Gene Disruption Technologies Have the Potential to Transform Stored Product Insect Pest Control
Abstract
:1. Introduction
2. Technology Background
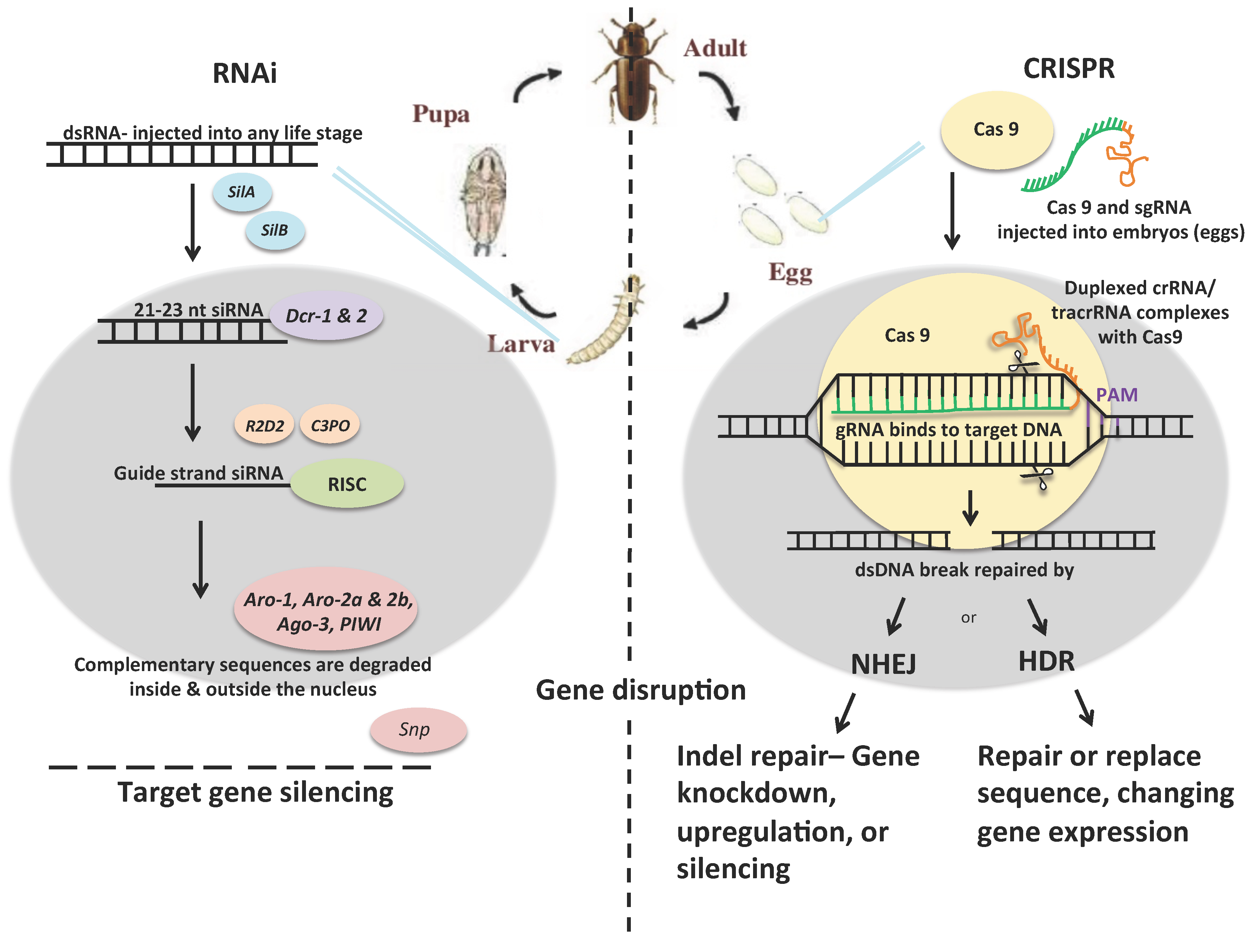
2.1. RNAi
2.2. CRISPR
2.3. RNAi vs. CRISPR as Insect Control Strategies
3. Application in Pest Management
3.1. Current Delivery Mechanisms
3.2. New Delivery Mechanisms
4. Complications
4.1. RNAi
4.2. CRISPR
4.3. Future Needs
5. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| RNAi | RNA interference |
| CRISPR | clustered regularly interspaced palindromic repeats |
| IPM | Integrated pest management |
| PH3 | phosphine |
| MeBr | methyl bromide |
| CRISPR-Cas9 | clustered regularly interspaced palindromic repeats and associated Cas9 protein |
| dsRNA | double stranded RNA |
| siRNA | short interfering RNA |
| RISC | RNAi-induced silencing complexes |
| CDA | chitin deacetylase |
| CHS | chitin synthase |
| ADC | aspartate 1-decarboxylase |
| RH | relative humidity |
| gRNA | guide RNA |
| sgRNA | single guide RNA |
| dsDNA | double stranded DNA |
| NHEJ | non-homologous end joining |
| HDR | homology-directed repair |
| INDELS | insertions/deletions |
| TALEN | transcription activator-like effector nucleases |
| CRISPRi | clustered regularly interspaced palindromic repeats interference |
| Cas9 | CRISPR associated protein 9 (endonuclease) |
| dCas9 | deactivated Cas9 |
| shRNA | short hairpin RNA |
| PCR | polymerase chain reaction |
| SNP | single nucleotide polymorphisms |
References
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Sadhu, M.J.; Bloom, J.S.; Day, L.; Druglyak, L. CRISPR-directied mitotic recombination enables genetic mapping without crosses. Science 2016, 352, 1113–1116. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Holper, S.; Kruger, M.; Stainier, D.Y. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 2015, 524, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.J.; Mahaffey, J.P.; Lorenzen, M.D.; Denell, R.E.; Mahaffey, J.W. Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol. Dev. 1999, 1, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of Coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 2010, 56, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Noh, M.Y.; Beeman, R.W.; Arakane, Y. RNAi-based functional genomics in Tribolium castaneum and possible application for controlling insect pests. Entomol. Res. 2012, 42, 1–10. [Google Scholar] [CrossRef]
- Hagstrum, D.W.; Phillips, T.W.; Cuperus, G. Stored Product Protection; Kansas State University: Manhattan, KS, USA, 2012. [Google Scholar]
- Collins, P.J.; Daglish, G.J.; Bengston, M.; Lambkin, T.M.; Pavic, H. Genetics of resistance to phosphine in Rhyzopertha dominica (Coleoptera: Bostrichidae). J. Econ. Entomol. 2002, 95, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Daglish, G.J.; Collins, P.J.; Pavic, H.; Kopittke, K.A. Effects of time and concentration on mortality of phosphine-resistant Sitophilus oryzae (L) fumigated with phosphine. Pest Manag. Sci. 2002, 58, 1015–1021. [Google Scholar] [CrossRef] [PubMed]
