The Effect of West Nile Virus Infection on the Midgut Gene Expression of Culex pipiens quinquefasciatus Say (Diptera: Culicidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus
2.2. Mosquitoes
2.3. Sequence Analysis
2.4. Mosquito Infection, Tissue Dissection and RNA Extraction
2.5. Generation of Antisense RNA and Microinjection of Mosquitoes
2.6. Semi-Quantitative RT-PCR and Quantitative RT-PCR
2.7. Statistical Analysis
3. Results
3.1. Sequence Analysis
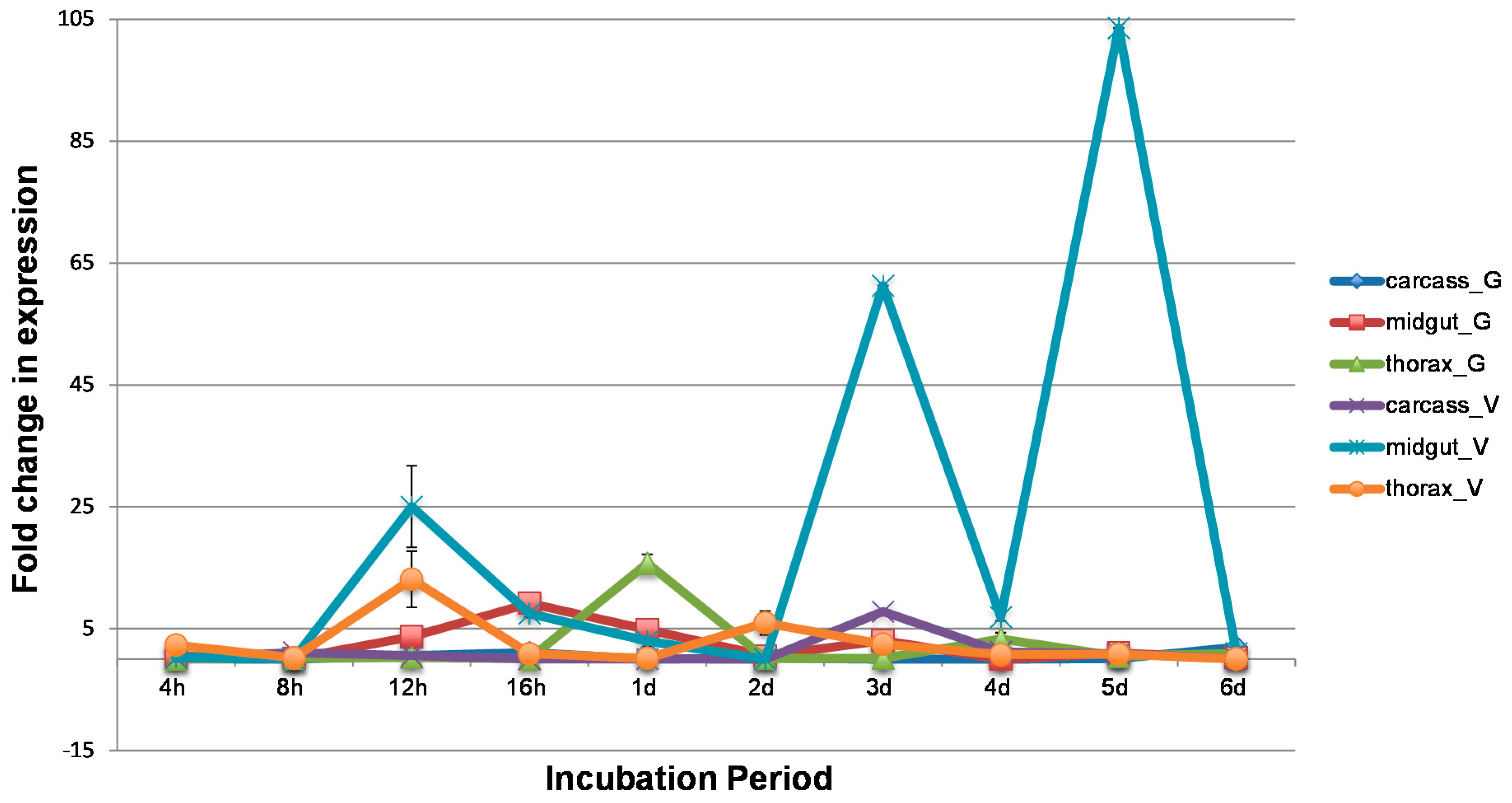
3.2. WNV Influences Temporal and Spatial Gene Expression
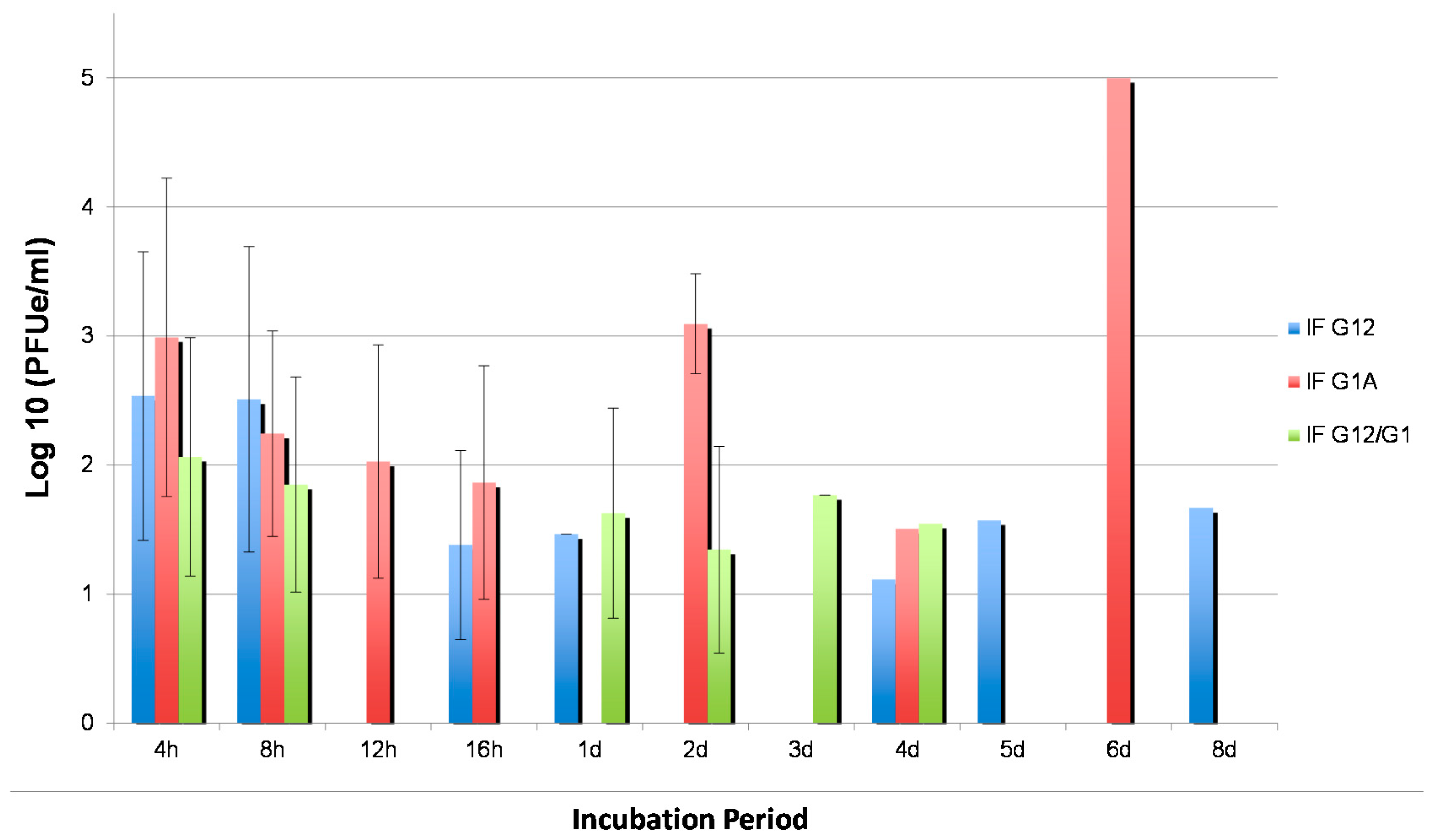
3.3. WNV Titers in Tissues from Two Cx. quinquefasciatus Populations
3.4. Suppression of Gene Expression Using RNAi
3.5. Effects of Silencing on WNV Replication
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lindsey, N.P.; Lehman, J.A.; Staples, J.E.; Fischer, M. West Nile Virus and Other Arboviral Diseases—United States, 2013. Morb. Mortal. Wkly. Rep. 2014, 63, 521–526. [Google Scholar]
- Lanciotti, R.S.; Roehrig, J.T.; Deubel, V.; Smith, J.; Parker, M.; Steele, K.; Crise, B.; Volpe, K.E.; Crabtree, M.B.; Scherret, J.H.; et al. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 1999, 286, 2333–2337. [Google Scholar] [CrossRef] [PubMed]
- Hayes, C.G. West Nile Fever. In The Arboviruses: Epidemiology and Ecology; Monath, T.P., Ed.; CRC Press: Boca Raton, FL, USA, 1989; Volume V, pp. 59–88. [Google Scholar]
- Turell, M.J.; Jones, J.W.; Sardelis, M.R.; Dohm, D.J.; Coleman, R.E.; Watts, D.M.; Fernandez, R.; Calampa, C.; Klein, T.A. Vector competence of Peruvian mosquitoes (Diptera: Culicidae) for epizootic and enzootic strains of Venezuelan equine encephalomyelitis virus. J. Med. Entomol. 2000, 37, 835–839. [Google Scholar] [CrossRef] [PubMed]
- Sardelis, M.R.; Turell, M.J.; Dohm, D.J.; O’Guinn, M.L. Vector competence of selected North American Culex and Coquillettidia mosquitoes for West Nile virus. Emerg. Infect. Dis. 2001, 7, 1018–1022. [Google Scholar] [CrossRef] [PubMed]
- Goddard, L.B.; Roth, A.E.; Reisen, W.K.; Scott, T.W. Vector competence of California mosquitoes for West Nile virus. Emerg. Infect. Dis. 2002, 8, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Blackmore, C.G.; Stark, L.M.; Jeter, W.C.; Oliveri, R.L.; Brooks, R.G.; Conti, L.A.; Wiersma, S.T. Surveillance results from the first West Nile virus transmission season in Florida, 2001. Am. J. Trop. Med. Hyg. 2003, 69, 141–150. [Google Scholar] [PubMed]
