Phylogeography of Saproxylic and Forest Floor Invertebrates from Tallaganda, South-eastern Australia
Abstract
:1. Introduction
1.1. Understanding the Past to Predict and Manage for the Future
1.2. Scaling-up from Single Species to Whole Communities
1.3. Saproxylic and Forest Floor Invertebrates as Models for Phylogeography
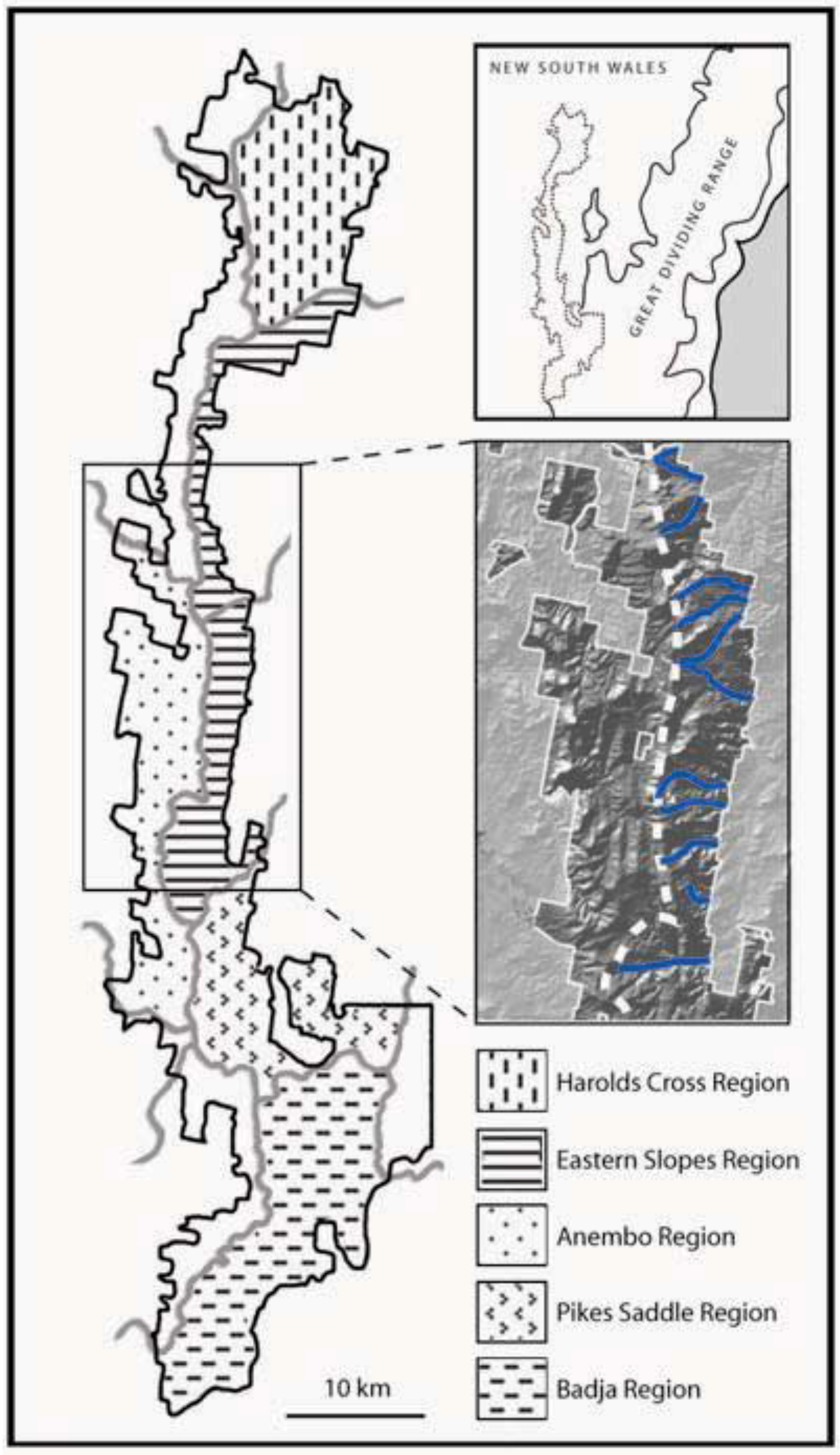
1.4. Landscape Context and Palaeoclimatic History of Tallaganda, South-Eastern Australia
2. Approach
2.1. Prior Expectations
- Prediction 1: Given the long-term stability of Tallaganda’s topography, five a priori regions delineated partly on the basis of drainage divisions will harbour distinct gene pools of low mobility forest floor invertebrates (see Figure 1 for geographic boundaries and nomenclature). These gene pools may be evident either as monophyletic haplotype clades on an estimated mitochondrial DNA (mtDNA) gene tree, or when diploid nuclear genetic markers are available, as natural genotypic clusters.