- Jagadeesan, R.; Collins, P.J.; Daglish, G.J.; Ebert, P.R.; Schlipalius, D.I. Phosphine resistance in the rust red flour beetle, Tribolium castaneum (Coleoptera: Tenebrionidae): Inheritance, gene interactions and fitness costs. PLoS ONE 2012, 7, e31582. [Google Scholar] [CrossRef] [PubMed]
- Lorini, I.; Collins, P.J.; Daglish, G.J.; Nayak, M.K.; Pavic, H. Detection and characterisation of strong resistance to phosphine in Brazilian Rhyzopertha dominica (F.) (Coleoptera: Bostrychidae). Pest Manag. Sci. 2007, 63, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Fields, P.G.; White, N.D.G. Alternatives to methyl bromide treatments for stored-product and quarantine insects. Annu. Rev. Entomol. 2002, 47, 331–359. [Google Scholar] [CrossRef] [PubMed]
- Boyer, S.; Zhang, H.; Lemperiere, G. A review of control methods and resistance mechanisms in stored-product insects. Bull. Entomol. Res. 2012, 102, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.F.; Arthur, F.H.; Zhu, K.Y. Spatial pattern in aerosol insecticide deposition inside a flour mill. J. Econ. Entomol. 2014, 107, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.G.S. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar]
- Stuart, J.J.; Ray, S.; Harrington, J.B.; Neal, J.J.; Beeman, R.W. Genetic mapping of a major locus controlling pyrethroid resistance in Tribolium castaneum (Coleoptera: Tenebrionidae). J. Econ. Entomol. 1998, 91, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Parthasarathy, R.; Bai, H.; Woithe, K.; Kaussmann, M.; Nauen, R.; Harrison, D.A.; Palli, S.R. A brain-specific cytochrome p450 responsible for the majority of deltamethrin resistance in the qtc279 strain of Tribolium castaneum. Proc. Natl. Acad. Sci. USA 2010, 107, 8557–8562. [Google Scholar] [CrossRef] [PubMed]
- Donitz, J.; Schmitt-Engel, C.; Grossmann, D.; Gerischer, L.; Tech, M.; Schoppmeier, M.; Klingler, M.; Bucher, G. iBeetle-base: A database for RNAi phenotypes in the red flour beetle Tribolium castaneum. Nucleic Acids Res. 2015, 43, D720–D725. [Google Scholar] [CrossRef] [PubMed]
- Gilles, A.F.; Schinko, J.B.; Averof, M. Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Tribolium castaneum. Development 2015, 142, 2832–2839. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.J. A new resistance to pyrethroids in Tribolium castaneum (Herbst). Pest Sci. 1990, 28, 101–115. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Tomoyasu, Y.; Miller, S.C.; Tomita, S.; Schoppmeier, M.; Grossmann, D.; Bucher, G. Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.G.; Michel, K.; Bartholomay, L.C.; Siegfried, B.D.; Hunter, W.B.; Smagghe, G.; Zhu, K.Y.; Douglas, A.E. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013, 59, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Kitzmann, P.; Schwirz, J.; Schmitt-Engel, C.; Bucher, G. RNAi phenotypes are influenced by the genetic background of the injected strain. BMC Genom. 2013. [Google Scholar] [CrossRef] [PubMed]
- Posnien, N.; Schinko, J.; Grossmann, D.; Shippy, T.D.; Konopova, B.; Bucher, G. RNAi in the red flour beetle (Tribolium). Cold Spring Harb. Protoc. 2009. [Google Scholar] [CrossRef] [PubMed]
