cDNA Cloning and Expression Analysis of Pattern Recognition Proteins from the Chinese Oak Silkmoth, Antheraea pernyi
Abstract
:1. Introduction
2. Results and Discussion
2.1. cDNA Cloning of ApβGRP, Aplectin-5 and ApCTL1



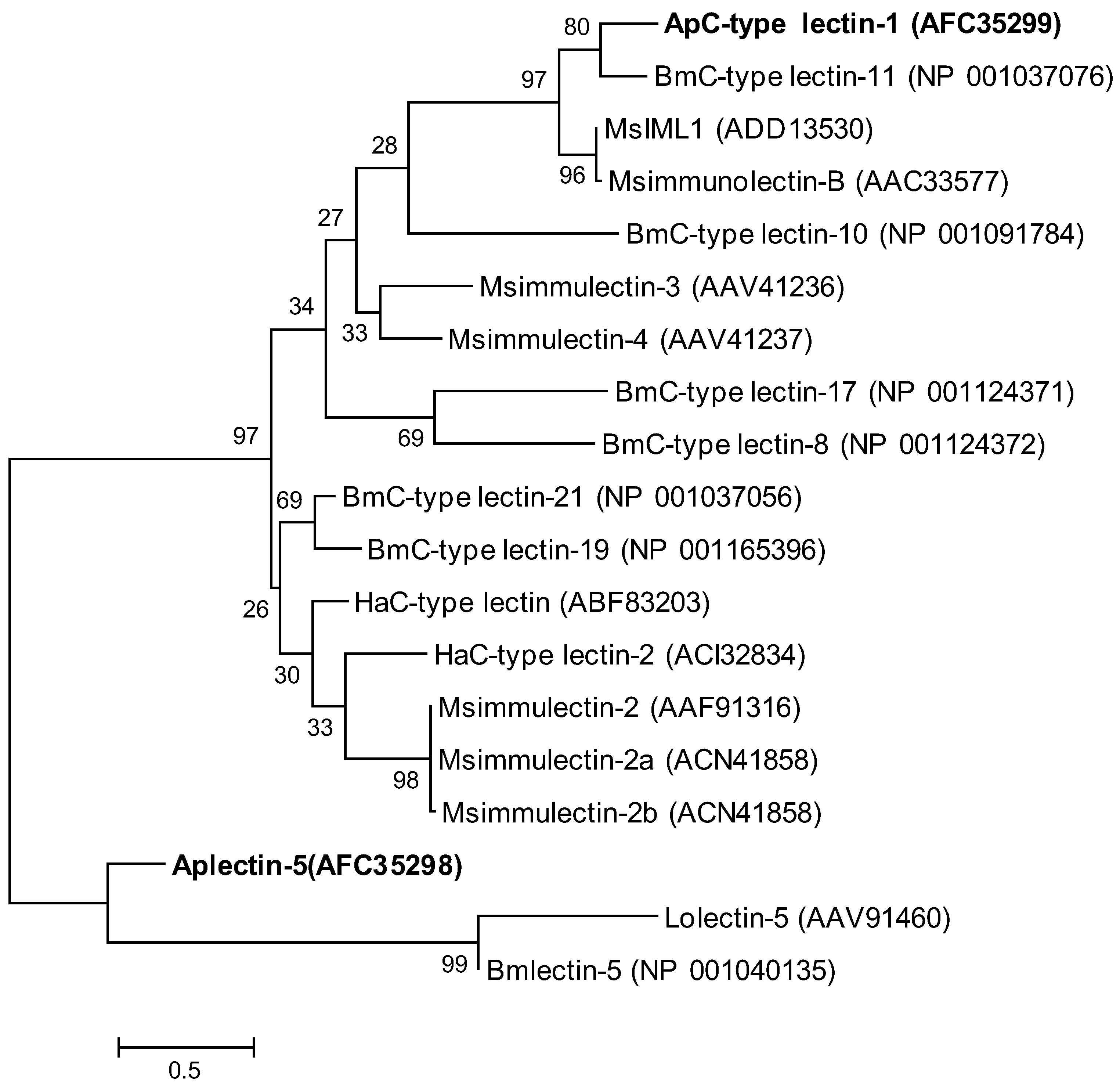
2.2. Phylogenetic Analysis of ApβGRP, Aplectin-5 and ApCTL1
