Molecular Quantification and Genetic Diversity of Toxigenic Fusarium Species in Northern Europe as Compared to Those in Southern Europe
Abstract
:1. Introduction
1.1. Type B Trichothecene Producers and Their Chemotypes

1.2. Historical Background
1.3. Type A Trichothecene Producers
1.4. Genetic Basis of Chemotypes
1.5. The Aim of the Work
2. Experimental Section
2.1. Fusarium Isolates
2.2. Mycotoxin Analyses
2.3. DNA Extraction
2.4. Quantitative PCR
3. Results and Discussion
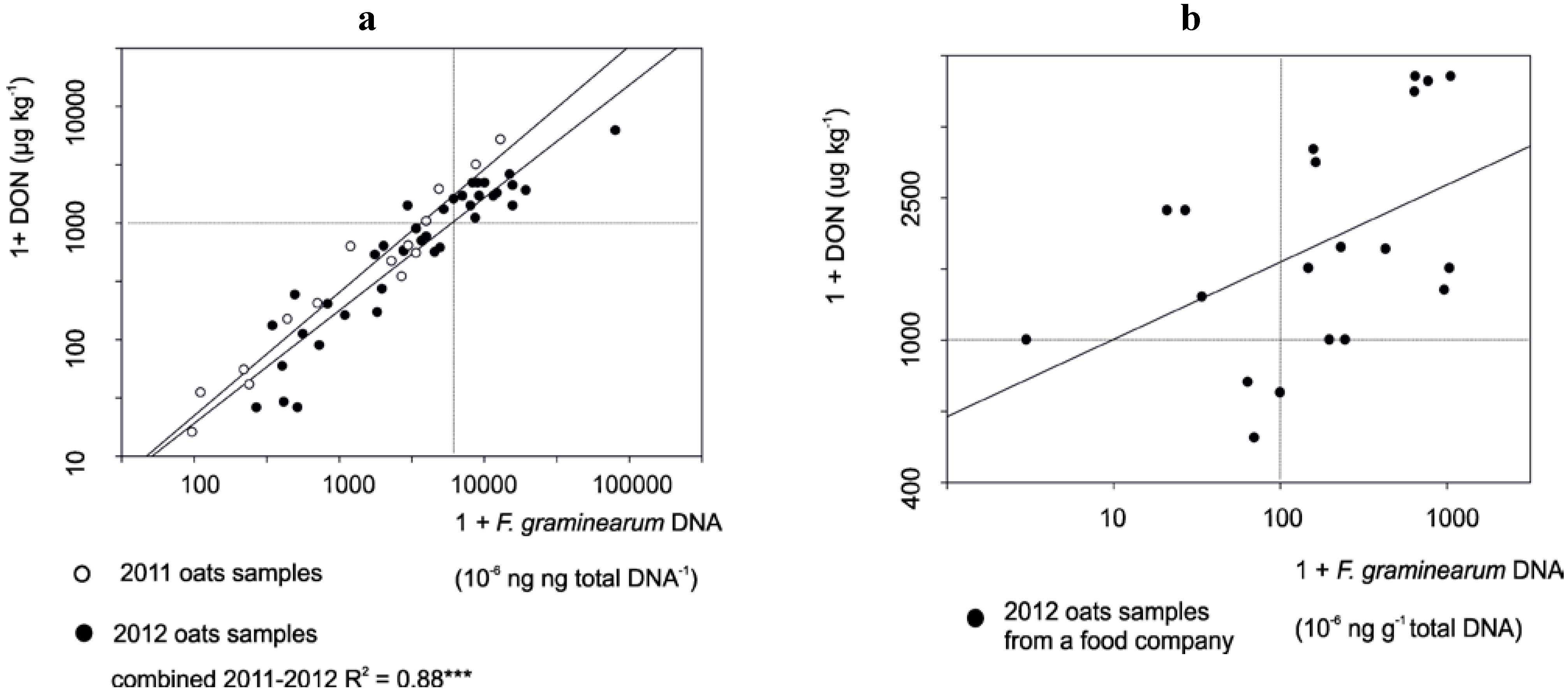
3.1. The Correlation between Mycotoxin and Fusarium DNA Levels in 2010–2012
3.2. Chemotypes of Fusarium Species in Finland during the Years 2010–2012

4. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Conflicts of Interest
References
- Ward, T.J.; Bielawski, J.P.; Kistler, H.C.; Sullivan, E.; O’Donnell, K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. USA 2002, 99, 9278–9283. [Google Scholar]
- Laday, M.; Juhasz, A.; Mule, G.; Moretti, A.; Logrieco, A. Mitochondrial DNA diversity and lineage determination of European isolates of Fusarium graminearum (Gibberella zeae). Eur. J. Plant Pathol. 2004, 110, 545–550. [Google Scholar] [CrossRef]
- Toth, B.; Mesterhazy, A.; Horvath, Z.; Bartok, T.; Varga, M.; Varga, J. Genetic variability of central European isolates of the Fusarium graminearum species complex. Eur. J. Plant Pathol. 2005, 113, 35–45. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Gagkaeva, T.; Ward, T.J.; Aoki, T.; Kistler, H.C.; O’Donnell, K. A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Far East of Russia. Mycologia 2009, 101, 841–852. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Gagkaeva, T. Molecular Chemotyping of Fusarium graminearum, F. culmorum and F. cerealis Isolates from Finland and Russia. In Molecular Identification of Fungi; Gherbawy, Y., Voigt, K., Eds.; Springer Verlag: New York, NY, USA, 2010; pp. 159–177. [Google Scholar]
