The Microenvironment in Epstein–Barr Virus-Associated Malignancies
Abstract
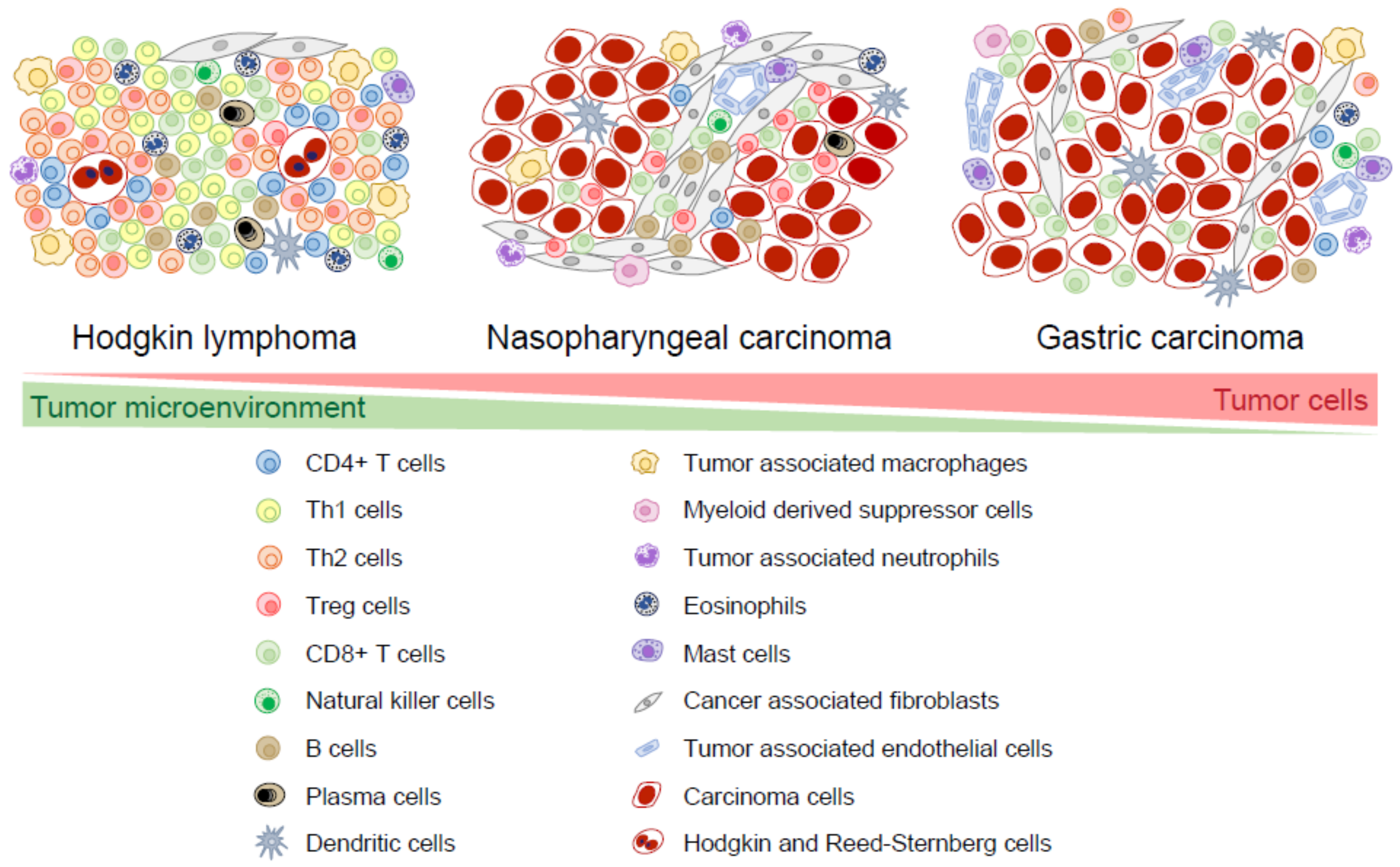
:1. Introduction
2. Composition of the Microenvironment
2.1. T Cells
2.2. Natural Killer Cells
2.3. B cells and Plasma Cells
2.4. Dendritic Cells
2.5. Tumor-Associated Macrophages
2.6. Myeloid-Derived Suppressor Cells
2.7. Granulocytic Cells
2.8. Cancer-Associated Fibroblasts
2.9. Endothelial Cells
3. Immune Escape Mechanisms
3.1. Latency Expression Patterns
3.2. Expression of HLA
3.3. Co-Stimulation
3.4. Excretion of Immunosuppressive Agents
3.5. Innate Immune Responses
4. Tumor Cell Promoting Mechanisms
4.1. Stimulation of NF-κB
4.2. Stimulation of JAK/STAT
4.3. Cytokine Receptors
4.4. Stimulation of MAPK/ERK
5. Susceptibility to EBV-Associated Malignancies
5.1. Genetic Associations in EBV+ HL
5.2. Genetic Associations in NPC
5.3. Genetic Associations in Gastric Carcinoma
6. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. (Eds.) WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; IARC Press: Lyon, France, 2017; ISBN 9789283244943. [Google Scholar]
- El-Naggar, A.K.; Chan, J.K.C.; Grandis, J.R.; Takata, T.S.P. (Eds.) WHO Classification of Head and Neck Tumours, 4th ed.; IARC Press: Lyon, France, 2017; ISBN 9789283224389. [Google Scholar]
- Shibata, D.; Weiss, L.M. Epstein-Barr virus-associated gastric adenocarcinoma. Am. J. Pathol. 1992, 140, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Yap, L.F.; Murray, P.G. Epstein–Barr virus: More than 50 years old and still providing surprises. Nat. Rev. Cancer 2016, 16, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W.; Gascoyne, R.D. The tumour microenvironment in B cell lymphomas. Nat. Publ. Gr. 2014, 14, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Van Beek, J.; Zur Hausen, A.; Snel, S.N.; Berkhof, J.; Kranenbarg, E.K.; Van De Velde, C.J.H.; Van Den Brule, A.J.C.; Middeldorp, J.M.; Meijer, C.J.L.M.; Bloemena, E. Morphological evidence of an activated cytotoxic T-cell infiltrate in EBV-positive gastric carcinoma preventing lymph node metastases. Am. J. Surg. Pathol. 2006, 30, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Alvaro, T. Outcome in Hodgkin’s Lymphoma Can Be Predicted from the Presence of Accompanying Cytotoxic and Regulatory T Cells. Clin. Cancer Res. 2005, 11, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Morales, O.; Mrizak, D.; François, V.; Mustapha, R.; Miroux, C.; Depil, S.; Decouvelaere, A.V.; Lionne-Huyghe, P.; Auriault, C.; de Launoit, Y.; et al. Epstein-Barr virus infection induces an increase of T regulatory type 1 cells in Hodgkin lymphoma patients. Br. J. Haematol. 2014, 166, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Marshall, N.A.; Christie, L.E.; Munro, L.R.; Culligan, D.J.; Johnston, P.W.; Barker, R.N.; Vickers, M.A. Immunosuppressive regulatory T cells are abundant in the reactive lymphocytes of Hodgkin lymphoma. Blood 2004, 103, 1755–1762. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, A.; Visser, L.; Poppema, S. High Expression of the CC Chemokine TARC in Reed-Sternberg Cells. Am. J. Pathol. 1999, 154, 1685–1691. [Google Scholar] [CrossRef]
- Baumforth, K.R.N.; Birgersdotter, A.; Reynolds, G.M.; Wei, W.; Kapatai, G.; Flavell, J.R.; Kalk, E.; Piper, K.; Lee, S.; Machado, L.; et al. Expression of the Epstein-Barr virus-encoded Epstein-Barr virus nuclear antigen 1 in Hodgkin’s lymphoma cells mediates up-regulation of CCL20 and the migration of regulatory T cells. Am. J. Pathol. 2008, 173, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Ishii, T.; Inagaki, A.; Yano, H.; Komatsu, H.; Iida, S.; Inagaki, H.; Ueda, R. Specific recruitment of CC chemokine receptor 4-positive regulatory T cells in Hodgkin lymphoma fosters immune privilege. Cancer Res. 2006, 66, 5716–5722. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Sattarzadeh, A.; Rutgers, B.; Diepstra, A.; Van Den Berg, A.; Visser, L. The microenvironment of classical Hodgkin lymphoma: Heterogeneity by Epstein-Barr virus presence and location within the tumor. Blood Cancer J. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.K.; Lambley, E.; Duraiswamy, J.; Dua, U.; Smith, C.; Elliott, S.; Gill, D.; Marlton, P.; Seymour, J.; Khanna, R. Expression of LAG-3 by tumor-infiltrating lymphocytes is coincident with the suppression of latent membrane antigen—Specific CD8 ϩ T-cell function in Hodgkin lymphoma patients. October 2006, 108, 2280–2289. [Google Scholar] [CrossRef]
- Barros, M.H.M.; Vera-Lozada, G.; Soares, F.A.; Niedobitek, G.; Hassan, R. Tumor microenvironment composition in pediatric classical Hodgkin lymphoma is modulated by age and Epstein-Barr virus infection. Int. J. Cancer 2012, 131, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Teichmann, M.; Meyer, B.; Beck, A.; Niedobitek, G. Expression of the interferon-inducible chemokine IP-10 (CXCL10), a chemokine with proposed anti-neoplastic functions, in Hodgkin lymphoma and nasopharyngeal carcinoma. J. Pathol. 2005, 206, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Teruya-Feldstein, J.; Tosato, G.; Jaffe, E.S. The Role of Chemokines in Hodgkin’s Disease. Leuk. Lymphoma 2000, 38, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.T.; Sheen, T.S.; Chen, C.L.; Lu, J.; Chang, Y.; Chen, J.Y.; Tsai, C.H. Profile of cytokine expression in nasopharyngeal carcinomas: A distinct expression of interleukin 1 in tumor and CD4+ T cells. Cancer Res. 1999, 59, 1599–1605. [Google Scholar] [PubMed]
- Zong, Y.S.; Lin, H.; Choy, D.T.K.; Sham, J.S.T.; Wei, W.; Chan, K.H.; Ng, M.H. Nasopharyngeal Carcinoma and Lymphoinfiltration. Oncology 1991, 48, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Ferradini, L.; Miescher, S.; Stoeck, M.; Busson, P.; Barras, C.; Cerf-Bensussan, N.; Lipinski, M.; von Fliedner, V.; Tursz, T. Cytotoxic potential despite impaired activation pathways in T lymphocytes infiltrating nasopharyngeal carcinoma. Int. J. Cancer 1991, 47, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Li, J.; Mo, H.Y.; Qiu, F.; Zheng, L.M.; Qian, C.N.; Zeng, Y.X. Different subsets of tumor infiltrating lymphocytes correlate with NPC progression in different ways. Mol. Cancer 2010, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hsin, L.-J.; Kao, H.-K.; Chen, I.-H.; Tsang, N.-M.; Hsu, C.-L.; Liu, S.-C.; Chang, Y.-S.; Chang, K.-P. Serum CXCL9 Levels Are Associated with Tumor Progression and Treatment Outcome in Patients with Nasopharyngeal Carcinoma. PLoS ONE 2013, 8, e80052. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Ou, N.; Li, C.; Lu, A.; Li, J.; Ma, L.; Zhong, W.; Gao, J.; Zheng, Y.; Cai, Y. Expression and Prognostic Significance of Macrophage Inflammatory Protein-3 Alpha and Cystatin A in Nasopharyngeal Carcinoma. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Mrizak, D.; Martin, N.; Barjon, C.; Jimenez-Pailhes, A.-S.; Mustapha, R.; Niki, T.; Guigay, J.; Pancré, V.; de Launoit, Y.; Busson, P.; et al. Effect of Nasopharyngeal Carcinoma-Derived Exosomes on Human Regulatory T Cells. JNCI J. Natl. Cancer Inst. 2015, 107, 363. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Pazolt, D.; Grabenbauer, G.G.; Nicholls, J.M.; Herbst, H.; Young, L.S.; Niedobitek, G. Expression of cytokine and chemokine genes in Epstein-Barr virus- associated nasopharyngeal carcinoma: Comparison with Hodgkin’s disease. J. Pathol. 2001, 194, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Haas, M.; Büttner, M.; Rau, T.T.; Fietkau, R.; Grabenbauer, G.G.; Distel, L.V. Inflammation in gastric adenocarcinoma of the cardia: How do EBV infection, Her2 amplification and cancer progression influence tumor-infiltrating lymphocytes? Virchows Arch. 2011, 458, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Feichtenbeiner, A.; Haas, M.; Büttner, M.; Grabenbauer, G.G.; Fietkau, R.; Distel, L.V. Critical role of spatial interaction between CD8+ and Foxp3 + cells in human gastric cancer: The distance matters. Cancer Immunol. Immunother. 2014, 63, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Kijima, Y.; Ishigami, S.; Hokita, S.; Koriyama, C.; Akiba, S.; Eizuru, Y.; Aikou, T. The comparison of the prognosis between Epstein-Barr virus (EBV)-positive gastric carcinomas and EBV-negative ones. Cancer Lett. 2003, 200, 33–40. [Google Scholar] [CrossRef]