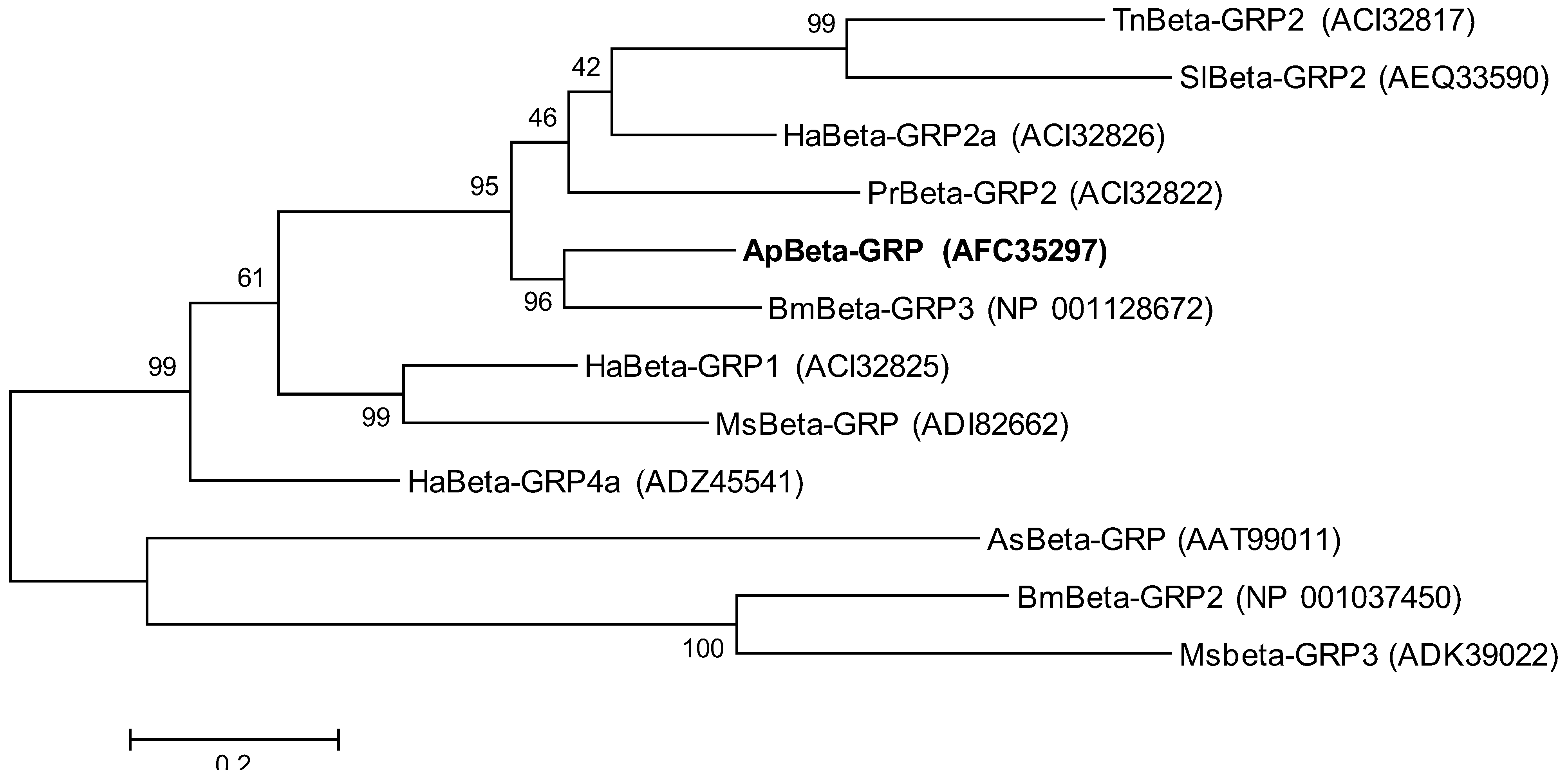
2.3. Expression Analysis of ApβGRP, Aplectin-5 and ApCTL1 in Different Tissues
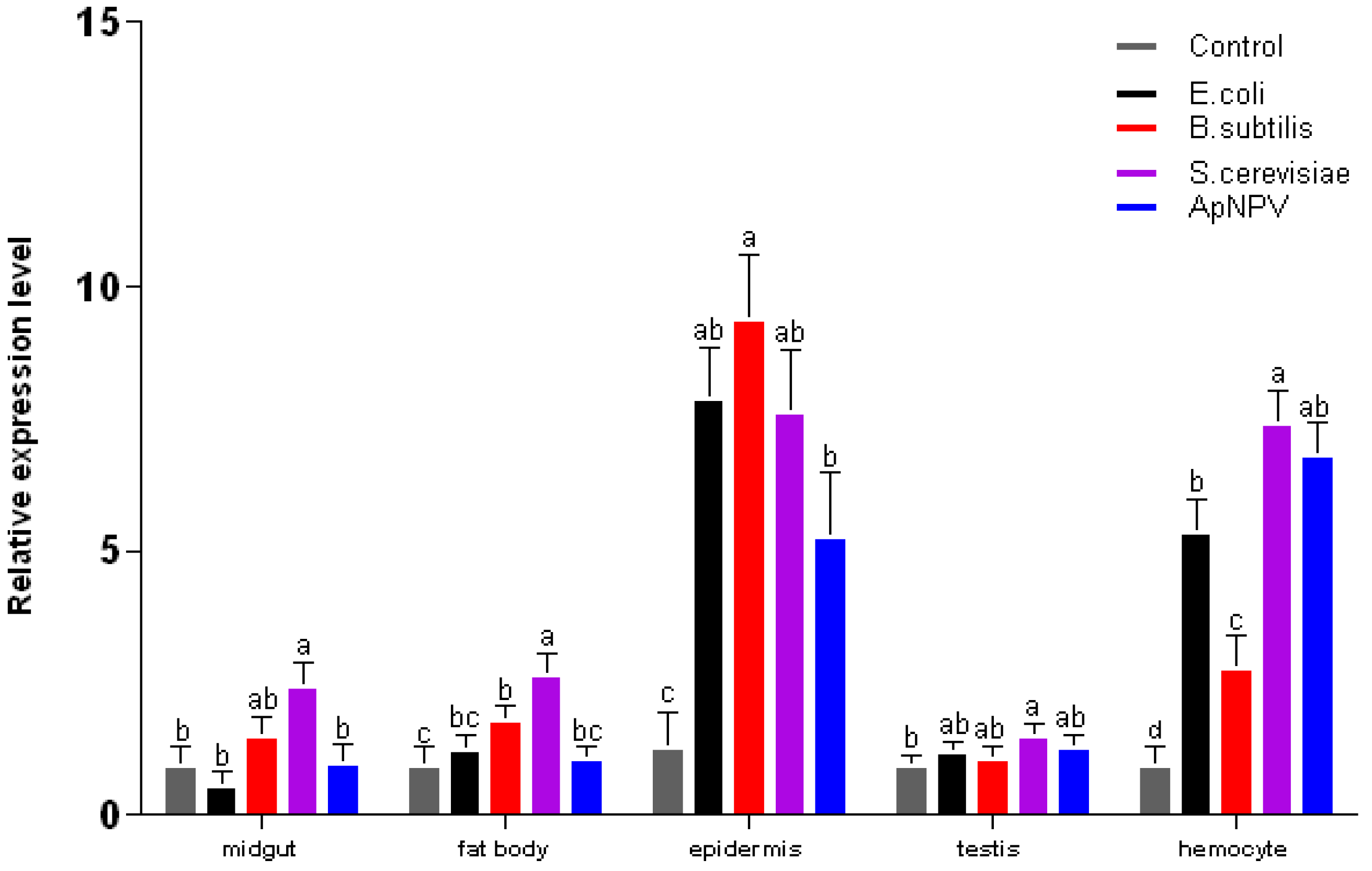

3. Experimental Section
3.1. Insects, Injection of Microorganisms and Tissue Sample Collections
3.2. Cloning of βGRP, Aplectin-5 and ApCTL1 Genes
- ApβGRP -F: 5’-CTCAACGAGGARATGGAAGGC-3’,
- ApβGRP -R: 5’-ACTGTCCAYTCYCCRTTATCCTG-3’,
- ApCTL1-F: 5’- CTACCGTTTATTTGCAAAGTGGAGG -3’,
- ApCTL1-R:5’-GTGCTATTTCCAACAGGTTTATCC-3’,
- Aplectin-5-F:5’-TCGGGTCCGATTACAATCTAGTG-3’,
- Aplectin-5-R: 5’-GTCATCACTATAGCCTTGTCCGT-3’.