- Aronstein, K.; Oppert, B.; Lorenzen, M. RNAi in agriculturally-important arthropods. In RNA Processing; Grabowski, P.P., Ed.; InTech: Rijeka, Croatia, 2011; pp. 157–180. [Google Scholar]
- Miller, S.C.; Miyata, K.; Brown, S.J.; Tomoyasu, Y. Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: Parameters affecting the efficiency of RNAi. PLoS ONE 2012, 7, e47431. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Dixit, R.; Begum, K.; Park, Y.; Specht, C.A.; Merzendorfer, H.; Kramer, K.J.; Muthukrishnan, S.; Beeman, R.W. Analysis of functions of the chitin deacetylase gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 2009, 39, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Muthukrishnan, S.; Kramer, K.J.; Specht, C.A.; Tomoyasu, Y.; Lorenzen, M.D.; Kanost, M.; Beeman, R.W. The tribolium chitin synthase genes tcCHS1 and tcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol. Biol. 2005, 14, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Lomakin, J.; Beeman, R.W.; Muthukrishnan, S.; Gehrke, S.H.; Kanost, M.R.; Kramer, K.J. Molecular and functional analyses of amino acid decarboxylases involved in cuticle tanning in Tribolium castaneum. J. Biol. Chem. 2009, 284, 16584–16594. [Google Scholar] [CrossRef] [PubMed]
- Noh, M.Y.; Kramer, K.J.; Muthukrishnan, S.; Beeman, R.W.; Kanost, M.R.; Arakane, Y. Loss of function of the yellow-e gene causes dehydration-induced mortality of adult Tribolium castaneum. Dev. Biol. 2015, 399, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, J.; Dao, V.A.; Majumdar, U.; Schmitt-Engel, C.; Schwirz, J.; Schultheis, D.; Strohlein, N.; Troelenberg, N.; Grossmann, D.; Richter, T.; et al. Large scale RNAi screen in Tribolium reveals novel target genes for pest control and the proteasome as prime target. BMC Genom. 2015. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, R.; Palli, S.R. Molecular analysis of nutritional and hormonal regulation of female reproduction in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2011, 41, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Broehan, G.; Arakane, Y.; Beeman, R.W.; Kramer, K.J.; Muthukrishnan, S.; Merzendorfer, H. Chymotrypsin-like peptidases from Tribolium castaneum: A role in molting revealed by RNA interference. Insect Biochem. Mol. Biol. 2010, 40, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Abdel-latief, M.; Hoffmann, K.H. Functional activity of allatotropin and allatostatin in the pupal stage of a holometablous insect, Tribolium castaneum (Coleoptera, Tenebrionidae). Peptides 2014, 53, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Perkin, L.C.; Elpidina, E.N.; Oppert, B. RNA interference and dietary inhibitors induce a similar compensation response in Tribolium castaneum larvae. Insect Mol. Biol. 2016, (in press). [Google Scholar]