- Chiaravalli, A.M.; Feltri, M.; Bertolini, V.; Bagnoli, E.; Furlan, D.; Cerutti, R.; Novario, R.; Capella, C. Intratumour T cells, their activation status and survival in gastric carcinomas characterised for microsatellite instability and Epstein-Barr virus infection. Virchows Arch. 2006, 448, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, J.; Hao, Y.; Nie, Y.; Li, Z.; Qian, M.; Liang, Q.; Yu, J.; Zeng, M.; Wu, K. Differentiated tumor immune microenvironment of Epstein–Barr virus-associated and negative gastric cancer: Implication in prognosis and immunotherapy. Oncotarget 2017, 8, 67094–67103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Chen, J.; Xiao, L.; Tang, F.; Zhang, Z.; Zhang, Y.; Feng, Z.; Jiang, Y.; Shao, C. Accumulation Mechanisms of CD4(+)CD25(+)FOXP3(+) Regulatory T Cells in EBV-associated Gastric Carcinoma. Sci. Rep. 2015, 5, 18057. [Google Scholar] [CrossRef] [PubMed]
- Strong, M.J.; Xu, G.; Coco, J.; Baribault, C.; Vinay, D.S.; Lacey, M.R.; Strong, A.L.; Lehman, T.A.; Seddon, M.B.; Lin, Z.; et al. Differences in Gastric Carcinoma Microenvironment Stratify According to EBV Infection Intensity: Implications for Possible Immune Adjuvant Therapy. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, H.; Nakayama, T.; Yoshie, O. In situ expression of the CCL20-CCR6 axis in lymphocyte-rich gastric cancer and its potential role in the formation of lymphoid stroma. Pathol. Int. 2011, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chen, X.-M.; Huang, H.-R.; Zhao, F.-P.; Wang, F.; Liu, X.; Li, X.-P. Detailed analysis of inflammatory cell infiltration and the prognostic impact on nasopharyngeal carcinoma. Head Neck 2018. [Google Scholar] [CrossRef] [PubMed]
- Miao, B.P.; Zhang, R.S.; Li, M.; Fu, Y.T.; Zhao, M.; Liu, Z.G.; Yang, P.C. Nasopharyngeal cancer-derived microRNA-21 promotes immune suppressive B cells. Cell. Mol. Immunol. 2015, 12, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Agathanggelou, A.; Niedobitek, G.; Chen, R.; Nicholls, J.; Yin, W.; Young, L.S. Expression of immune regulatory molecules in Epstein-Barr virus-associated nasopharyngeal carcinomas with prominent lymphoid stroma. Evidence for a functional interaction between epithelial tumor cells and infiltrating lymphoid cells. Am. J. Pathol. 1995, 147, 1152–1160. [Google Scholar] [PubMed]
- Braz-Silva, P.H.; Vitale, S.; Butori, C.; Guevara, N.; Santini, J.; Magalhães, M.; Hofman, P.; Doglio, A. Specific infiltration of langerin-positive dendritic cells in EBV-infected tonsil, Hodgkin lymphoma and nasopharyngeal carcinoma. Int. J. Cancer 2011, 128, 2501–2508. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Zhang, C.; Zhang, F.; Kuan, J.; Chen, M.; Feng, K.; Yu, Z. Infiltrating Lymphocytes and Accessory Cells in Nasopharyngeal Carcinoma 1. Jpn. J. Cancer Res. 1993, 84, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Giannini, A.; Bianchi, S.; Messerini, L.; Gallo, O.; Gallina, E.; Asprella Libonati, G.; Olmi, P.; Zampi, G. Prognostic Significance of Accessory Cells and Lymphocytes in Nasopharyngeal Carcinoma. Pathol. Res. Pract. 1991, 187, 496–502. [Google Scholar] [CrossRef]
- Chapel, F.; Fabiani, B.; Davi, F.; Raphael, M.; Tepper, M.; Champault, G.; Guettier, C. Epstein-Barr virus and gastric carcinoma in Western patients: Comparison of pathological parameters and p53 expression in EBV-positive and negative tumours. Histopathology 2000, 36, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.C.; Yip, T.C.; Ngan, K.C.; Cheng, W.W.; Law, C.K.; Chan, P.S.; Chan, K.C.; Wong, C.K.C.; Wong, R.N.S.; Lo, K.W.; et al. Role of MIF/CXCL8/CXCR2 signaling in the growth of nasopharyngeal carcinoma tumor spheres. Cancer Lett. 2013, 335, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.F.; Tan, S.Y.; Chan, S.H.; Chong, S.M.; Loh, K.S.; Tan, L.K.S.; Hu, H. A distinct expression of CC chemokines by macrophages in nasopharyngeal carcinoma: Implication for the intense tumor infiltration by T lymphocytes and macrophages. Hum. Pathol. 2001, 32, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Hollander, P.; Rostgaard, K.; Smedby, K.E.; Molin, D.; Loskog, A.; de Nully Brown, P.; Enblad, G.; Amini, R.M.; Hjalgrim, H.; Glimelius, I. An anergic immune signature in the tumor microenvironment of classical Hodgkin lymphoma is associated with inferior outcome. Eur. J. Haematol. 2017, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Zameer, M.A.L.; Premalata, C.S.; Arunakumari, B.; Appaji, L.; Rama Rao, C. Pediatric hodgkin lymphoma in a south indian regional cancer center: Its immunomorphology, tumor-Associated macrophages, and association with epstein-barr virus. Pediatr. Hematol. Oncol. 2015, 32, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Kamper, P.; Bendix, K.; Hamilton-Dutoit, S.; Honore, B.; Nyengaard, J.R.; D’Amore, F. Tumor-infiltrating macrophages correlate with adverse prognosis and Epstein-Barr virus status in classical Hodgkin’s lymphoma. Haematologica 2011, 96, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Barros, M.H.M.; Hassan, R.; Niedobitek, G. Tumor-associated macrophages in pediatric classical Hodgkin lymphoma: Association with Epstein-Barr virus, lymphocyte subsets, and prognostic impact. Clin. Cancer Res. 2012, 18, 3762–3771. [Google Scholar] [CrossRef] [PubMed]
- Barros, M.H.M.; Segges, P.; Vera-Lozada, G.; Hassan, R.; Niedobitek, G. Macrophage polarization reflects T cell composition of tumor microenvironment in pediatric classical Hodgkin lymphoma and has impact on survival. PLoS ONE 2015, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ooft, M.L.; van Ipenburg, J.A.; Sanders, M.E.; Kranendonk, M.; Hofland, I.; de Bree, R.; Koljenović, S.; Willems, S.M. Prognostic role of tumour-associated macrophages and regulatory T cells in EBV-positive and EBV-negative nasopharyngeal carcinoma. J. Clin. Pathol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Ding, T.; Zheng, L.-M.; Shao, J.-Y. Influence of tumor-associated macrophages on progression and prognosis of nasopharyngeal carcinoma. Ai Zheng 2006, 25, 1340–1345. [Google Scholar] [PubMed]