- ApβGRP, 5’RACE outer primer: 5’-CTCGGGATATCACAAAAGCGAAGGA-3’
- ApβGRP, 5’RACE inner primer: 5’-GCAATACACCGTACCACCAGCAAAA-3’
- ApβGRP, 3’RACE outer primer: 5’- GTACCACCAGCAAAACTAGAAGC-3’
- ApβGRP, 3’RACE inner primer: 5’- GGTCTCGGGATATCACAAAAGC-3’
- Aplectin-5, 5’RACE outer primer: 5’-AACGTTCCTCCATGTTGTATAATTT-3’
- Aplectin-5, 5’RACE inner primer: 5’-ACCAAATTATTTATACCTAGCCATG-3’
- Aplectin-5, 3’RACE outer primer: 5’-ACAGGTGGGCTGGACTTAACGG-3’
- Aplectin-5, 3’RACE inner primer: 5’-AACATGGAGGAACGTTGATGGTGA-3’
- ApCTL1, 5’RACE outer primer: 5’-GTTCATCATAAGGAGCATCTTTGGC-3’
- ApCTL1, 5’RACE inner primer: 5’-GCCTGTTCCAAAAACGTGACAATGT-3’
- ApCTL1, 3’RACE outer primer: 5’-TTTGCGTGGTTTTTCTATGCTGGTT-3’
- ApCTL1, 3’RACE inner primer: 5’-ACGCTCATCTACTGGTCTTGAACTC-3’
3.3. Phylogenetic Analysis and Protein Structure Prediction
3.4. Transcript Profiles of ApβGRP, Aplectin-5 and ApCTL1
- Apβ-actin-F: 5’-ACCAACTGGGACGACATGGAGAAA-3’
- Apβ-actin-R: 5’-TCTCTCTGTTGGCCTTTGGGTTGA-3’
- ApβGRP-F: 5’- ATACGTGGACGAGCAGGGAAATGT-3’
- ApβGRP-R: 5’-TTTACCACGACCTTGCACGACTGT-3’
- Aplectin-5-F: 5’-CTTCAAACGAGGCGTGAAGAAGCA-3’
- Aplectin-5-R: 5’- AATTTGTAACCGTGACCGACTGCG -3’
- ApCTL1-F: 5’- AGTGGAGGCCAAAGATGCTCCTTA -3’
- ApCTL1-R: 5’- TACGCTTCGTTCCACGAGTACACA -3’
4. Conclusions
Acknowledgments
References
- Jiang, H.; Vilcinskas, A.; Kanost, M.R. Immunity in lepidopteran insects. Adv. Exp. Med. Biol. 2010, 708, 181–204. [Google Scholar] [CrossRef]
- Yu, X.Q.; Zhu, Y.F.; Ma, C.; Fabrick, J.A.; Kanost, M.R. Pattern recognition proteins in Manduca sexta plasma. Insect Biochem. Mol. Biol. 2002, 32, 1287–1293. [Google Scholar] [CrossRef]
- Ochiai, M.; Ashida, M. Purification of a beta-1,3-glucan recognition protein in the prophenoloxidase activating system from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 1988, 263, 12056–12062. [Google Scholar]
- Jiang, H.B.; Ma, C.C.; Lu, Z.Q.; Kanost, M.R. B-1,3-glucan recognition protein-2 (beta grp-2) from Manduca sexta: An acute-phase protein that binds beta-1,3-glucan and lipoteichoic acid to aggregate fungi and bacteria and stimulate prophenoloxidase activation. Insect Biochem. Mol. Biol. 2004, 34, 89–100. [Google Scholar] [CrossRef]
- Fabrick, J.A.; Baker, J.E.; Kanost, M.R. cDNA cloning, purification, properties, and function of a beta-1,3-glucan recognition protein from a pyralid moth, Plodia interpunctella. Insect Biochem. Mol. Biol. 2003, 33, 579–594. [Google Scholar] [CrossRef]
- Duvic, B.; Söderhäll, K. Purification and characterization of a beta-1,3-glucan binding-protein from plasma of the crayfish Pacifastacus leniusculus. J. Biol. Chem. 1990, 265, 9327–9332. [Google Scholar]