- Fraga, A.; Ribeiro, L.; Lobato, M.; Santos, V.; Silva, J.R.; Gomes, H.; da Cunha Moraes, J.L.; de Souza Menezes, J.; de Oliveira, C.J.; Campos, E.; et al. Glycogen and glucose metabolism are essential for early embryonic development of the red flour beetle Tribolium castaneum. PLoS ONE 2013, 8, e65125. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, C.G.; van der Zee, M. Immune competence in insect eggs depends on the extraembryonic serosa. Dev. Comp. Immunol. 2013, 41, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Diez-Villasenor, C.; Garcia-Martinez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.E.; Dupuis, M.; Viooion, M.; Romero, D.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadan, A.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Haur, M.; Doudna, J.A.; Carpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.B.; Root, D.E. Resources for the design of CRISPR gene editing experiments. Genome Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Travis, J. Making the cut: CRISPR genome-editing technology shows its power. Science 2015, 350, 1456–1457. [Google Scholar] [CrossRef] [PubMed]
- Gilles, A.F.; Averof, M. Functional genetics for all: Engineered nucleases, CRISPR and the gene editing revolution. EvoDevo 2014. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, Z.; Rong, Y.S. Genome editing: From Drosophila to non-model insects and beyond. J. Genet. Genom. 2016, 43, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Bassett, A.R.; Tibbit, C.; Ponting, C.P.; Liu, J. Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biol. Open 2013, 3, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Kistler, K.E.; Vosshall, L.B.; Matthews, B.J. Genome engineering with CRISPR-Cas9 in the mosquito Aedes Aegypti. Cell Rep. 2015, 11, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Scott, M.J. CRISPR/Cas9 mediated mutagenesis of the white and sex lethal loci in the invasive pest, Drosophila suzukii. J. Biochem. Biophys. Res. Commun. 2016, 469, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Xu, J.; Cui, Z.; Dong, X.; Ye, Z.; Niu, D.; Hauang, Y.; Dong, S. Functional characterization of slitpbp3 in Spodoptera litura by CRISPR/Cas9 mediated genome editing. Insect Biochem. Mol. Biol. 2016, 75, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kwak, S.; Kim, J.; Kim, A.; Noh, H.M.; Kim, J.; Yu, K. RNA-guided genome editing in Drosophila with the purified Cas9 protein. G3 Genes Genomes Genet. 2014, 4, 1291–1295. [Google Scholar]
- Zeng, B.; Zhan, S.; Wang, Y.; Huang, Y.; Xu, J.; Liu, Q.; Li, Z.; Huang, Y.; Tan, A. Expansion of CRISPR targeting sites in Bombyx mori. Insect Biochem. Mol. Biol. 2016, 72, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Bucher, G.; Scholten, J.; Klingler, M. Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 2002, 12, R85–R86. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dai, Z.; Liang, Y.; Yin, M.; Ma, K.; He, M.; Ouyang, H.; Teng, C. Sequence-specific inhibition of microrna via CRISPR/CRISPRi system. Sci. Rep. 2014. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.M.; Suckling, D.M.; Byers, J.A.; Jang, E.B.; Wearing, C.H. Potential of “lure and kill” in long-term management and eradication of invasive spcies. J. Econ. Entomol. 2009, 102, 815–835. [Google Scholar] [CrossRef] [PubMed]