- Talas, F.; Parzies, H.K.; Miedaner, T. Diversity in genetic structure and chemotype composition of Fusarium graminearum sensu stricto populations causing wheat head blight in individual fields in Germany. Eur. J. Plant Pathol. 2011, 131, 39–48. [Google Scholar] [CrossRef]
- Sarver, B.A.J.; Ward, T.J.; Gale, L.R.; Broz, K.; Kistler, H.C.; Aoki, T.; Nicholson, P.; Carter, J.; O’Donnell, K. Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet. Biol. 2011, 48, 1096–1107. [Google Scholar] [CrossRef]
- Chandler, E.A.; Simpson, D.R.; Sthonsett, M.A.; Nicholson, P. Development of PCR assays to Tri7 and Tri13 trichothecene biosynthetic genes, and characterisation of chemotypes of Fusarium graminearum and Fusarium cerealis. Phys. Mol. Plant Pathol. 2003, 62, 355–367. [Google Scholar] [CrossRef]
- Mert-Türk, F.; Gencer, R. Distribution of the 3-AcDON, 15-AcDON and NIV chemotypes of Fusarium culmorum in the North-West of Turkey. Plant Prot. Sci. 2013, 49, 57–64. [Google Scholar]
- Stepien, L.; Popiel, D.; Koczyk, G.; Chelkowsky, J. Wheat-infecting Fusarium species in Poland—Their chemotypes and frequencies revealed by PCR assay. J. Appl. Genet. 2008, 49, 433–441. [Google Scholar] [CrossRef]
- Ward, T.J.; Clear, R.; Rooney, A.; O’Donnell, K.; Gaba, D.; Patrick, S.; Starkey, D.; Gilbert, J.; Geiser, D.; Nowicki, T. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet. Biol. 2008, 45, 473–484. [Google Scholar] [CrossRef]
- Nielsen, L.K.; Jensen, J.D.; Nielsen, G.C.; Jensen, J.E.; Spliid, N.H.; Thomsen, I.K.; Justesen, A.F.; Collinge, D.B.; Jorgensen, L.N. Fusarium head blight of cereals in Denmark: Species complex and related mycotoxins. Phytopathology 2011, 101, 960–969. [Google Scholar] [CrossRef]
- Hietaniemi, V. Cerveg Database (in Finnish). MTT Agrifood Research Finland: Jokioinen, Finland, 2013. Available online: https://portal.mtt.fi/portal/page/portal/kasper/pelto/peltopalvelut/cerveg (accessed on 26 November 2013).