- Beschin, A.; Bilej, M.; Hanssens, F.; Raymakers, J.; Van Dyck, E.; Revets, H.; Brys, L.; Gomez, J.; De Baetselier, P.; Timmermans, M. Identification and cloning of a glucan- and liopoplysaccharide-binding protein from Eisenia foetida earthworm involved in the activation of prophenoloxidase cascade. J. Biol. Chem. 1998, 273, 24948–24954. [Google Scholar]
- Kim, Y.S.; Ryu, J.H.; Han, S.J.; Choi, K.H.; Nam, K.B.; Jang, I.H.; Lemaitre, B.; Brey, P.T.; Lee, W.J. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and beta-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J. Biol. Chem. 2000, 275, 32721–32727. [Google Scholar]
- Vasta, G.R.; Quesenberry, M.; Ahmed, H.; O'Leary, N. C-type lectins and galectins mediate innate and adaptive immune functions: Their roles in the complement activation pathway. Dev. Comp. Immunol. 1999, 23, 401–420. [Google Scholar] [CrossRef]
- Weis, W.I.; Taylor, M.E.; Drickamer, K. The C-type lectin superfamily in the immune system. Immunol. Rev. 1998, 163, 19–34. [Google Scholar] [CrossRef]
- Ling, E.J.; Ao, J.Q.; Yu, X.Q. Nuclear translocation of immulectin-3 stimulates hemocyte proliferation. Mol. Immunol. 2008, 45, 2598–2606. [Google Scholar] [CrossRef]
- Ling, E.J.; Yu, X.Q. Cellular encapsulation and melanization are enhanced by immulectins, pattern recognition receptors from the tobacco hornworm manduca sexta. Dev. Comp. Immunol. 2006, 30, 289–299. [Google Scholar] [CrossRef]
- Takase, H.; Watanabe, A.; Yoshizawa, Y.; Kitami, M.; Sato, R. Identification and comparative analysis of three novel C-type lectins from the silkworm with functional implications in pathogen recognition. Dev. Comp. Immunol. 2009, 33, 789–800. [Google Scholar] [CrossRef]
- Yu, X.Q.; Gan, H.; Kanost, M.R. Immulectin, an inducible C-type lectin from an insect, Manduca sexta, stimulates activation of plasma prophenol oxidae. Insect Biochem. Mol. Biol. 1999, 29, 585–597. [Google Scholar] [CrossRef]
- Yu, X.Q.; Kanost, M.R. Immulectin-2, a lipopolysaccharide specific lectin from an insect, Manduca sexta, is induced in response to Gram-negative bacteria. J. Biol. Chem. 2000, 275, 37373–37381. [Google Scholar]
- Koizumi, N.; Imamura, M.; Kadotani, T.; Yaoi, K.; Iwahana, H.; Sato, R. The lipopolysaccharide-binding protein participating in hemocyte nodule formation in the silkworm Bombyx mori is a novel member of the C-type lectin superfamily with two different tandem carbohydrate-recognition domains. FEBS. Lett. 1999, 443, 139–143. [Google Scholar] [CrossRef]
- Shin, S.W.; Park, D.S.; Kim, S.C.; Park, H.Y. Two carbohydrate recognition domains of Hyphantria cunea lectin bind to bacterial lipopolysaccharides through o-specific chain. FEBS. Lett. 2000, 467, 70–74. [Google Scholar] [CrossRef]
- Chothia, C.; Lesk, A.M. The relation between the divergence of sequence and structure in proteins. EMBO. J. 1986, 5, 823–826. [Google Scholar]