- Duehl, A.; Cohnstaedt, L.; Arbogast, R.; Teal, P. Evaluating light attraction to increase trap efficiency for Tribolium castaneum (Coleoptera: Tenebrionidae). J. Econ. Entomol. 2011, 104, 1430–1435. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.F. Attraction of walking Tribolium castaneum adults to traps. J. Stored Prod. Res. 2012, 51, 11–22. [Google Scholar] [CrossRef]
- Diaz-Montano, J.; Campbell, J.F.; Phillips, T.W.; Cohnstaedt, L.W.; Throne, J.E. Evaluation of light attraction for the stored-product psocid, Liposcelis bostrychophila. J. Pest Sci. 2016, 89, 923–930. [Google Scholar] [CrossRef]
- Nansen, C.; Barton, L.E.E.; Nansen, M. Uptake, retention, and repellency of a potential carrier of active ingredients in crack and crevice treatments for stored-grain beetles. J. Stored Prod. Res. 2007, 43, 417–424. [Google Scholar] [CrossRef]
- Rogers, C.D.; Armsworth, C.G.; Poppy, G.M. Conspecific transmission of insecticidal adhesive powder through mating in the mediterranean fruit fly, Ceratitis capitata. J. Pest Sci. 2013, 87, 361–369. [Google Scholar] [CrossRef]
- Abdel-latief, M. A family of chemoreceptors in Tribolium castaneum (Tenebrionidae: Coleoptera). PLoS ONE 2007, 2, e1319. [Google Scholar] [CrossRef] [PubMed]
- Mandiana Diakite, M.; Wang, J.; Ali, S.; Wang, M.-Q. Identification of chemosensory gene families in Rhyzopertha dominica (Coleoptera: Bostrichidae). Can. Entomol. 2015, 148, 8–21. [Google Scholar] [CrossRef]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Whitten, M.M.; Facey, P.D.; Del Sol, R.; Fernandez-Martinez, L.T.; Evans, M.C.; Mitchell, J.J.; Bodger, O.G.; Dyson, P.J. Symbiont-mediated RNA interference in insects. Proc. R. Soc. B Biol. Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Arista, M.; Smith, R.; Olalde-Portugal, V.; Hinojosa, R.; Hernandez-Delgadillo, R.; Blanco-Labra, A. Cellulolytic bacteria in the digestive system of Prostephanus truncatus (Coleoptera: Bostrichidae). J. Econ. Entomol. 1997, 90, 1371–1376. [Google Scholar] [CrossRef]
- Van Wyk, J.; Hodson, A.; Christensen, C. Microflora associated with the confused flour beetle, Tribolium confusum. Ann. Entomol. Soc. Am. 1959, 52, 452–463. [Google Scholar] [CrossRef]
- Shen, S.; Dowd, P. Detoxification spectrum of the cigarette beetle symbiont Symbiotaphrina kochii in culture. Entomol. Exp. Appl. 1991, 60, 51–59. [Google Scholar] [CrossRef]
- Bachman, P.M.; Bolognesi, R.; Moar, W.J.; Mueller, G.M.; Paradise, M.S.; Ramaseshadri, P.; Tan, J.; Uffman, J.P.; Warren, J.; Wiggins, B.E.; et al. Characterization of the spectrum of insecticidal activity of a double-stranded RNA with targeted activity against western corn rootworm (Diabrotica virgifera virgifera leconte). Transgenic Res. 2013, 22, 1207–1222. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M. Transgenic cereals: Current status and future prospects. J. Cereal Sci. 2014, 59, 419–434. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Bachman, P.M.; Huizinga, K.M.; Jensen, P.D.; Mueller, G.; Tan, J.; Uffman, J.P.; Levine, S.L. Ecological risk assessment for dvSnf7 RNA: A plant-incorporated protectant with targeted activity against western corn rootworm. Regul. Toxicol. Pharmacol. 