- Huang, H.; Liu, X.; Zhao, F.; Lu, J.; Zhang, B.; Peng, X.; Zhang, M.; Chen, X.; Li, G.; Li, X. M2-polarized tumour-associated macrophages in stroma correlate with poor prognosis and Epstein–Barr viral infection in nasopharyngeal carcinoma. Acta Otolaryngol. 2017, 137, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Buettner, M.; Meyer, B.; Schreck, S.; Niedobitek, G. Expression of RANTES and MCP-1 in epithelial cells is regulatedvia LMP1 and CD40. Int. J. Cancer 2007, 121, 2703–2710. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, T.; Abe, H.; Morikawa, T.; Yamashita, H.; Ishikawa, S.; Ushiku, T.; Seto, Y.; Fukayama, M. Low density of CD204-positive M2-type tumor-associated macrophages in Epstein-Barr virus–associated gastric cancer: A clinicopathologic study with digital image analysis. Hum. Pathol. 2016, 56, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Marini, O.; Spina, C.; Mimiola, E.; Cassaro, A.; Malerba, G.; Todeschini, G.; Perbellini, O.; Scupoli, M.; Carli, G.; Facchinelli, D.; et al. Identification of granulocytic myeloid-derived suppressor cells (G-MDSCs) in the peripheral blood of Hodgkin and non-Hodgkin lymphoma patients. Oncotarget 2016, 7, 27676–27688. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Parrinello, N.L.; Vetro, C.; Forte, S.; Chiarenza, A.; Figuera, A.; Motta, G.; Palumbo, G.A.; Ippolito, M.; Consoli, U.; et al. Di Circulating myeloid-derived suppressor cells correlate with clinical outcome in Hodgkin Lymphoma patients treated up-front with a risk-adapted strategy. Br. J. Haematol. 2015, 168, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-L.; Ye, S.-B.; OuYang, L.-Y.; Zhang, H.; Chen, Y.-S.; He, J.; Chen, Q.-Y.; Qian, C.-N.; Zhang, X.-S.; Cui, J.; et al. COX-2 promotes metastasis in nasopharyngeal carcinoma by mediating interactions between cancer cells and myeloid-derived suppressor cells. Oncoimmunology 2015, 4, e1044712. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.C.; Liu, L.Q.; Wang, B.H.; Wang, C.M.; Ma, L.; Cao, W.M.; Dai, E.X. Effect of concurrent chemoradiotherapy and radiotherapy alone on peripheral myeloid-derived suppressor and T regulatory cells in patients with nasopharyngeal cancer. Zhonghua Zhong Liu Za Zhi 2017, 39, 579–583. [Google Scholar] [PubMed]
- Gabitass, R.F.; Annels, N.E.; Stocken, D.D.; Pandha, H.A.; Middleton, G.W. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol. Immunother. 2011, 60, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Shoji, H.; Tada, K.; Kitano, S.; Nishimura, T.; Shimada, Y.; Nagashima, K.; Aoki, K.; Hiraoka, N.; Honma, Y.; Iwasa, S.; et al. The peripheral immune status of granulocytic myeloid-derived suppressor cells correlates the survival in advanced gastric cancer patients receiving cisplatin-based chemotherapy. Oncotarget 2017, 8, 95083–95094. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chang, E.W.Y.; Wong, S.C.; Ong, S.-M.; Chong, D.Q.Y.; Ling, K.L. Increased Myeloid-Derived Suppressor Cells in Gastric Cancer Correlate with Cancer Stage and Plasma S100A8/A9 Proinflammatory Proteins. J. Immunol. 2013, 190, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Ha, S.Y.; Kim, H.-M.; Ahn, S.M.; Kang, M.-S.; Kim, K.-M.; Choi, M.G.; Lee, J.H.; Sohn, T.S.; Bae, J.M.; et al. The prognostic effects of tumor infiltrating regulatory T cells and myeloid derived suppressor cells assessed by multicolor flow cytometry in gastric cancer patients. Oncotarget 2016, 7, 7940–7951. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Li, J.; Liu, S.-M.; Feng, X.-Y.; Chen, S.; Chen, Y.-B.; Zhang, X.-S. CD33+/p-STAT1+ double-positive cell as a prognostic factor for stage IIIa gastric cancer. Med. Oncol. 2013, 30, 442. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.T.; Ye, S.B.; Liu, Y.N.; He, J.; Chen, Q.Y.; Mai, H.Q.; Zhang, C.X.; Cui, J.; Zhang, X.S.; Busson, P.; et al. LMP1-mediated glycolysis induces myeloid-derived suppressor cell expansion in nasopharyngeal carcinoma. PLoS Pathog. 2017, 13, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Wang, L.J.; Tsang, N.M.; Ojcius, D.M.; Chen, C.C.; Ouyang, C.N.; Hsueh, C.; Liang, Y.; Chang, K.P.; Chen, C.C.; et al. Tumour inflammasome-derived IL-1β recruits neutrophils and improves local recurrence-free survival in EBV-induced nasopharyngeal carcinoma. EMBO Mol. Med. 2012, 4, 1276–1293. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Morikawa, T.; Saito, R.; Yamashita, H.; Seto, Y.; Fukayama, M. In Epstein–Barr virus-associated gastric carcinoma a high density of CD66b-positive tumor-associated neutrophils is associated with intestinal-type histology and low frequency of lymph node metastasis. Virchows Arch. 2016, 468, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Samoszuk, M. lnvited Revie w Eosinophils and human cancer. Histol. Histopathol. 1987, 12, 807–881. [Google Scholar]
- Glimelius, I.; Rubin, J.; Rostgaard, K.; Amini, R.M.; Simonsson, M.; Sorensen, K.M.; Smedby, K.E.; Venge, P.; Hjalgrim, H.; Molin, D.; et al. Predictors of histology, tissue eosinophilia and mast cell infiltration in Hodgkin’s Lymphoma—A population-based study. Eur. J. Haematol. 2011, 87, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Von Wasielewski, R.; Seth, S.; Franklin, J.; Fischer, R.; Hübner, K.; Hansmann, M.L.; Diehl, V.; Georgii, A. Tissue eosinophilia correlates strongly with poor prognosis in nodular sclerosing Hodgkin’s disease, allowing for known prognostic factors. Blood 2000, 95, 1207–1213. [Google Scholar] [PubMed]
- Looi, L.-M. Tumor-associated tissue eosinophilia in nasopharyngeal carcinoma. A pathologic study of 422 primary and 138 metastatic tumors. Cancer 1987, 59, 466–470. [Google Scholar] [CrossRef]
- Liu, C.M.; Ko, J.J.; Shun, C.T.; Hsiao, T.Y.; Sheen, T.S. Soluble adhesion molecules and cytokines in tumor-associated tissue eosinophilia of nasopharyngeal carcinoma. Acta Otolaryngol. 2001, 121, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Yamashita, T.; Ishiguro, R.; Tashiro, M. Significance of epidermal growth factor receptor and tumor associated tissue eosinophilia in the prognosis of patients with nasopharyngeal carcinoma. Auris Nasus Larynx 2002, 29, 175–181. [Google Scholar] [CrossRef]
- Caruso, R.A.; Parisi, A.; Quattrocchi, E.; Scardigno, M.; Branca, G.; Parisi, C.; Lucianò, R.; Paparo, D.; Fedele, F. Ultrastructural descriptions of heterotypic aggregation between eosinophils and tumor cells in human gastric carcinomas. Ultrastruct. Pathol. 2011, 35, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.D.; Kamper, P.; Nielsen, P.S.; Bendix, K.; Riber-Hansen, R.; Steiniche, T.; Hamilton-Dutoit, S.; Clausen, M.; d’Amore, F. Tumour-associated mast cells in classical Hodgkin’s lymphoma: Correlation with histological subtype, other tumour-infiltrating inflammatory cell subsets and outcome. Eur. J. Haematol. 2016, 96, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Molin, D.; Edstrom, A.; Glimelius, I.; Glimelius, B.; Nilsson, G.; Sundstrom, C.; Enblad, G. Mast cell infiltration correlates with poor prognosis in Hodgkin’s lymphoma. Br. J. Haematol. 2002, 119, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Hügle, T. Beyond allergy: The role of mast cells in fibrosis. Swiss Med. Wkly. 2014, 144, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, X.; Zhao, F.; Huang, H.; Lu, J.; Liu, X. Distribution and prognostic significance of tumor-infiltrating mast cells in nasopharyngeal carcinoma. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 2015, 50, 306–311. [Google Scholar] [PubMed]