- Yli-Mattila, T.; Parikka, P.; Lahtinen, T.; Rämö, S.; Kokkonen, M.; Rizzo, A.; Jestoi, M.; Hietaniemi, V. Fusarium DNA Levels in Finnish Cereal Grains. In Current Advances in Molecular Mycology; Gherbawy, Y., Mach, R.L., Rai, M., Eds.; Nova Science Publishers, Inc.: New York, NY, USA, 2009; pp. 107–138. [Google Scholar]
- Yli-Mattila, T.; Paavanen-Huhtala, S.; Parikka, P.; Konstantinova, P.; Gagkaeva, T.; Eskola, M.; Rizzo, A. Occurrence of Fusarium fungi and their toxins in Finnish cereals in 1998 and 2000. J. Appl. Genet. 2002, 43A, 207–214. [Google Scholar]
- Yli-Mattila, T.; Gagkaeva, T. Fusarium Toxins in Cereals in Northern Europe and Asia. In Fungi and Their Applications; Misra, J.K., Tewari, J.P., Deshmukh, S.K., Eds.; CRC press: Boca Raton, FL, USA, 2013; in press. [Google Scholar]
- Mugrabi de Kuppler, A.L.; Steiner, U.; Sulyok, M.; Krska, R.; Oerke, E.-C. Genotyping and phenotyping of Fusarium graminearum isolates from Germany related to their mycotoxin biosynthesis. Int. J. Food Microbiol. 2011, 151, 78–86. [Google Scholar] [CrossRef]
- Jennings, P.; Coates, M.E.; Walsh, K.; Turner, J.A.; Nicholson, P. Determination of deoxynivalenol- and nivalenol-producing chemotypes of Fusarium graminearum isolates from wheat crops in England and Wales. Plant Pathol. 2004, 53, 643–652. [Google Scholar] [CrossRef]
- Pasquali, M.; Giraud, F.; Brochot, C.; Cocco, E.; Hoffmann, L.; Bohn, T. Genetic Fusarium chemotyping as a useful tool for predicting nivalenol contamination in winter wheat. Int. J. Food Microbiol. 2011, 137, 246–253. [Google Scholar]
- Prodi, A.; Salomoni, D.; Bertacchini, E.; Alkadri, D.; Pisi, A.; Tonti, S.; Alberti, I.; Dal Prà, M.; Pancaldi, D.; Covarelli, L.; et al. Determination of deoxynivalenol and nivalenol producing chemotypes of Fusarium graminearum isolated from durum wheat in different Italian regions. Plant Breed. Seed Sci. 2011, 64, 75–80. [Google Scholar]
- Nielsen, L.K.; Jensen, J.D.; Rodríguez, A.; Jørgensen, L.N.; Justesen, A.F. TRI12 based quantitative real-time PCR assays reveal the distribution of trichothecene genotypes of F. graminearum and F. culmorum isolates in Danish small grain cereals. Int. J. Food Microbiol. 2012, 157, 384–392. [Google Scholar] [CrossRef]
- Waalwijk, C.; Kastelein, P.; de Vries, I.; Kerenyi, Z.; van der Lee, T.; Hesselink, T.; Kohl, J.; Kema, G. Major changes in Fusarium spp. in the Netherlands. Eur. J. Plant Pathol. 2003, 109, 743–754. [Google Scholar] [CrossRef]
- Nicholson, P.; Chandler, E.; Draeger, R.C.; Gosman, N.E.; Simpson, D.R.; Thomsett, M.; Wilson, A.H. Molecular tools to study epidemiology and toxicology of Fusarium head blight of cereals. Eur. J. Plant Pathol. 2003, 109, 691–703. [Google Scholar]
- Rainio, A.J. Punahome Fusarium roseum Link—Gibberella saubinetii (Mont.) Sacc. ja sen aiheuttamat myrkytykset kaurassa. In Valtion maatalouskoetoiminnan Julkaisuja; (in Finnish). Valtioneuvoston Kirjap: Helsinki, Finland, 1932; Volumn 50, pp. 1–45. [Google Scholar]
- Uoti, J.; Ylimäki, A. The occurrence of Fusarium species in cereal grain in Finland. Ann. Agric. Fenn. 1974, 13, 5–17. [Google Scholar]
- Ylimäki, A. The Mycoflora of cereal seeds and some feedstuffs. Ann. Agric. Fenn. 1981, 20, 74–78. [Google Scholar]
- Ylimäki, A.; Koponen, H.; Hintikka, E.-L.; Nummi, M.; Niku-Paavola, M.-L.; Ilus, T.; Enari, T.M. Mycoflora and Occurrence of Fusarium Toxins in Finnish Grain. In Materials and Processing Technology; Technical Research Centre of Finland: Espoo, Finland, 1979; Volume 21, pp. 1–28. [Google Scholar]
- Rizzo, A.F. Determination of Major Naturally Occurring Fusarium Toxins in Finnish Grains and Feeds. The Haemolytic Activity of DON and T-2 Toxin, and the Lipid Peroxidation induced in Experimental Animals. Ph.D. Thesis, University of Helsinki, Helsinki, Finland, 24 March 1993. [Google Scholar]
- Eskola, M.; Parikka, P.; Rizzo, A. Trichothecenes, ochratoxin A and zearalenone contamination and Fusarium infection in Finnish cereal samples in 1998. Food Addit. Contam. 2001, 18, 707–718. [Google Scholar]
- Yli-Mattila, T. Ecology and evolution of toxigenic Fusarium species in cereals in northern Europe and Asia. J. Plant Pathol. 2010, 92, 7–18. [Google Scholar]
- Suproniene, S.; Justesen, A.F.; Nicolaisen, M.; Mankeviciene, A.; Dabkevicius, Z.; Semaskiene, R.; Leistrumait, A. Distribution of trichothecene and zearalenone producing Fusarium species in grain of different cereal species and cultivars grown under organic farming conditions in Lithuania. Ann. Agric. Environ. Med. 2010, 17, 73–80. [Google Scholar]
- Yli-Mattila, T.; Rämö, S.; Tanner, R.; Loiveke, H.; Hietaniemi, V. Fusarium DNA levels as compared to mycotoxin levels in Finnish and Estonian grain samples. Plant Breed. Seed Sci. 2011, 64, 131–140. [Google Scholar]
- Aamot, H.U.; Hofgaard, I.S.; Brodal, G.; Ward, T.; Elameen, T.; Vrålstad, T.; Larsen, G.; Clasen, P.E.; Elen, O.; Klemsdal, S. Genetic and Phenotypic Diversity of Fusarium gramienarum and Interactions between Fusarium Species in Oats. In Proceedings of the 12th European Fusarium Seminar, Bordeaux, France, 12–16 May 2013.