2016, 81, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Sedlacek, J.D.; Komaravalli, S.R.; Hanley, A.M.; Price, B.D.; Davis, P.M. Life history attributes of Indian meal moth (Lepidoptera: Pyralidae) and angoumois grain moth (Lepidoptera: Gelechiidae) reared on transgenic corn kernels. J. Econ. Entomol. 2001, 94, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Bosch, M.; Wright, L.P.; Gershenzon, J.; Wasternack, C.; Hause, B.; Schaller, A.; Stintzi, A. Jasmonic acid and its precursor 12-oxophytodienoic acid control different aspects of constitutive and induced herbivore defenses in tomato. Plant Physiol. 2014, 166, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Xu, J.; Palli, R.; Ferguson, J.; Palli, S.R. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 2011, 67, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.A.; Tabuloc, C.A.; Cervantes, K.R.; Chiu, J.C. Ingestion of genetically modified yeast symbiont reduces fitness of an insect pest via RNA interference. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Zelikin, A.N.; Becker, A.L.; Johnston, A.P.R.; Wark, K.L.; Turatti, F.; Carsuso, F. A general approach for DNA encapsulation in degradable polymer microcapsules. ACS Nano 2007, 1, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, J.; Zhu, K.Y. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 2010, 19, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Champer, J.; Buchman, A.; Akbari, O. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 2016, 17, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Gantz, V.M.; Jasinskiene, N.; Tatarenkova, O.; Fazekas, A.; Macias, V.M.; Bier, E.; James, A.A. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 2015, 112, E6736–E6743. [Google Scholar] [CrossRef] [PubMed]
- Hammond, A.; Galizi, R.; Kyrou, K.; Simoni, A.; Siniscalchi, C.; Katsanos, D.; Gribble, M.; Baker, D.; Marois, E.; Russell, S.; et al. A CRISIPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anophelies gambiae. Nat. Biotechnol. 2016, 34, 78–83. [Google Scholar] [CrossRef] [PubMed]
- DiCarlo, J.E.; Chavez, A.; Dietz, S.L.; Esvelt, K.M.; Church, G.M. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 2015, 33, 1250–1255. [Google Scholar] [CrossRef] [PubMed]
- Gantz, V.M.; Bier, E. Genome editing. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science 2015, 348, 442–444. [Google Scholar] [CrossRef] [PubMed]
- Port, F.; Muschalik, N.; Bullock, L.S. Systematic evaluation of Drosophila CRISPR tools reveals safe and robust alternatives to autonomous gene drives in basic research. G3 Genes Genomes Genet. 2015, 5, 1493–1502. [Google Scholar]