- Ribatti, D.; Guidolin, D.; Marzullo, A.; Nico, B.; Annese, T.; Benagiano, V.; Crivellato, E. Mast cells and angiogenesis in gastric carcinoma. Int. J. Exp. Pathol. 2010, 91, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Ammendola, M.; Sacco, R.; Zuccalà, V.; Luposella, M.; Patruno, R.; Gadaleta, P.; Zizzo, N.; Gadaleta, C.D.; De Sarro, G.; Sammarco, G.; et al. Mast cells density positive to tryptase correlate with microvascular density in both primary gastric cancer tissue and loco-regional lymph node metastases from patients that have undergone radical surgery. Int. J. Mol. Sci. 2016, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ma, N.; Kawanishi, S.; Hiraku, Y.; Oikawa, S.; Xie, Y.; Zhang, Z.; Huang, G.; Murata, M. Relationships of alpha-SMA-positive fibroblasts and SDF-1-positive tumor cells with neoangiogenesis in nasopharyngeal carcinoma. Biomed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yang, P.; Xiao, Y.; Zhang, Y.; Liu, J.; Xie, D.; Cai, M.; Zhang, X. Overexpression of α-sma-positive fibroblasts (CAFs) in Nasopharyngeal Carcinoma Predicts Poor Prognosis. J. Cancer 2017, 8, 3897–3902. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.F.; Zhang, L.H.; Shan, L.H.; Sun, W.G.; Chai, C.C.; Wu, H.M.; Ibla, J.C.; Wang, L.F.; Liu, J.R. Effects of the fibroblast activation protein on the invasion and migration of gastric cancer. Exp. Mol. Pathol. 2013, 95, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.M.; Fusenig, N.E. Friends or foes - Bipolar effects of the tumour stroma in cancer. Nat. Rev. Cancer 2004, 4, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Michiels, C. Endothelial cell functions. J. Cell. Physiol. 2003, 196, 430–443. [Google Scholar] [CrossRef] [PubMed]
- Doussis-Anagnostopoulou, I.A.; Talks, K.L.; Turley, H.; Debnam, P.; Tan, D.C.; Mariatos, G.; Gorgoulis, V.; Kittas, C.; Gatter, K.C. Vascular endothelial growth factor (VEGF) is expressed by neoplastic Hodgkin-Reed-Sternberg cells in Hodgkin’s disease. J. Pathol. 2002, 197, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Murono, S.; Inoue, H.; Tanabe, T.; Joab, I.; Yoshizaki, T.; Furukawa, M.; Pagano, J.S. Induction of cyclooxygenase-2 by Epstein-Barr virus latent membrane protein 1 is involved in vascular endothelial growth factor production in nasopharyngeal carcinoma cells. Proc. Natl. Acad. Sci. USA 2001, 98, 6905–6910. [Google Scholar] [CrossRef] [PubMed]
- Fhu, C.W.; Graham, A.M.; Yap, C.T.; Al-Salam, S.; Castella, A.; Chong, S.M.; Lim, Y.C. Reed-Sternberg cell-derived lymphotoxin-alpha activates endothelial cells to enhance T-cell recruitment in classical Hodgkin lymphoma. Blood 2014, 124, 2973–2982. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Y.; Zeng, S.; Hu, X. LMP1 expression is positively associated with metastasis of nasopharyngeal carcinoma: Evidence from a meta-analysis. J. Clin. Pathol. 2012, 65, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.; Oliveira, C.; Sousa, H. Epstein—Barr virus gene expression and latency pattern in gastric carcinomas: A systematic review. Future Oncol. 2017, 13, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yang, L.; Middeldorp, J.; Sheen, T.S.; Chen, J.Y.; Fukayama, M.; Eizuru, Y.; Ooka, T.; Takada, K. Epstein-Barr virus (EBV)-encoded BARF1 gene is expressed in nasopharyngeal carcinoma and EBV-associated gastric carcinoma tissues in the absence of lytic gene expression. J. Med. Virol. 2005, 76, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Brink, A.A.T.P.; Vervoort, M.B.H.J.; Middeldorp, J.M.; Meijer, C.J.L.M.; Van Den Brule, A.J.C. Nucleic acid sequence-based amplification, a new method for analysis of spliced and unspliced Epstein-Barr virus latent transcripts, and its comparison with reverse transcriptase PCR. J. Clin. Microbiol. 1998, 36, 3164–3169. [Google Scholar] [PubMed]
- Blake, N.; Lee, S.; Redchenko, I.; Thomas, W.; Steven, N.; Leese, A.; Steigerwald-Mullen, P.; Kurilla, M.G.; Frappier, L.; Rickinson, A. Human CD8+T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity 1997, 7, 791–802. [Google Scholar] [CrossRef]
- Jones, K.; Nourse, J.P.; Morrison, L.; Nguyen-Van, D.; Moss, D.J.; Burrows, S.R.; Gandhi, M.K. Expansion of EBNA1-specific effector T cells in posttransplantation lymphoproliferative disorders. Blood 2010, 116, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Long, H.M.; Chagoury, O.L.; Leese, A.M.; Ryan, G.B.; James, E.; Morton, L.T.; Abbott, R.J.M.; Sabbah, S.; Kwok, W.; Rickinson, A.B. MHC II tetramers visualize human CD4 + T cell responses to Epstein–Barr virus infection and demonstrate atypical kinetics of the nuclear antigen EBNA1 response. J. Exp. Med. 2013, 210, 933–949. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Gudgeon, N.H.; Hui, E.P.; Jia, H.; Qun, X.; Taylor, G.S.; Barnardo, M.C.N.M.; Lin, C.K.; Rickinson, A.B.; Chan, A.T.C. CD4 and CD8 T cell responses to tumour-associated Epstein-Barr virus antigens in nasopharyngeal carcinoma patients. Cancer Immunol. Immunother. 2008, 57, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Nijland, M.; Veenstra, R.N.; Visser, L.; Xu, C.; Kushekhar, K.; van Imhoff, G.W.; Kluin, P.M.; van den Berg, A.; Diepstra, A. HLA dependent immune escape mechanisms in B-cell lymphomas: Implications for immune checkpoint inhibitor therapy? Oncoimmunology 2017, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rowe, M.; Khanna, R.; Jacob, C.A.; Argaet, V.; Kelly, A.; Powis, S.; Belich, M.; Groom-Carter, D.; Lee, S.; Burrows, S.R.; et al. Restoration of endogenous antigen processing in Burkitt’s lymphoma cells by Epstein-Barr virus latent membrane protein-1: Coordinate up-regulation of peptide transporters and HLA-class I antigen expression. Eur. J. Immunol. 1995, 25, 1374–1384. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; van den Berg, A.; Veenstra, R.; Rutgers, B.; Nolte, I.; van Imhoff, G.; Visser, L.; Diepstra, A. PML Nuclear Bodies and SATB1 Are Associated with HLA Class I Expression in EBV+ Hodgkin Lymphoma. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Ogino, T.; Moriai, S.; Ishida, Y.; Ishii, H.; Katayama, A.; Miyokawa, N.; Harabuchi, Y.; Ferrone, S. Association of immunoescape mechanisms with Epstein-Barr virus infection in nasopharyngeal carcinoma. Int. J. Cancer 2007, 120, 2401–2410. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.-X.; Yang, J.; Zhang, L.-J.; Sun, R.-M.; Zhao, L.-F.; Zhang, M.; Chen, Y.; Ma, J.; Qiao, K.; Sun, Q.-M.; et al. Downregulation of expression of transporters associated with antigen processing 1 and 2 and human leukocyte antigen I and its effect on immunity in nasopharyngeal carcinoma patients. Mol. Clin. Oncol. 2014, 2, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Chapuy, B.; Ouyang, J.; Sun, H.H.; Roemer, M.G.M.; Xu, M.L.; Yu, H.; Fletcher, C.D.M.; Freeman, G.J.; Shipp, M.A.; et al. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clin. Cancer Res. 2013, 19, 3462–3473. [Google Scholar] [CrossRef] [PubMed]
