MUC6, MUC19 | Secreted | Involved in dimerization. |
| Von Willebrand Factor D (D1, D2, D’, D3) | MUC2, MUC5AC, MUC5B, MUC6, MUC19 | Secreted | Mediate oligomerisation located at N- & C-terminus D3 is directly active in polymerization. |
| Von Willebrand Factor D (D4) | MUC2, MUC5AC, MUC5B, MUC6 MUC4 | Secreted & Membrane-associated | Located N-terminally to the D4 is located C-terminally to the VNTR domains, contains the GDPH autocatalytic cleavage site. |
| Cytoplasmic Tail | MUC1, MUC3A/B, MUC12, MUC13, MUC16, MUC17, MUC21 | Membrane-associated | Located on the cytoplasmic side of the cell surface membrane. Contains phosphorylation sites involved in signaling. MUC3, MUC12, and MUC17 have PDZ binding motifs |
| SEA (Sperm protein, Enterokinase & Agrin) | MUC3A/B, MUC4, MUC12, MUC13, MUC17, MUC21 | Membrane-associated | Protein binding properties. Contains autocatalytic proteolytic cleavage site. |
| EGF (Epidermal Growth Factor) | MUC1, MUC3A/B, MUC12, MUC13, MUC17 | Membrane-associated | Mediate interactions between mucin subunits and ERBB receptors. |
| Transmembrane | MUC1, MUC3A/B, MUC4, MUC12, MUC13, MUC16, MUC17, MUC20, MUC21 | Membrane-associated | Membrane-spanning sequence typical for membrane proteins |
| GDPH autocatalytic proteolytic site | MUC2, MUC4, MUC5AC | Secreted & Membrane-associated | Autocatalytic site cleaving between GD and PH residues |
| Proteolytic cleavage site | MUC1, MUC3A/B, MUC4, MUC12, MUC13, MUC16, MUC17 | Membrane-associated | Found in MUCs with the SEA domain and in MUC16 |
| Core Type | Structure |
|---|---|
| 1 |  Galβ1-3GalNAc |
| 2 |  Galβ1-3(GlcNAcβ1-6)GalNAc |
| 3 |  GlcNAcβ1-3GalNAc |
| 4 |  GlcNAcβ1-3(GlcNAcβ1-6)GalNAc |
| Backbone Repeat | Structure |
| Type 1 |  Galβ1-3GlcNAc |
| Type 2 |  Galβ1-4GlcNAc |
| Poly N-acetyllactosamine type 2 |  (Galβ1-4GlcNAcβ1-3-)n |
| Branched N-acetyllactosamine type 2 |  Galβ1-4GlcNAcβ1-6 Galβ1 Galβ1-4GlcNAcβ1-3 |
| Protein Carrier | Glycan Structure |
|---|---|
| Glycoproteins N-Glycans |   Mannose 6-phosphate glycans |
| Glycoproteins O-Glycans |  Mucin type O-Glycans Linear sialylated core 3 Branched sialylated core 4 |
| Glycoproteins O-GlcNAcylation |  |
| Glycoproteins C-Mannose |  |
| Type of Glycan | Structure |
|---|---|
| Blood group H type 1  (Galβ1-3GlcNAc) |  Fucα1-2Galβ1-3GlcNAcβ1- |
| Blood group A type 2  (Galβ1-4GlcNAc) |  GalNAcα1-3Galβ1-4GlcNAcβ1- | α1-2 Fuc |
| Blood group B type 1  (Galβ1-3GlcNAc) |  Galα1-3Galβ1-3GlcNAcβ1- | α1-2 Fuc |
| Lewisa and Lewisx |  Galα1-3GlcNAcβ1- | α1-4 Fuc  Galβ1-4GlcNAcβ1- | α1-3 Fuc |
| Lewisb |  Galβ1-3GlcNAcβ1- | α1-2 | α1-4 Fuc Fuc |
| Lewisy |  Galβ1-4GlcNAcβ1- | α1-2 | α1-3 Fuc Fuc |
| Sialyl Lewisa and Sialyl Lewisx |  Neu5Acα2-3Galβ1-3GlcNAcβ1- | α1-4 Fuc  Neu5Acα2-3Galβ1-4GlcNAcβ1- | α1-3 Fuc |
| Sialyl-Tn |  Neu5Acα2-6GalNAc-α-O-Ser/Thr |
| Monosialylated-T-antigen |  Neu5Acα2-3Galβ1-3GalNAc-α-O-Ser/Thr  Neu5Ac | α2-6 Galβ1-3GalNAc-α-O-Ser/Thr |
| Monosialylated core 3 |  Neu5Ac | α2-6 Galβ1-4GlcNAcβ1-3 GalNAc |
| Sda antigen Type 1 & 2 chains |  Neu5Ac | α2-3 GalNAcβ1-4Galβ1-3GlcNAcαβ1-3GalNAc  Neu5Ac | α2-3 GalNAcβ1-4Galβ1-4GlcNAcαβ1-3GalNAc- |
| Transfer Moiety | Nucleotide | Nucleotide Transport ER | Nucleotide Transport Golgi | Comment |
|---|---|---|---|---|
| Neu5Ac | CMP-Neu5Ac | − | + | Golgi location. Also transfers Neu5Gc |