- Takahasi, K.; Ochiai, M.; Horiuchi, M.; Kumeta, H.; Ogura, K.; Ashida, M.; Inagaki, F. Solution structure of the silkworm betagrp/gnbp3 n-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition. P. Natl. Acad. Sci. USA 2009, 106, 11679–11684. [Google Scholar]
- Felsenstein, J. Confidence-limits on phylogenies - an approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(t)(-delta delta c) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hoffmann, J.A. The immune response of drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C.A. Decoding the patterns of self and nonself by the innate immune system. Science 2002, 296, 298–300. [Google Scholar] [CrossRef]
- Xu, P.-Z.; Zhang, M.-R. Molecular cloning and expression profile analysis of genes encoding pattern recognition receptors pgrp and βgrp in the silk-worm Bombyx mori. Sci. Sericulture 2010, 36, 383–390. [Google Scholar]
- Yu, X.Q.; Kanost, M.R. Manduca sexta lipopolysaccharide-specific immulectin-2 protects larvae from bacterial infection. Dev. Comp. Immunol. 2003, 27, 189–196. [Google Scholar] [CrossRef]
- Li, W.; Terenius, O.; Hirai, M.; Nilsson, A.S.; Faye, I. Cloning, expression and phylogenetic analysis of hemolin, from the chinese oak silkmoth, Antheraea pernyi. Dev. Comp. Immunol. 2005, 29, 853–864. [Google Scholar] [CrossRef]
- Bettencourt, R.; Terenius, O.; Faye, I. Hemolin gene silencing by ds-RNA injected into cecropia pupae is lethal to next generation embryos. Insect Mol. Biol. 2002, 11, 267–271. [Google Scholar] [CrossRef]
- Samakovlis, C.; Kylsten, P.; Kimbrell, D.A.; Engstrom, A.; Hultmark, D. The andropin gene and its product, a male-specific antibacterial peptide in Drosophila melanogaster. EMBO. J. 1991, 10, 163–169. [Google Scholar]
- Phatsara, C.; Jennen, D.G.J.; Ponsuksili, S.; Murani, E.; Tesfaye, D.; Schellander, K.; Wimmers, K. Molecular genetic analysis of porcine mannose-binding lectin genes, mbl1 and mbl2, and their association with complement activity. Int. J. Immunogenet. 2007, 34, 55–63. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, F.; Terenius, O.; Li, Y.; Fang, S.; Li, W. cDNA Cloning and Expression Analysis of Pattern Recognition Proteins from the Chinese Oak Silkmoth, Antheraea pernyi. Insects 2012, 3, 1093-1104. https://doi.org/10.3390/insects3041093
Li F, Terenius O, Li Y, Fang S, Li W. cDNA Cloning and Expression Analysis of Pattern Recognition Proteins from the Chinese Oak Silkmoth, Antheraea pernyi. Insects. 2012; 3(4):1093-1104. https://doi.org/10.3390/insects3041093
Chicago/Turabian StyleLi, Fengjuan, Olle Terenius, Yuan Li, Suyun Fang, and Wenli Li. 2012. "cDNA Cloning and Expression Analysis of Pattern Recognition Proteins from the Chinese Oak Silkmoth, Antheraea pernyi" Insects 3, no. 4: 1093-1104. https://doi.org/10.3390/insects3041093