- Derks, S.; Liao, X.; Chiaravalli, A.M.; Xu, X.; Camargo, M.C.; Solcia, E.; Sessa, F.; Fleitas, T.; Freeman, G.J.; Rodig, S.J.; et al. Abundant PD-L1 expression in Epstein-Barr Virus-infected gastric cancers. Oncotarget 2016, 7, 32925–32932. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Abe, H.; Kunita, A.; Yamashita, H.; Seto, Y.; Fukayama, M. Overexpression and gene amplification of PD-L1 in cancer cells and PD-L1 + immune cells in Epstein-Barr virus-associated gastric cancer: The prognostic implications. Mod. Pathol. 2017, 30, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Zhang, J.; Hong, S.; Zhan, J.; Chen, N.; Qin, T.; Tang, Y.; Zhang, Y.; Kang, S.; Zhou, T.; et al. EBV-driven LMP1 and IFN-γ up-regulate PD-L1 in nasopharyngeal carcinoma: Implications for oncotargeted therapy. Oncotarget 2014, 5, 12189–12202. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Rodig, S.; Juszczynski, P.; Ouyang, J.; Sinha, P.; O’Donnell, E.; Neuberg, D.; Shipp, M.A. Constitutive AP-1 Activity and EBV Infection Induce PD-L1 in Hodgkin Lymphomas and Posttransplant Lymphoproliferative Disorders: Implications for Targeted Therapy. Clin. Cancer Res. 2012, 18, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.-Y.; Guo, S.-S.; Zhang, Y.; Lu, J.-B.; Chen, Q.-Y.; Tang, L.-Q.; Zhang, L.; Liu, L.-T.; Zhang, L.; Mai, H.-Q. Tumor CTLA-4 overexpression predicts poor survival in patients with nasopharyngeal carcinoma. Oncotarget 2016, 7, 13060–13068. [Google Scholar] [CrossRef] [PubMed]
- Herling, M.; Rassidakis, G.Z.; Medeiros, L.J.; Vassilakopoulos, T.P.; Kliche, K.; Nadali, G.; Viviani, S.; Bonfante, V.; Giardini, R.; Chilosi, M.; et al. Expression of Epstein-Barr Virus Latent Membrane Protein-1 in Hodgkin and Reed-Sternberg Cells of Classical Hodgkin â€TM s Lymphoma : Associations with Presenting Features, Serum Interleukin 10 Levels, and Clinical Outcome Expression of Epstein-Barr Virus. Afr. Health Sci. 2003, 9, 2114–2120. [Google Scholar]
- Ohshima, K.; Suzumiya, J.; Akamatu, M.; Takeshjta, M.; Kikuchi, M. Human and viral interleukin-10 in Hodgkin’s disease, and its influence on CD4+ and CD8+ T lymphocytes. Int. J. Cancer 1995, 62, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Ohshima, K.; Suzumiya, J.; Kume, T.; Shiroshita, T.O.; Kikuchi, M. Interleukin-10 expression and cytotoxic-T-cell response in Epstein-Barr-virus-associated nasopharyngeal carcinoma. Int. J. Cancer 1997, 72, 398–402. [Google Scholar] [CrossRef]
- Pachnia, D.; Drop, B.; Dworzańska, A.; Kliszczewska, E.; Polz-Dacewicz, M. Transforming Growth Factor-β, Interleukin-10, and Serological Markers in EBV-associated Gastric Carcinoma. Anticancer Res. 2017, 37, 4853–4858. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Saito, H.; Tatebe, S.; Tsujitani, S.; Ozaki, M.; Ito, H.; Ikeguchi, M. Interleukin-10 expression significantly correlates with minor CD8 + T-cell infiltration and high microvessel density in patients with gastric cancer. Int. J. Cancer 2006, 118, 1909–1914. [Google Scholar] [CrossRef] [PubMed]
- Incrocci, R.; Barse, L.; Stone, A.; Vagvala, S.; Montesano, M.; Subramaniam, V.; Swanson-Mungerson, M. Epstein-Barr Virus Latent Membrane Protein 2A (LMP2A) enhances IL-10 production through the activation of Bruton’s tyrosine kinase and STAT3. Virology 2017, 500, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Chao, P.-Z.; Hsieh, M.-S.; Cheng, C.-W.; Hsu, T.-J.; Lin, Y.-T.; Lai, C.-H.; Liao, C.-C.; Chen, W.-Y.; Leung, T.-K.; Lee, F.-P.; et al. Dendritic cells respond to nasopharygeal carcinoma cells through annexin A2-recognizing DC-SIGN. Oncotarget 2015, 6, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of Tumor-Associated Neutrophil Phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Maggio, E.; van den Berg, A.; Diepstra, A.; Kluiver, J.; Visser, L.; Poppema, S. Chemokines, cytokines and their receptors in Hodgkin’s lymphoma cell lines and tissues. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2002, 13 (Suppl 1), 52–56. [Google Scholar] [CrossRef]
- Umansky, V.; Blattner, C.; Gebhardt, C.; Utikal, J. The Role of Myeloid-Derived Suppressor Cells (MDSC) in Cancer Progression. Vaccines 2016, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.-E.; Tan, T.; Li, C.; Chen, Z.-C.; Ruan, L.; Wang, H.-H.; Su, T.; Zhang, P.-F.; Xiao, Z.-Q. Identification of Galectin-1 as a novel biomarker in nasopharyngeal carcinoma by proteomic analysis. Oncol. Rep. 2010, 24, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.K.; Moll, G.; Smith, C.; Dua, U.; Lambley, E.; Ramuz, O.; Gill, D.; Marlton, P.; Seymour, J.F.; Khanna, R. Brief report Galectin-1 mediated suppression of Epstein-Barr virus—Specific T-cell immunity in classic Hodgkin lymphoma. Boold 2015, 110, 1326–1330. [Google Scholar] [CrossRef]
- Klibi, J.; Niki, T.; Riedel, A.; Pioche-durieu, C.; Souquere, S.; Rubinstein, E.; Moulec, S.L.; Hirashima, M.; Guemira, F.; Adhikary, D.; et al. Blood diffusion and Th1-suppressive effects of galectin-9—Containing exosomes released by Epstein-Barr virus—Infected nasopharyngeal carcinoma cells. Blood 2009, 113, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Diepstra, A.; Van Imhoff, G.W.; Schaapveld, M.; Karim-Kos, H.; Van Den Berg, A.; Vellenga, E.; Poppema, S. Latent Epstein-Barr virus infection of tumor cells in classical Hodgkin’s lymphoma predicts adverse outcome in older adult patients. J. Clin. Oncol. 2009, 27, 3815–3821. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.B.; Han, H.Q.; Bei, J.X.; Liu, C.C.; Lei, J.J.; Cui, Q.; Feng, Q.S.; Wang, H.Y.; Zhang, J.X.; Liang, Y.; et al. Expression of human leukocyte antigen G is associated with prognosis in nasopharyngeal carcinoma. Int. J. Biol. Sci. 2012, 8, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.; Ernst, D.M.; Keating, A. Acquired natural killer cell dysfunction in the tumor microenvironment of classic Hodgkin lymphoma. Front. Immunol. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Krappmann, D.; Emmerich, F.; Kordes, U.; Scharschmidt, E.; Do, B.; Scheidereit, C. Molecular mechanisms of constitutive NF-kB/Rel activation in Hodgkin. Oncogene 1999, 18, 943. [Google Scholar] [CrossRef] [PubMed]
- Cabannes, E.; Khan, G.; Aillet, F.; Jarrett, R.F.; Hay, R.T. Mutations in the IkBa gene in Hodgkin’s disease suggest a tumour suppressor role for IκBα. Oncogene 1999, 18, 3063–3070. [Google Scholar] [CrossRef] [PubMed]
- Joos, S.; Menz, C.K.; Wrobel, G.; Siebert, R.; Gesk, S.; Ohl, S.; Mechtersheimer, G.; Trümper, L.; Moller, P.; Lichter, P.; et al. Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood 2002, 99, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, J.; Hiramoto, N.; Kato, M.; Sanada, M.; Maeshima, A.M.; Taniguchi, H.; Hosoda, F.; Asakura, Y.; Munakata, W.; Sekiguchi, N.; et al. Deletion of the TNFAIP3/A20gene detected by FICTION analysis in classical Hodgkin lymphoma. BMC Cancer 2012, 12, 457. [Google Scholar] [CrossRef] [PubMed]
- Chung, G.T.Y.; Lou, W.P.K.; Chow, C.; To, K.F.; Choy, K.W.; Leung, A.W.C.; Tong, C.Y.K.; Yuen, J.W.F.; Ko, C.W.; Yip, T.T.C.; et al. Constitutive activation of distinct NF-κB signals in EBV-associated nasopharyngeal carcinoma. J. Pathol. 2013, 231, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Chow, Y.P.; Tan, L.P.; Chai, S.J.; Abdul Aziz, N.; Choo, S.W.; Lim, P.V.H.; Pathmanathan, R.; Kornain, N.K.M.; Lum, C.L.; Pua, K.C.; et al. Exome Sequencing Identifies Potentially Druggable Mutations in Nasopharyngeal Carcinoma. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Heussinger, N.; Büttner, M.; Ott, G.; Brachtel, E.; Pilch, B.Z.; Kremmer, E.; Niedobitek, G. Expression of the Epstein-Barr virus (EBV)-encoded latent membrane protein 2A (LMP2A) in EBV-associated nasopharyngeal carcinoma. J. Pathol. 2004, 203, 696–699. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.L.; Hu, L.J.; Cao, J.Y.; Huang, Y.J.; Xu, L.H.; Liang, Y.; Xiong, D.; Guan, S.; Guo, B.H.; Mai, H.Q.; et al. Epstein-barr virus-encoded LMP2A induces an epithelial- mesenchymal transition and increases the number of side population stem-like cancer cells in nasopharyngeal carcinoma. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Wang, Y.; Wang, X.-F.; Liang, H.; Yan, L.-P.; Huang, B.-H.; Zhao, P. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinomas. World J. Gastroenterol. 2005, 11, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, K.; Haraoka, S.; Sugihara, M.; Suzumiya, J.; Kawasaki, C.; Kanda, M.; Kikuchi, M. Amplification and expression of a decoy receptor for Fas ligand (DcR3) in virus (EBV or HTLV-I) associated lymphomas. Cancer Lett. 2000, 160, 89–97. [Google Scholar] [CrossRef]