- Rutledge, C.R.; Day, J.F.; Lord, C.C.; Stark, L.M.; Tabachnick, W.J. West Nile virus infection rates in Culex nigripalpus (Diptera: Culicidae) do not reflect transmission rates in Florida. J. Med. Entomol. 2003, 40, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.L.; Lord, C.C.; Pesko, K.N.; Tabachnick, W.J. Environmental and biological factors influencing Culex pipiens quinquefasciatus (Diptera: Culicidae) vector competence for West Nile Virus. Am. J. Trop. Med. Hyg. 2010, 83, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.L.; Lord, C.C.; Pesko, K.; Tabachnick, W.J. Environmental and biological factors influencing Culex pipiens quinquefasciatus Say (Diptera: Culicidae) vector competence for Saint Louis encephalitis virus. Am. J. Trop. Med. Hyg. 2009, 81, 264–272. [Google Scholar] [PubMed]
- Hardy, J.L.; Houk, E.J.; Kramer, L.D.; Reeves, W.C. Intrinsic factors affecting vector competence of mosquitoes for arboviruses. Annu. Rev. Entomol. 1983, 28, 229–262. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, R.W.; Sudia, W.D. Mechanism of transmission of viruses by mosquitoes. Annu. Rev. Entomol. 1961, 6, 371–390. [Google Scholar] [CrossRef] [PubMed]
- Beerntsen, B.T.; James, A.A.; Christensen, B.M. Genetics of mosquito vector competence. Microbiol. Mol. Biol. Rev. 2000, 64, 115–137. [Google Scholar] [CrossRef] [PubMed]
- Tabachnick, W.J. Genetics of insect vector competence for arboviruses. In Advances in Disease Vector Research; Harris, K.F., Ed.; Springer: New York, NY, USA, 1994; Volume 10, pp. 93–108. [Google Scholar]
- Bosio, C.F.; Beaty, B.J.; Black, W.C.T. Quantitative genetics of vector competence for dengue-2 virus in Aedes aegypti. Am. J. Trop. Med. Hyg. 1998, 59, 965–970. [Google Scholar] [PubMed]
- Bosio, C.F.; Fulton, R.E.; Salasek, M.L.; Beaty, B.J.; Black, W.C.T. Quantitative trait loci that control vector competence for dengue-2 virus in the mosquito Aedes aegypti. Genetics 2000, 156, 687–698. [Google Scholar] [PubMed]