- Prediction 2: Based on topographic and hydrological characteristics of Tallaganda, the Eastern Slopes Region is predicted to have contained the highest concentration of suitable refuges for wet-adapted forest floor invertebrates during periglacial periods (Figure 1, lower inset). Thus, within Eastern Slopes Region we expect high genetic diversity, deep molecular divergences among sampled haplotypes, and early-branching DNA sequences or lineages on an outgroup-rooted gene tree to be concentrated here.
- Prediction 3: Owing to the repetitive nature of historical climatic cycles, their impacts will be detectable on multiple timescales. When inferences are based on DNA sequence data, molecular dating approaches should reveal two or more mostly non-overlapping timescales of lineage divergence and/or demographic changes.
- Prediction 4: The strongest evidence for postglacial range expansion is expected within catchments that experienced the greatest retraction of moist forests during Pleistocene periglaciation. Harolds Cross Region, Anembo Region, Pikes Saddle Region, and Badja Region are predicted to have been dominated by dry woodland or treeless steppe during cool dry periods. Accordingly, these regions are expected to show gene tree topologies that are indicative of recent population growth and geographic expansion, and reduced genetic diversity compared to Eastern Slopes Region, the area of relative stability and/or putative geographic source of recolonists.
2.2. Focal Invertebrates

3. Spatial Patterns of Intraspecific Diversity
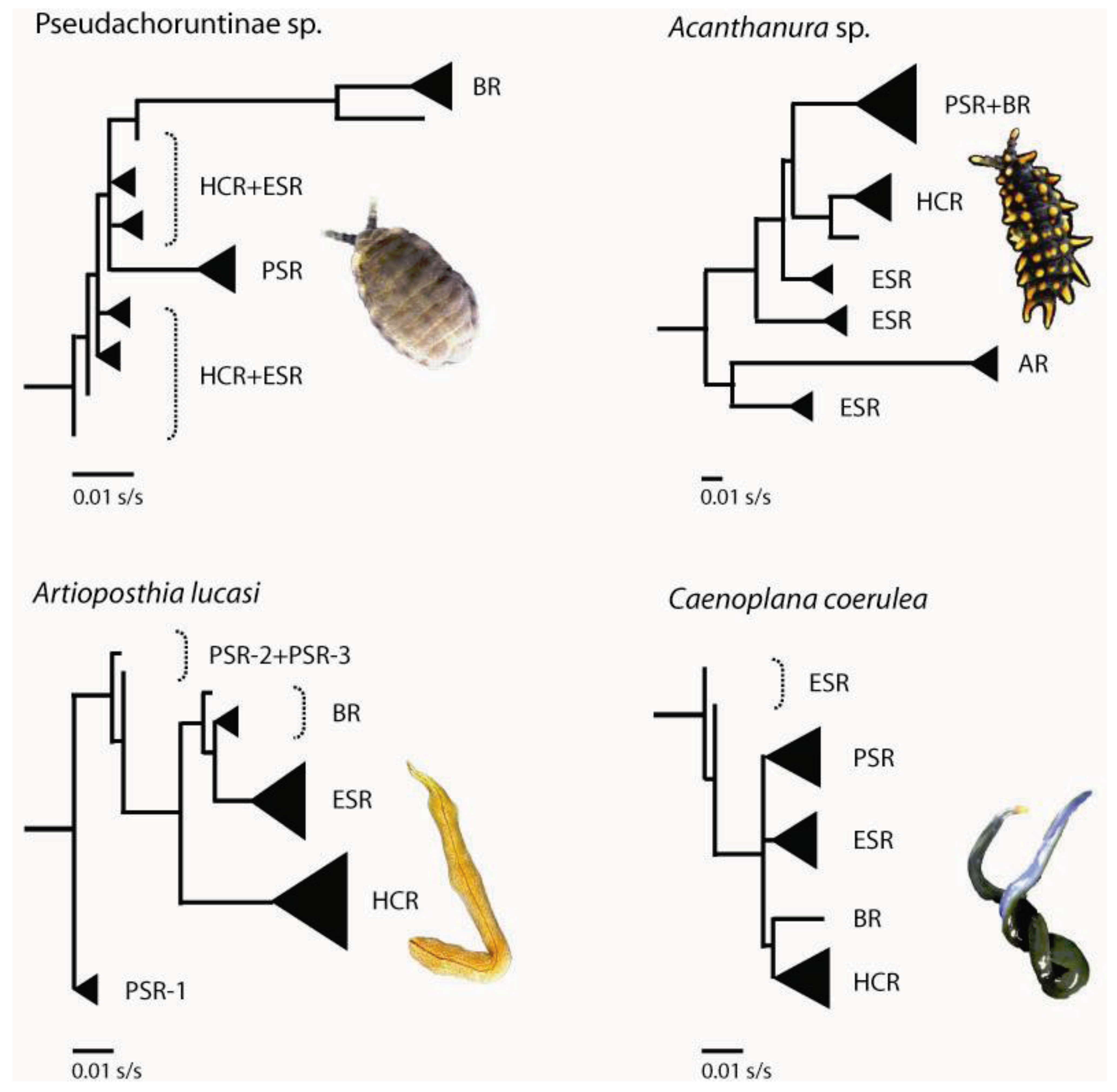
3.1. Geographically Localized Genetic Lineages
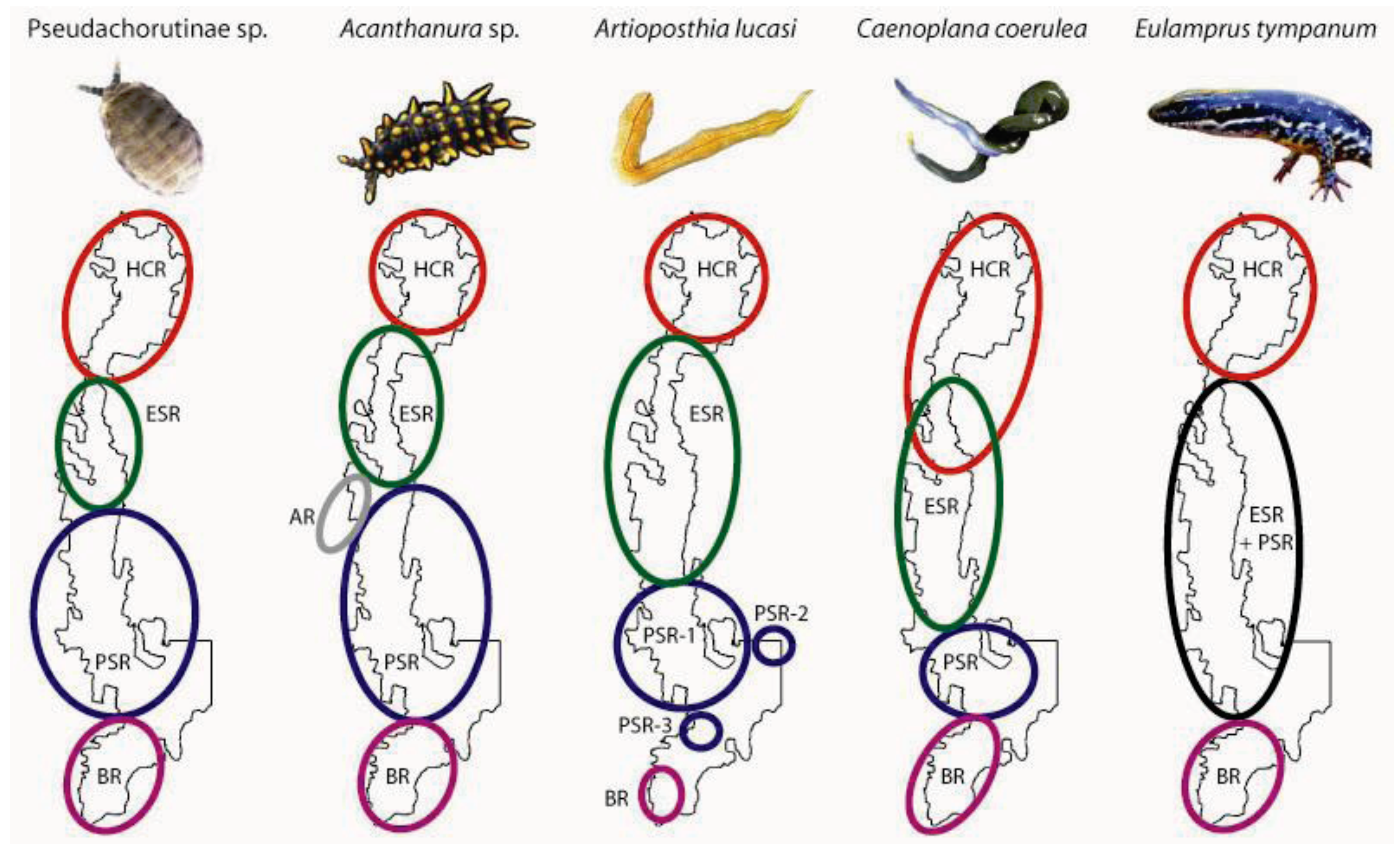