- Ho, C.-H.; Chen, C.-L.; Li, W.-Y.; Chen, C.-J. Decoy receptor 3, upregulated by Epstein-Barr virus latent membrane protein 1, enhances nasopharyngeal carcinoma cell migration and invasion. Carcinogenesis 2009, 30, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Stein, H.; Pileri, S.; Canino, S.; Farabbi, R.; Martelli, M.F.; Grignani, F.; Fagioli, M.; Minelli, O.; Ciani, C. Expression of lymphoid-associated antigens on Hodgkin’s and Reed-Sternberg cells of Hodgkin’s disease. An immunocytochemical study on lymph node cytospins using monoclonal antibodies. Histopathology 1987, 11, 1229–1242. [Google Scholar] [CrossRef] [PubMed]
- Horie, R.; Watanabe, T.; Morishita, Y.; Ito, K.; Ishida, T.; Kanegae, Y.; Saito, I.; Higashihara, M.; Mori, S.; Kadin, M.E.; et al. Ligand-independent signaling by overexpressed CD30 drives NF-κB activation in Hodgkin-Reed-Sternberg cells. Oncogene 2002, 21, 2493–2503. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.; Aldinucci, D.; Gloghini, A.; Zagonel, V.; Degan, M.; Improta, S.; Juzbasic, S.; Todesco, M.; Perin, V.; Gattei, V.; et al. Human eosinophils express functional CD30 ligand and stimulate proliferation of a Hodgkin’s disease cell line. Blood 1996, 88, 3299–3305. [Google Scholar]
- Molin, D.; Fischer, M.; Xiang, Z.; Larsson, U.; Harvima, I.; Venge, P.; Nilsson, K.; Sundström, C.; Enblad, G.; Nilsson, G. Mast cells express functional CD30 ligand and are the predominant CD30L-positive cells in Hodgkin’s disease. Br. J. Haematol. 2001, 114, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Gruss, H.J.; Hirschstein, D.; Wright, B.; Ulrich, D.; Caligiuri, M.A.; Barcos, M.; Strockbine, L.; Armitage, R.J.; Dower, S.K. Expression and function of CD40 on Hodgkin and Reed-Sternberg cells and the possible relevance for Hodgkin’s disease. Blood 1994, 84, 2305–2314. [Google Scholar] [PubMed]
- O’Grady, J.T.; Stewart, S.; Lowrey, J.; Howie, S.E.; Krajewski, A.S. CD40 expression in Hodgkin’s disease. Am. J. Pathol. 1994, 144, 21–26. [Google Scholar] [PubMed]
- Li, R.; Chen, W.-C.; Pang, X.-Q.; Hua, C.; Li, L.; Zhang, X.-G. Expression of CD40 and CD40L in Gastric Cancer Tissue and Its Clinical Significance. Int. J. Mol. Sci. 2009, 10, 3900–3917. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, K.; Karube, K.; Hamasaki, M.; Suefuji, H.; Tutiya, T.; Yamaguchi, T.; Suzumiya, J.; Kikuchi, M. Imbalances of chemokines, chemokine receptors and cytokines in Hodgkin lymphoma: Classical Hodgkin lymphomavs. Hodgkin-like ATLL. Int. J. Cancer 2003, 106, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Imadome, K.I.; Shimizu, N.; Yajima, M.; Watanabe, K.; Nakamura, H.; Takeuchi, H.; Fujiwara, S. CD40 signaling activated by Epstein-Barr virus promotes cell survival and proliferation in gastric carcinoma-derived human epithelial cells. Microbes Infect. 2009, 11, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Gires, O.; Zimber-Strobl, U.; Gonnella, R.; Ueffing, M.; Marschall, G.; Zeidler, R.; Pich, D.; Hammerschmidt, W. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997, 16, 6131–6140. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, B.F. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 2002, 99, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Joos, S.; Küpper, M.; Ohl, S.; Cells, C.D.H.; Ku, M.; Von Bonin, F.; Mechtersheimer, G.; Bentz, M.; Marynen, P.; Mo, P.; et al. Genomic Imbalances Including Amplification of the Tyrosine Kinase Gene JAK2 in CD30+ Hodgkin Cells Advances in Brief Genomic Imbalances Including Amplification of the Tyrosine Kinase Gene JAK2 in cd30+ hodgkin cells. Cancer Res. 2000, 63, 549–552. [Google Scholar]
- Gunawardana, J.; Chan, F.C.; Telenius, A.; Woolcock, B.; Kridel, R.; Tan, K.L.; Ben-Neriah, S.; Mottok, A.; Lim, R.S.; Boyle, M.; et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat. Genet. 2014, 46, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Weniger, M.A.; Melzner, I.; Menz, C.K.; Wegener, S.; Bucur, A.J.; Dorsch, K.; Mattfeldt, T.; Barth, T.F.E.; Möller, P. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene 2006, 25, 2679–2684. [Google Scholar] [CrossRef] [PubMed]
- Gires, O.; Kohlhuber, F.; Kilger, E.; Baumann, M.; Kieser, A.; Kaiser, C.; Zeidler, R.; Scheffer, B.; Ueffing, M.; Hammerschmidt, W. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 1999, 18, 3064–3073. [Google Scholar] [CrossRef] [PubMed]
- Stewart, S.; Dawson, C.W.; Takada, K.; Curnow, J.; Moody, C.A.; Sixbey, J.W.; Young, L.S. Epstein-Barr virus-encoded LMP2A regulates viral and cellular gene expression by modulation of the NF-kappaB transcription factor pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 15730–15735. [Google Scholar] [CrossRef] [PubMed]
- Herbst, H.; Samol, J.; Foss, H.D.; Raff, T.; Niedobitek, G. Modulation of interleukin-6 expression in Hodgkin and Reed-Sternberg cells by Epstein-Barr virus. J. Pathol. 1997, 182, 299–306. [Google Scholar] [CrossRef]
- Liu, Y.; Sattarzadeh, A.; Diepstra, A.; Visser, L.; Van Den Berg, A. The microenvironment in classical Hodgkin lymphoma: An actively shaped and essential tumor component. Semin. Cancer Biol. 2014, 24, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Kis, L.L.; Gerasimčik, N.; Salamon, D.; Persson, E.K.; Nagy, N.; Klein, G.; Severinson, E.; Klein, E. STAT6 signaling pathway activated by the cytokines IL-4 and IL-13 induces expression of the Epstein-Barr virus-encoded protein LMP-1 in absence of EBNA-2: Implications for the type II EBV latent gene expression in Hodgkin lymphoma. Blood 2011, 117, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Tempera, I.; De Leo, A.; Kossenkov, A.V.; Cesaroni, M.; Song, H.; Dawany, N.; Showe, L.; Lu, F.; Wikramasinghe, P.; Lieberman, P.M. Identification of MEF2B, EBF1, and IL6R as Direct Gene Targets of Epstein-Barr Virus (EBV) Nuclear Antigen 1 Critical for EBV-Infected B-Lymphocyte Survival. J. Virol. 2016, 90, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Tsang, C.M.; Deng, W.; Yip, Y.L.; Lui, V.W.-Y.; Wong, S.C.C.; Cheung, A.L.-M.; Hau, P.M.; Zeng, M.; Lung, M.L.; et al. Enhanced IL-6/IL-6R signaling promotes growth and malignant properties in EBV-infected premalignant and cancerous nasopharyngeal epithelial cells. PLoS ONE 2013, 8, e62284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Tsang, N. Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. J. Clin. Investig. 2013, 123, 5269–5283. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wu, Q.-L.; Fang, Y.; Mai, H.-Q.; Zeng, M.-S.; Shen, G.-P.; Hou, J.-H.; Zeng, Y.