- Mercado-Curiel, R.F.; Black, W.C.T.; Munoz Mde, L. A dengue receptor as possible genetic marker of vector competence in Aedes aegypti. BMC Microbiol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Sanders, H.R.; Foy, B.D.; Evans, A.M.; Ross, L.S.; Beaty, B.J.; Olson, K.E.; Gill, S.S. Sindbis virus induces transport processes and alters expression of innate immunity pathway genes in the midgut of the disease vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2005, 35, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008, 4, e1000098. [Google Scholar] [CrossRef] [PubMed]
- Smartt, C.T.; Richards, S.L.; Anderson, S.L.; Erickson, J.S. West Nile virus infection alters midgut gene expression in Culex pipiens quinquefasciatus Say (Diptera: Culicidae). Am. J. Trop. Med. Hyg. 2009, 81, 258–263. [Google Scholar] [PubMed]
- Fragkoudis, R.; Attarzadeh-Yazdi, G.; Nash, A.A.; Fazakerley, J.K.; Kohl, A. Advances in dissecting mosquito innate immune responses to arbovirus infection. J. Gen. Virol. 2009, 90, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Merkling, S.H.; van Rij, R.P. Beyond rnai: Antiviral defense strategies in drosophila and mosquito. J. Insect Physiol. 2013, 59, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.T.; Ebel, G.D.; Lanciotti, R.S.; Brault, A.C.; Guzman, H.; Siirin, M.; Lambert, A.; Parsons, R.E.; Beasley, D.W.; Novak, R.J.; et al. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: Evidence for the emergence of a dominant genotype. Virology 2005, 342, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Chisenhall, D.M.; Mores, C.N. Diversification of West Nile virus in a subtropical region. Virol. J. 2009, 6, 106. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.L.; Mores, C.N.; Lord, C.C.; Tabachnick, W.J. Impact of extrinsic incubation temperature and virus exposure on vector competence of Culex pipiens quinquefasciatus Say (Diptera: Culicidae) for West Nile virus. Vector Borne Zoonotic Dis. 2007, 7, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.L.; Richards, S.L.; Tabachnick, W.J.; Smartt, C.T. Effects of West Nile virus dose and extrinsic incubation temperature on temporal progression of vector competence in Culex pipiens quinquefasciatus. J. Am. Mosq. Control Assoc. 2010, 26, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Arensburger, P.; Megy, K.; Waterhouse, R.M.; Abrudan, J.; Amedeo, P.; Antelo, B.; Bartholomay, L.; Bidwell, S.; Caler, E.; Camara, F.; et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science 2010, 330, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Nene, V.; Wortman, J.R.; Lawson, D.; Haas, B.; Kodira, C.; Tu, Z.J.; Loftus, B.; Xi, Z.; Megy, K.; Grabherr, M.; et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 2007, 316, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Holt, R.A.; Subramanian, G.M.; Halpern, A.; Sutton, G.G.; Charlab, R.; Nusskern, D.R.; Wincker, P.; Clark, A.G.; Ribeiro, J.M.; Wides, R.; et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science 2002, 298, 129–149. [Google Scholar] [CrossRef] [PubMed]
- Mongin, E.; Louis, C.; Holt, R.A.; Birney, E.; Collins, F.H. The Anopheles gambiae genome: An update. Trends Parasitol. 2004, 20, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Ishibashi, J.; Fujita, K.; Nakajima, Y.; Sagisaka, A.; Tomimoto, K.; Suzuki, N.; Yoshiyama, M.; Kaneko, Y.; Iwasaki, T.; et al. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1087–1110. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. Cdd: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Civana, A.; Acevedo, C.; Smartt, C.T. Transcriptomics of differential vector competence: West Nile virus infection in two populations of Culex pipiens quinquefasciatus linked to ovary development. BMC Genom. 2014. [Google Scholar] [CrossRef] [PubMed]
- Bridi, L.C.; Rafael, M.S. Gnbp domain of Anopheles darlingi: Are polymorphic inversions and gene variation related to adaptive evolution? Genetica 2016, 144, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Ryu, J.H.; Han, S.J.; Choi, K.H.; Nam, K.B.; Jang, I.H.; Lemaitre, B.; Brey, P.T.; Lee, W.J. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and beta-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J. Biol. Chem. 2000, 275, 32721–32727. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, T.; Leandro, D.C.; Ayres, C.F. Knock-down of REL2, but not defensin A, augments Aedes aegypti susceptibility to Bacillus subtilis and Escherichia coli. Acta Trop. 2010, 113, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Warr, E.; Das, S.; Dong, Y.; Dimopoulos, G. The Gram-negative bacteria-binding protein gene family: Its role in the innate immune system of Anopheles gambiae and in anti-Plasmodium defence. Insect Mol. Biol. 2008, 17, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Christophides, G.K.