- Itokawa, K.; Komagata, O.; Kasai, S.; Ogawa, K.; Tomita, T. Testing the causality between cypm10 and pyrethroid resistance using the TALEN and CRISPR/Cas9 technologies. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Schlipalius, D.I.; Valmas, N.; Tuck, A.G.; Jagadeesan, R.; Ma, L.; Kaur, R.; Goldinger, A.; Anderson, C.; Kuang, J.; Zuryn, S.; et al. A core metabolic enzyme mediates resistance to phosphine gas. Science 2012, 338, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Goptar, I.A.; Semashko, T.A.; Danilenko, S.A.; Lysogorskaya, E.N.; Oksenoit, E.S.; Zhuzhikov, D.P.; Belozersky, M.A.; Dunaevsky, Y.E.; Oppert, B.; Filippova, I.Y.; et al. Cysteine digestive peptidases function as post-glutamine cleaving enzymes in tenebrionid stored-product pests. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012, 161, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Goptar, I.A.; Shagin, D.A.; Shagina, I.A.; Mudrik, E.S.; Smirnova, Y.A.; Zhuzhikov, D.P.; Belozersky, M.A.; Dunaevsky, Y.E.; Oppert, B.; Filippova, I.Y.; et al. A digestive prolyl carboxypeptidase in Tenebrio molitor larvae. Insect Biochem. Mol. Biol. 2013, 43, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Tereshchenkova, V.F.; Goptar, I.A.; Kulemzina, I.A.; Zhuzhikov, D.P.; Serebryakova, M.V.; Belozersky, M.A.; Dunaevsky, Y.E.; Oppert, B.; Filippova, I.Y.; Elpidina, E.N. Dipeptidyl peptidase 4—An important digestive peptidase in Tenebrio molitor larvae. Insect Biochem. Mol. Biol. 2016, 76, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.L.; Walker, W.B., 3rd. Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J. Insect Physiol. 2012, 58, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Ledford, H. Safety Upgrade Found for Gene-Editing Technique. Available online: http://www.nature.com/news/safety-upgrade-found-for-gene-editing-technique-1.18799 (accessed on 6 July 2016).
- Pennisi, E.U.S. Academies Gives Cautions Go-Ahead to Gene Drive. Available online: http://www.sciencemag.org/news/2016/06/us-academies-give-cautious-go-head-gene-drive (accessed on 6 July 2016).
| Tc Strain | Resistance | Source |
|---|---|---|
| A20 Rdiel | Dieldrin | n/a |
| QTC-279 | Pyrethroid | Collins, P.J. (Australia) [22] |
| Rdiel BC9 Lab-S | Dieldrin | n/a |
| BRZ-4, BRZ-5 | Organophosphate, Phosphine | Rice patty, Pacheco, (Brazil, September 1987) |
| Tc Allele | Name/Alias | Origin |
|---|---|---|
| B | Black | Alexander Sokoloff, University of California, Berkeley |
| B (eve) | Black (eve) | n/a |
| B (i-2) | Black (i-2) | Jeff Stuart, Purdue |
| B (New) | Black (New) | n/a |
| B (ST) | Black (Scott Thomson) | M. Scott Thomson, University of Wisconsin, Riverside |
| B (t) 2 | Black (tawny) | C.E. Dyte and Miss Dorothy G. Blackman Ministry of Agriculture, Fisheries and Food, England |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perkin, L.C.; Adrianos, S.L.; Oppert, B. Gene Disruption Technologies Have the Potential to Transform Stored Product Insect Pest Control. Insects 2016, 7, 46. https://doi.org/10.3390/insects7030046
Perkin LC, Adrianos SL, Oppert B. Gene Disruption Technologies Have the Potential to Transform Stored Product Insect Pest Control. Insects. 2016; 7(3):46. https://doi.org/10.3390/insects7030046
Chicago/Turabian StylePerkin, Lindsey C., Sherry L. Adrianos, and Brenda Oppert. 2016. "Gene Disruption Technologies Have the Potential to Transform Stored Product Insect Pest Control" Insects 7, no. 3: 46. https://doi.org/10.3390/insects7030046