-X. Expression of chemokine receptor CXCR4 in nasopharyngeal carcinoma: Pattern of expression and correlation with clinical outcome. J. Transl. Med. 2005, 3, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Deng, X.; Tang, M.; Li, L.; Xiao, L.; Yang, L.; Zhong, J.; Bode, A.M.; Dong, Z.; Tao, Y.; et al. Tyrosylprotein Sulfotransferase-1 and Tyrosine Sulfation of Chemokine Receptor 4 Are Induced by Epstein-Barr Virus Encoded Latent Membrane Protein 1 and Associated with the Metastatic Potential of Human Nasopharyngeal Carcinoma. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, X.S.; Xie, D.; Deng, H.X.; Gao, Y.F.; Chen, Q.Y.; Huang, W.L.; Masucci, M.G.; Zeng, Y.X. Expression of immune-related molecules in primary EBV positive Chinese nasopharyngeal carcinoma: Associated with latent membrane protein 1 (LMP1) expression. Cancer Biol. Ther. 2007, 6, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Sakuma, K.; Sudo, M.; Osawa, T.; Ohara, E.; Uozaki, H.; Shibahara, J.; Kuroiwa, K.; Tominaga, S.; Hippo, Y.; et al. Interleukin-1beta expression in human gastric carcinoma with Epstein-Barr virus infection. J. Virol. 2002, 76, 6825–6831. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; MacIsaac, K.D.; Zhou, T.; Huang, P.-Y.; Xin, C.; Dobson, J.R.; Yu, K.; Chiang, D.Y.; Fan, Y.; Pelletier, M.R.; et al. Genomic Analysis of Nasopharyngeal Carcinoma Reveals TME-based Subtypes. Mol. Cancer Res. 2017, molcanres.0134.2017. [Google Scholar] [CrossRef] [PubMed]
- Renné, C.; Hinsch, N.; Willenbrock, K.; Fuchs, M.; Klapper, W.; Engert, A.; Küppers, R.; Hansmann, M.L.; Bräuninger, A. The aberrant coexpression of several receptor tyrosine kinases is largely restricted to EBV-negative cases of classical Hodgkin’s lymphoma. Int. J. Cancer 2007, 120, 2504–2509. [Google Scholar] [CrossRef] [PubMed]
- Renné, C.; Willenbrock, K.; Küppers, R.; Hansmann, M.L.; Bräuninger, A. Autocrine- and paracrine-activated receptor tyrosine kinases in classic Hodgkin lymphoma. Blood 2005, 105, 4051–4059. [Google Scholar] [CrossRef] [PubMed]
- Cader, F.Z.; Vockerodt, M.; Bose, S.; Nagy, E.; Brundler, M.A.; Kearns, P.; Murray, P.G. Lymphoid neoplasia: The EBV oncogene LMP1 protects lymphoma cells from cell death through the collagen-mediated activation of DDR1. Blood 2013, 122, 4237–4245. [Google Scholar] [CrossRef] [PubMed]
- Teofili, L.; Di Febo, A.L.; Pierconti, F.; Maggiano, N.; Bendandi, M.; Rutella, S.; Cingolani, A.; Di Renzo, N.; Musto, P.; Pileri, S.; et al. Expression of the c-met proto-oncogene and its ligand, hepatocyte growth factor, in Hodgkin disease. Blood 2001, 97, 1063–1069. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Plattel, W.; van den Berg, A.; Rüther, N.; Huang, X.; Wang, M.; de Jong, D.; Vos, H.; van Imhoff, G.; Viardot, A.; et al. Expression of the c-Met oncogene by tumor cells predicts a favorable outcome in classical Hodgkin’s lymphoma. Haematologica 2012, 97, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Guo, X.; Cao, B.; Kort, E.J.; Lee, C.; Chen, J.; Wang, L.; Mai, W.; Min, H.; Hong, M.; et al. Met Protein Expression Level Correlates with Survival in Patients with Late-stage Nasopharyngeal Carcinoma Met Protein Expression Level Correlates with Survival in Patients with Late-stage Nasopharyngeal Carcinoma. Cancer Res. 2002, 589–596. [Google Scholar]
- Luo, B.; Wang, Y.; Wang, X.F.; Gao, Y.; Huang, B.H.; Zhao, P. Correlation of Epstein-Barr virus and its encoded proteins with Helicobacter pylori and expression of c-met and c-myc in gastric carcinoma. World J. Gastroenterol. 2006, 12, 1842–1848. [Google Scholar] [CrossRef] [PubMed]
- Weimar, I.S.; Weijer, K.; Van Den Berk, P.C.M.; Muller, E.J.; Miranda, N.; Bakker, A.Q.; Heemskerk, M.H.M.; Hekman, A.; De Gast, G.C.; Gerritsen, W.R. HCF/SF and its receptor c-MET play a minor role in the dissemination of human B-lymphoma cells in SCID mice. Br. J. Cancer 1999, 81, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Lam, B.Q.; Dai, L.; Qin, Z. The role of HGF/c-MET signaling pathway in lymphoma. J. Hematol. Oncol. 2016, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Van Der Voort, B.R.; Taher, T.E.I.; Keehnen, R.M.J.; Smit, L.; Groenink, M.; Pals, S.T. Paracrine Regulation of Germinal Center B Cell Adhesion through the c-Met—Hepatocyte Growth Factor/Scatter Factor Pathway. J. Exp. Med. 1997, 185, 2121–2131. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Diepstra, A.; Xu, C.; Van Imhoff, G.; Plattel, W.; Van Den Berg, A.; Visser, L. Insulin-like growth factor 1 receptor is a prognostic factor in classical hodgkin lymphoma. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, D.; Sheen, T.S.; Chen, J.Y.; Huang, D.P.; Takada, K. Epstein-Barr virus-encoded small RNA induces insulin-like growth factor 1 and supports growth of nasopharyngeal carcinoma-derived cell lines. Oncogene 2005, 24, 1767–1773. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, D. Autocrine Growth of Epstein-Barr Virus-Positive Gastric Carcinoma Cells Mediated by an Epstein-Barr Virus-Encoded Small RNA Advances in Brief Autocrine Growth of Epstein-Barr Virus-Positive Gastric Carcinoma Cells. Cancer Res. 2003, 63, 7062–7067. [Google Scholar] [PubMed]
- Houldcroft, C.J.; Kellam, P. Host genetics of Epstein-Barr virus infection, latency and disease. Rev. Med. Virol. 2015, 25, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Yasui, Y.; Hamajima, N.; Nakamura, T.; El-Din, N.S.; Tajima, K.; Potter, J.D. Association of Epstein-Barr virus antibody titers with a human IL-10 promoter polymorphism in Japanese women. J. Autoimmune Dis. 2008, 5, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Rubicz, R.; Yolken, R.; Drigalenko, E.; Carless, M.A.; Dyer, T.D.; Bauman, L.; Melton, P.E.; Kent, J.W.; Harley, J.B.; Curran, J.E.; et al. A Genome-Wide Integrative Genomic Study Localizes Genetic Factors Influencing Antibodies against Epstein-Barr Virus Nuclear Antigen 1 (EBNA-1). PLoS Genet. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Pedergnana, V.; Syx, L.; Cobat, A.; Guergnon, J.; Brice, P.; Fermé, C.; Carde, P.; Hermine, O.; Le- Pendeven, C.; Amiel, C.; et al. Combined linkage and association studies show that HLA class II variants control levels of antibodies against Epstein-Barr virus antigens. PLoS ONE 2014, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Mandage, R.; Telford, M.; Rodríguez, J.A.; Farré, X.; Layouni, H.; Marigorta, U.M.; Cundiff, C.; Heredia-Genestar, J.M.; Navarro, A.; Santpere, G. Genetic factors affecting EBV copy number in lymphoblastoid cell lines derived from the 1000 Genome Project samples. PLoS ONE 2017, 12, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Diepstra, A.; Niens, M.; Vellenga, E.; Van Imhoff, G.W.; Nolte, I.M.; Schaapveld, M.; Van Der Steege, G.; Van Den Berg, A.; Kibbelaar, R.E.; Te Meerman, G.J.; et al. Association with HLA class I in Epstein-Barr-virus-positive and with HLA class III in Epstein-Barr-virus-negative Hodgkin’s lymphoma. Lancet 2005, 365, 2216–2224. [Google Scholar] [CrossRef]
- Urayama, K.Y.; Jarrett, R.F.; Hjalgrim, H.; Diepstra, A.; Kamatani, Y.; Chabrier, A.; Gaborieau, V.; Boland, A.; Nieters, A.; Becker, N.; et al. Genome-Wide Association Study of Classical Hodgkin Lymphoma and Epstein–Barr Virus Status–Defined Subgroups. J. Natl. Cancer Inst. 2012, 104, 240–253. [Google Scholar] [CrossRef] [PubMed]
- Cozen, W.; Li, D.; Best, T.; Van Den Berg, D.J.; Gourraud, P.; Victoria, K.; Skol, A.D.; Mack, T.M.; Glaser, S.L.; Weiss, L.M.; et al. A genome-wide meta-analysis of nodular sclerosis Hodgkin lymphoma identifies risk loci at 6p21. 32. Boold 2011, 119, 1–3. [Google Scholar] [CrossRef]