| Fuc | GDP-Fuc | + | + | ER and Golgi location |
| Gal | UDP-Gal | − | + | Only in Golgi |
| Man | GDP-Man | − | + | Only in Golgi |
| GlcNAc | UDP-GlcNAc | + | + | ER and Golgi location |
| GalNAc | UDP-GalNAc | + | + | ER and Golgi location |
| Sulphate | PAPS * | − | + | Only in Golgi |
| Acetate | Acetyl-CoA | + | + | ER and Golgi location |
| Acyl | Acyl-CoA | ? | ? | Not known |
| Methyl | S-adenosyl-methionine | ? | ? | Not known |
| Phosphate | ATP | + | + | ER and Golgi location |
| Structure | Reagent | Reference | Non-Diverted n = 50 | Diverted n = 35 | p |
|---|---|---|---|---|---|
| Mucins | |||||
| MUC1 | HMFG2 | [345] | strong | no change | NS |
| MUC2 | LUM2-3 | [346] | strong | no change | NS |
| MUC3A/B | EU MUC3 | European Union consortium | weak | no change | NS |
| MUC4 | M4.275 | [347] | strong | no change | NS |
| MUC5AC | 21M1 | [348] | negative | no change | NS |
| MUC5B | LUM5B-2 | [349] | weak | no change | NS |
| MUC12 | M11.123 | [350] | strong | no change | NS |
| MUC13 | M13.234 | [351] | strong | no change | NS |
| Glycosylation | |||||
| Antibodies | |||||
| Tn | IE3 | [352] | weak | no change | NS |
| Sialyl-Tn | TKH2 | [353] | weak | strong | <0.001 |
| Sialyl-Lea | CA19.9 | [354] | weak | strong | <0.01 |
| Sialyl-Lex | AM3 | [355] | strong | strong | NS |
| Sulpho-Lea | 91.9H | [356] | strong | strong | NS |
| Sulpho-Lea | F2 | [357] | strong | strong | NS |
| Sda | KM694 | Kyowa Hakko Kogyo Co. Ltd., Tokyo, Japan | strong | negative | <0.001 |
| O-Acetyl sialic acid 1 | PR3A5 | [358] | strong | strong | NS |
| O-Acetyl sialic acid 2 | 6G4 | [359] | strong | negative | <0.001 |
| Lectins | |||||
| Sialyl-α2-3 | MALII | [360] | strong | strong | NS |
| Sialyl-α2-6 | SNA | [361] | strong | strong | NS |
| GalNAc-protein | VVA | [362] | strong | strong | NS |
| Fucosyl-α1-2 | UEA1 | [363] | weak | weak | NS |
| Fucosyl-α1-6 | AAA | [363] | strong | strong | NS |
| Galβ1-3GalNAc | PNA | [364] | strong | strong | NS |
| Galβ1-4 GlcNAc | MALI | [365] | strong | strong | NS |
| GalNAcα1-3GalAc | DBA | [366] | strong | negative | <0.001 |
| β1-3Gal | |||||
| GalNAcβ1-4 | |||||
| (Neu5Acα2-3)Gal | [367] | ||||
| Patient Group | mPAS | PR3A5 | 6G4 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Positive | Non-Acetylator | Total | Positive | p | Total | Positive | p | |
| UC | 30 | 25 | 5 | 30 | 25 | 25 | |||
| UC div | 17 | 14 | 3 | 17 | 14 | >0.05 | 14 | 0.0003 | |
| CD | 9 | 19 | 0 | 9 | 8 | 9 | |||
| CD div | 17 | 15 | 2 | 17 | 15 | >0.05 | 17 | 0.0112 | |
| Non IBD | 19 | 16 | 3 | 19 | 16 | 9 | 13 | ||
| Non IBD div | 15 | 15 | 0 | 15 | 15 | >0.05 | 15 | 5 | 0.0113 |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corfield, A.P. The Interaction of the Gut Microbiota with the Mucus Barrier in Health and Disease in Human. Microorganisms 2018, 6, 78. https://doi.org/10.3390/microorganisms6030078
Corfield AP. The Interaction of the Gut Microbiota with the Mucus Barrier in Health and Disease in Human. Microorganisms. 2018; 6(3):78. https://doi.org/10.3390/microorganisms6030078
Chicago/Turabian StyleCorfield, Anthony P. 2018. "The Interaction of the Gut Microbiota with the Mucus Barrier in Health and Disease in Human" Microorganisms 6, no. 3: 78. https://doi.org/10.3390/microorganisms6030078