- Yoruk, M.; Albayrak, G. Chemotyping of Fusarium graminearum and F. culmorum isolates from Turkey by PCR assay. Mycopathologia 2012, 173, 53–61. [Google Scholar] [CrossRef]
- Davari, M.; Wei, S.H.; Babay-Ahari, A.; Arzanlou, M.; Waalwijk, C.; van der Lee, T.A.J.; Zare, R.; Gerrits van den Ende, A.H.G.; de Hoog, G.S.; van Diepeningen, A.D. Geographic differences in trichothecene chemotypes of Fusarium graminearum in the Northwest and North of Iran. World Mycotoxin J. 2013, 6, 137–150. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Ward, T.; O’Donnell, K.; Proctor, R.H.; Burkin, A.; Kononenko, G.; Gavrilova, O.; Aoki, T.; McCormick, S.P.; Gagkaeva, T. F. sibiricum sp. nov, a novel type A trichothecene-producing Fusarium from northern Asia closely related to F. sporotrichioides and F. langsethiae. Int. J. Food Microbiol. 2011, 147, 58–68. [Google Scholar] [CrossRef]
- Kachuei, R.; Yadegari, M.H.; Rezaie, S.; Allameh, A; Safaie, N.; Zaini, F.; Khanezad Yadzi, F. Investigation of stored wheat mycoflora, reporting the Fusarium cf. langsethiae in three provinces of Iran during 2007. Ann. Microbiol. 2009, 59, 383–390. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Paavanen-Huhtala, S.; Jestoi, M.; Parikka, P.; Hietaniemi, V.; Gagkaeva, T.; Sarlin, T.; Haikara, A.; Laaksonen, S.; Rizzo, A. Real-time PCR detection and quantification of Fusarium poae, F. graminearum, F. sporotrichioides and F. langsethiae in cereal grains in Finland and Russia. Arch. Phytopathol. Plant Prot. 2008, 41, 243–260. [Google Scholar] [CrossRef]
- Pettersson, H. Nivalenol production by Fusarium poae. Mycotoxin Res. 1991, 7A, 26–30. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Paavanen-Huhtala, S.; Parikka, P.; Hietaniemi, V.; Jestoi, M.; Rizzo, A. Real-Time PCR Detection and Quantification of Fusarium poae as Compared to Mycotoxin Production in Grains in Finland. In Proceedings of the 2nd International Symposium on Fusarium Head Blight, Orlando, FL, USA, 11–15 December 2004.