- Niens, M.; Jarrett, R.F.; Hepkema, B.; Nolte, I.M.; Diepstra, A.; Platteel, M.; Kouprie, N.; Delury, C.P.; Gallagher, A.; Visser, L.; et al. HLA-A*02 is associated with a reduced risk and HLA-A*01 with an increased risk of developing EBV+ Hodgkin lymphoma. Blood 2007, 110, 3310–3315. [Google Scholar] [CrossRef] [PubMed]
- Hjalgrim, H.; Rostgaard, K.; Johnson, P.C.D.; Lake, A.; Shield, L.; Little, A.-M.; Ekstrom-Smedby, K.; Adami, H.-O.; Glimelius, B.; Hamilton-Dutoit, S.; et al. HLA-A alleles and infectious mononucleosis suggest a critical role for cytotoxic T-cell response in EBV-related Hodgkin lymphoma. Proc. Natl. Acad. Sci. USA 2010, 107, 6400–6405. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.C.D.; McAulay, K.A.; Montgomery, D.; Lake, A.; Shield, L.; Gallagher, A.; Little, A.M.; Shah, A.; Marsh, S.G.E.; Taylor, G.M.; et al. Modeling HLA associations with EBV-positive and -negative Hodgkin lymphoma suggests distinct mechanisms in disease pathogenesis. Int. J. Cancer 2015, 137, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.; Wockner, L.; Brennan, R.M.; Keane, C.; Chattopadhyay, P.K.; Roederer, M.; Price, D.A.; Cole, D.K.; Hassan, B.; Beck, K.; et al. The impact of HLA class I and EBV latency-II antigen-specific CD8 + T cells on the pathogenesis of EBV + Hodgkin lymphoma. Clin. Exp. Immunol. 2016, 183, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hepkema, B.; Nolte, I.; Kushekhar, K.; Jongsma, T.; Veenstra, R.; Poppema, S.; Gao, Z.; Visser, L.; Diepstra, A.; et al. HLA-A*02:07 is a protective allele for EBV negative and a susceptibility allele for EBV positive classical Hodgkin lymphoma in China. PLoS ONE 2012, 7, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Bei, J.-X.; Li, Y.; Jia, W.-H.; Feng, B.-J.; Zhou, G.; Chen, L.-Z.; Feng, Q.-S.; Low, H.-Q.; Zhang, H.; He, F.; et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat. Genet. 2010, 42, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Chin, Y.M.; Mushiroda, T.; Takahashi, A.; Kubo, M.; Krishnan, G.; Yap, L.F.; Teo, S.H.; Lim, P.V.H.; Yap, Y.Y.; Pua, K.C.; et al. HLA-A SNPs and amino acid variants are associated with nasopharyngeal carcinoma in Malaysian Chinese. Int. J. Cancer 2015, 136, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.J. Linkage of a nasopharyngeal carcinoma susceptibility locus to the HLA region. Nature 1990, 346, 470–471. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Cai, H.; Li, X.; Zheng, H.; Yang, X.; Fang, W.; Zhang, L.; Wei, G.; Li, M.; Yao, K.; et al. Further evidence for the existence of major susceptibility of nasopharyngeal carcinoma in the region near HLA-A locus in Southern Chinese. J. Transl. Med. 2012, 10. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Lautenberger, J.A.; Gao, X.; Sezgin, E.; Hendrickson, S.L.; Troyer, J.L.; David, V.A.; Guan, L.; McIntosh, C.E.; Guo, X.; et al. The principal genetic determinants for nasopharyngeal carcinoma in China involve the HLA class I antigen recognition groove. PLoS Genet. 2012, 8, e1003103. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.L.; Tse, K.P.; Liang, S.; Chien, Y.C.; Su, W.H.; Yu, K.J.; Cheng, Y.J.; Tsang, N.M.; Hsu, M.M.; Chang, K.P.; et al. Evaluation of human leukocyte antigen-a (HLA-A), other NON-HLA markers on chromosome 6p21 and risk of nasopharyngeal carcinoma. PLoS ONE 2012, 7, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Tse, K.-P.; Su, W.-H.; Chang, K.-P.; Tsang, N.-M.; Yu, C.-J.; Tang, P.; See, L.-C.; Hsueh, C.; Yang, M.-L.; Hao, S.-P.; et al. Genome-wide Association Study Reveals Multiple Nasopharyngeal Carcinoma-Associated Loci within the HLA Region at Chromosome 6p21.3. Am. J. Hum. Genet. 2009, 85, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.C.; Yew, P.Y.; Puah, S.M.; Krishnan, G.; Yap, L.F.; Teo, S.H.; Lim, P.V.H.; Govindaraju, S.; Ratnavelu, K.; Sam, C.K.; et al. A genome-wide association study identifies ITGA9 conferring risk of nasopharyngeal carcinoma. J. Hum. Genet. 2009, 54, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Hutt-Fletcher, L.M.; Chesnokova, L.S. Integrins as triggers of Epstein-Barr virus fusion and epithelial cell infection. Virulence 2010, 1, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Sousa, H.; Breda, E.; Santos, A.M.; Catarino, R.; Pinto, D.; Canedo, P.; Machado, J.C.; Medeiros, R. IL-1RN VNTR polymorphism as a susceptibility marker for nasopharyngeal carcinoma in Portugal. Arch. Oral Biol. 2013, 58, 1040–1046. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Feng, Q.-S.; Mo, H.-Y.; Sun, J.; Xia, Y.-F.; Zhang, H.; Foo, J.N.; Guo, Y.-M.; Chen, L.-Z.; Li, M.; et al. An extended genome-wide association study identifies novel susceptibility loci for nasopharyngeal carcinoma. Hum. Mol. Genet. 2016, 25, 3626–3634. [Google Scholar] [CrossRef] [PubMed]
- Bei, J.X.; Su, W.H.; Ng, C.C.; Yu, K.; Chin, Y.M.; Lou, P.J.; Hsu, W.L.; McKay, J.D.; Chen, C.J.; Chang, Y.S.; et al. A GWAS meta-analysis and replication study identifies a novel locus within CLPTM1L/TERT associated with nasopharyngeal carcinoma in individuals of Chinese ancestry. Cancer Epidemiol. Biomarkers Prev. 2016, 25, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zheng, H.; Cheung, A.K.L.; Tang, C.S.; Ko, J.M.Y.; Wong, B.W.Y.; Leong, M.M.L.; Sham, P.C.; Cheung, F.; Kwong, D.L.-W.; et al. Whole-exome sequencing identifies MST1R as a genetic susceptibility gene in nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA 2016, 113, 3317–3322. [Google Scholar] [CrossRef] [PubMed]
- Koriyama, C.; Shinkura, R.; Hamasaki, Y.; Fujiyoshi, T.; Eizuru, Y.; Tokunaga, M. Human leukocyte antigens related to Epstein-Barr virus-associated gastric carcinoma in Japanese patients. Eur. J. Cancer Prev. 2001, 10, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-S.; Huang, S.-P.; Chang, Y.-T.; Shun, C.-T.; Chang, M.-C.; Lin, M.-T.; Wang, H.-P.; Lin, J.-T. Tumor necrosis factor-α and interleukin-10 promoter polymorphisms in Epstein-Barr virus-associated gastric carcinoma. J. Infect. Dis. 2002, 185, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Kuang, X.; Sun, L.; Liu, S.; Zhao, Z.; Zhao, D.; Liu, S.; Luo, B. Association of single nucleotide polymorphism rs2065955 of the filaggrin gene with susceptibility to Epstein-Barr virus-associated gastric carcinoma and EBV-negative gastric carcinoma. Virol. Sin. 2016, 31, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Liu, S.; Sun, L.; Zhao, Z.; Liu, S.; Kuang, X.; Shu, J.; Luo, B. Glypican-4 gene polymorphism (rs1048369) and susceptibility to Epstein-Barr virus-associated and -negative gastric carcinoma. Virus Res. 2016, 220, 52–56. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, G.W.; Visser, L.; Tan, L.P.; Berg, A.V.d.; Diepstra, A. The Microenvironment in Epstein–Barr Virus-Associated Malignancies. Pathogens 2018, 7, 40. https://doi.org/10.3390/pathogens7020040
Tan GW, Visser L, Tan LP, Berg AVd, Diepstra A. The Microenvironment in Epstein–Barr Virus-Associated Malignancies. Pathogens. 2018; 7(2):40. https://doi.org/10.3390/pathogens7020040
Chicago/Turabian StyleTan, Geok Wee, Lydia Visser, Lu Ping Tan, Anke Van den Berg, and Arjan Diepstra. 2018. "The Microenvironment in Epstein–Barr Virus-Associated Malignancies" Pathogens 7, no. 2: 40. https://doi.org/10.3390/pathogens7020040