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G.; et al. Immunity-related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, R.M.; Kriventseva, E.V.; Meister, S.; Xi, Z.; Alvarez, K.S.; Bartholomay, L.C.; Barillas-Mury, C.; Bian, G.; Blandin, S.; Christensen, B.M.; et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 2007, 316, 1738–1743. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Lee, J.D.; Kravchenko, V.V.; Ulevitch, R.J.; Brey, P.T. Purification and molecular cloning of an inducible gram-negative bacteria-binding protein from the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. USA 1996, 93, 7888–7893. [Google Scholar] [CrossRef] [PubMed]
- Girard, Y.A.; Klingler, K.A.; Higgs, S. West Nile virus dissemination and tissue tropisms in orally infected Culex pipiens quinquefasciatus. Vector Borne Zoonotic Dis. 2004, 4, 109–122. [Google Scholar] [CrossRef] [PubMed]
- McLean, D. Multiplication of viruses in mosquitoes following feeding and injection into the body cavity. Aust. J. Exp. Biol. Med. Sci. 1955, 33, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Mellor, P.S. Replication of arboviruses in insect vectors. J. Comp. Pathol. 2000, 123, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, G.; Richman, A.; Muller, H.M.; Kafatos, F.C. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc. Natl. Acad. Sci. USA 1997, 94, 11508–11513. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.L.; Dimopoulos, G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev. Comp. Immunol. 2010, 34, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.; Jupatanakul, N.; Dimopoulos, G. Mosquito immunity against arboviruses. Viruses 2014, 6, 4479–4504. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smartt, C.T.; Shin, D.; Anderson, S.L. The Effect of West Nile Virus Infection on the Midgut Gene Expression of Culex pipiens quinquefasciatus Say (Diptera: Culicidae). Insects 2016, 7, 76. https://doi.org/10.3390/insects7040076
Smartt CT, Shin D, Anderson SL. The Effect of West Nile Virus Infection on the Midgut Gene Expression of Culex pipiens quinquefasciatus Say (Diptera: Culicidae). Insects. 2016; 7(4):76. https://doi.org/10.3390/insects7040076
Chicago/Turabian StyleSmartt, Chelsea T., Dongyoung Shin, and Sheri L. Anderson. 2016. "The Effect of West Nile Virus Infection on the Midgut Gene Expression of Culex pipiens quinquefasciatus Say (Diptera: Culicidae)" Insects 7, no. 4: 76. https://doi.org/10.3390/insects7040076