- Sugiura, Y.; Fukasaku, K.; Tanaka, T.; Matsui, Y.; Ueno, Y. Fusarium poae and Fusarium crookwellense, fungi responsible for the natural occurrence of nivalenol in Hokkaido. Appl. Environ. Microbiol. 1993, 59, 3334–3338. [Google Scholar]
- Pettersson, H.; Hedman, R.; Engstrom, B.; Elwinger, K.; Fossum, O. Nivalenol in Swedish cereals—Occurrence, production and toxicity towards chickens. Food Addit. Contam. 1995, 12, 373–376. [Google Scholar] [CrossRef]
- Alexander, N.J.; Proctor, R.H.; McCormick, S.P. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusariu. Toxin Rev. 2009, 28, 198–215. [Google Scholar] [CrossRef]
- McCormick, S.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: From simple to complex mycotoxins. Toxins 2011, 3, 802–814. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Waalwijk, C.; van der Lee, T.; Proctor, R.H. The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium. Fungal Genet. Biol. 2011, 38, 485–495. [Google Scholar]
- Hietaniemi, V.; Kontturi, M.; Rämö, S.; Eurola, M.; Kangas, A.; Niskanen, M.; Saastamoinen, M. Contents of trichothecenes in oats during official variety, organic cultivation and nitrogen fertilization trials in Finland. Agric. Food Sci. 2004, 13, 54–67. [Google Scholar] [CrossRef]
- Halstensen, A.S.; Nordby, K.C.; Eduard, W.; Klemsdal, S.S. Real-time PCR detection of toxigenic Fusarium in airborne and settled grain dust and associations with trichothecene mycotoxins. J. Environ. Monitor. 2006, 8, 1235–1241. [Google Scholar] [CrossRef]
- Waalwijk, C.; van der Heide, R.; de Vries, I.; van der Lee, T.; Schoen, C.; Costrel-de Corainville, G.; Häuser-Hahn, I.; Kastelein, P.; Köhl, J.; Lonnet, P.; et al. Quantitative detection of Fusarium species in wheat using TaqMan. Eur. J. Plant Pathol. 2004, 110, 481–494. [Google Scholar] [CrossRef]
- Rauvola, M.; Hovinen, T.; Hietaniemi, V.; Kaitaranta, J.; Rämö, S. Screening Deoxynivalenol in Oat using a Quickmethod with Comparison to a Quantitative GC-MS Analysis. In Proceedings of the 12th European Fusarium Seminar, Bordeaux, France, 12–16 May 2013; p. 127.
- Rauvola, M. Determination of deoxynivalenol in grain by semi-quantitative quick test. Bachelor’s Thesis, Turku University of Applied Sciences, Turku, Finland, 2013. [Google Scholar]
- Hakulin, S. Miten homemyrkkyjä jaetaan viljaketjussa? Proceedings of the Producer Seminar in Hämeenlinna, 31 January 2013; Available online: http://www.vyr.fi/www/fi/tapahtumat/menneet_tapahtumat/viljelijaseminaari_31012013.php (accessed on 26 November 2013). (in Finnish).
- European Commission. Commission Regulation (EC) No. 1881/2006 setting maximum levels of certain contaminants in foodstuffs. Off. J. Eur. Union 2006, L364, 5–24. [Google Scholar]
- Gagkaeva, T.; Gavrilova, O.P.; Yli-Mattila, T.; Loskutov, I.G. Sources of resistance to Fusarium head blight in VIR oat collection. Euphytica 2013, 3, 355–364. [Google Scholar]
- Rämö, S.; Hietaniemi, V.; Parikka, P.; Hankoniemi, J. Lajittelu ja kuorinta vähentävät viljojen hometoksiineja. (in Finnish). Maaseudun Tiede. 2008. Available online: http://www.mtt.fi/maaseuduntiede/pdf/mtt-mt-v65n03s16a.pdf (accessed on 26 November 2013).
- Parikka, P.; Rämö, S.; Hietaniemi, V. Fusarium infection and mycotoxins in Finnish cereals in 2005–2006. J. Plant Pathol. 2008, 90, 56–57. [Google Scholar]
- Parikka, P.; Hakala, K.; Tiilikkala, K. Expected shifts in Fusarium species’ composition on cereal grain in Northern Europe due to climatic change. Food Addit. Contam. 2012, 29, 1543–1555. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yli-Mattila, T.; Rämö, S.; Hietaniemi, V.; Hussien, T.; Carlobos-Lopez, A.L.; Cumagun, C.J.R. Molecular Quantification and Genetic Diversity of Toxigenic Fusarium Species in Northern Europe as Compared to Those in Southern Europe. Microorganisms 2013, 1, 162-174. https://doi.org/10.3390/microorganisms1010162
Yli-Mattila T, Rämö S, Hietaniemi V, Hussien T, Carlobos-Lopez AL, Cumagun CJR. Molecular Quantification and Genetic Diversity of Toxigenic Fusarium Species in Northern Europe as Compared to Those in Southern Europe. Microorganisms. 2013; 1(1):162-174. https://doi.org/10.3390/microorganisms1010162
Chicago/Turabian StyleYli-Mattila, Tapani, Sari Rämö, Veli Hietaniemi, Taha Hussien, Ana Liza Carlobos-Lopez, and Christian Joseph R. Cumagun. 2013. "Molecular Quantification and Genetic Diversity of Toxigenic Fusarium Species in Northern Europe as Compared to Those in Southern Europe" Microorganisms 1, no. 1: 162-174. https://doi.org/10.3390/microorganisms1010162