| Pseudachorutinae sp. | Acanthanura sp. | Artioposthia lucasi* | Caenoplana coerulea | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic population | N (Nhap) | Hd | p-dist. | N (Nhap) | Hd | p-dist. | N (Nhap) | Hd | p-dist. | N (Nhap) | Hd | p-dist. |
| Harolds Cross Region | 81 (12) | 0.795 | 0.003 | 25 (11) | 0.880 | 0.013 | 64 (6) | 0.379 | 0.006 | 80 (3) | 0.251 | 0.001 |
| Eastern Slopes Region | 61 (7) | 0.538 | 0.001 | 19 (7) | 0.819 | 0.052 | 105 (5) | 0.527 | 0.006 | 47 (5) | 0.375 | 0.001 |
| Pikes Saddle Region | 112 (11) | 0.280 | 0.004 | 66 (25) | 0.891 | 0.023 | 21 (2) | 0.095 | 0.001 | 47 (4) | 0.537 | 0.002 |
| Badja Region | 64 (9) | 0.792 | 0.009 | 78 (21) | 0.903 | 0.010 | – | – | – | – | – | – |
4. Past Events that Shaped Spatial-genetic Patterns
4.1. Long-term Persistence at Tallaganda
4.2. Repeated Population Isolation and Expansion
4.3. Number and Locations of Moist Forest Refuges at Tallaganda
4.4. Pseudo-Congruence and Incongruence
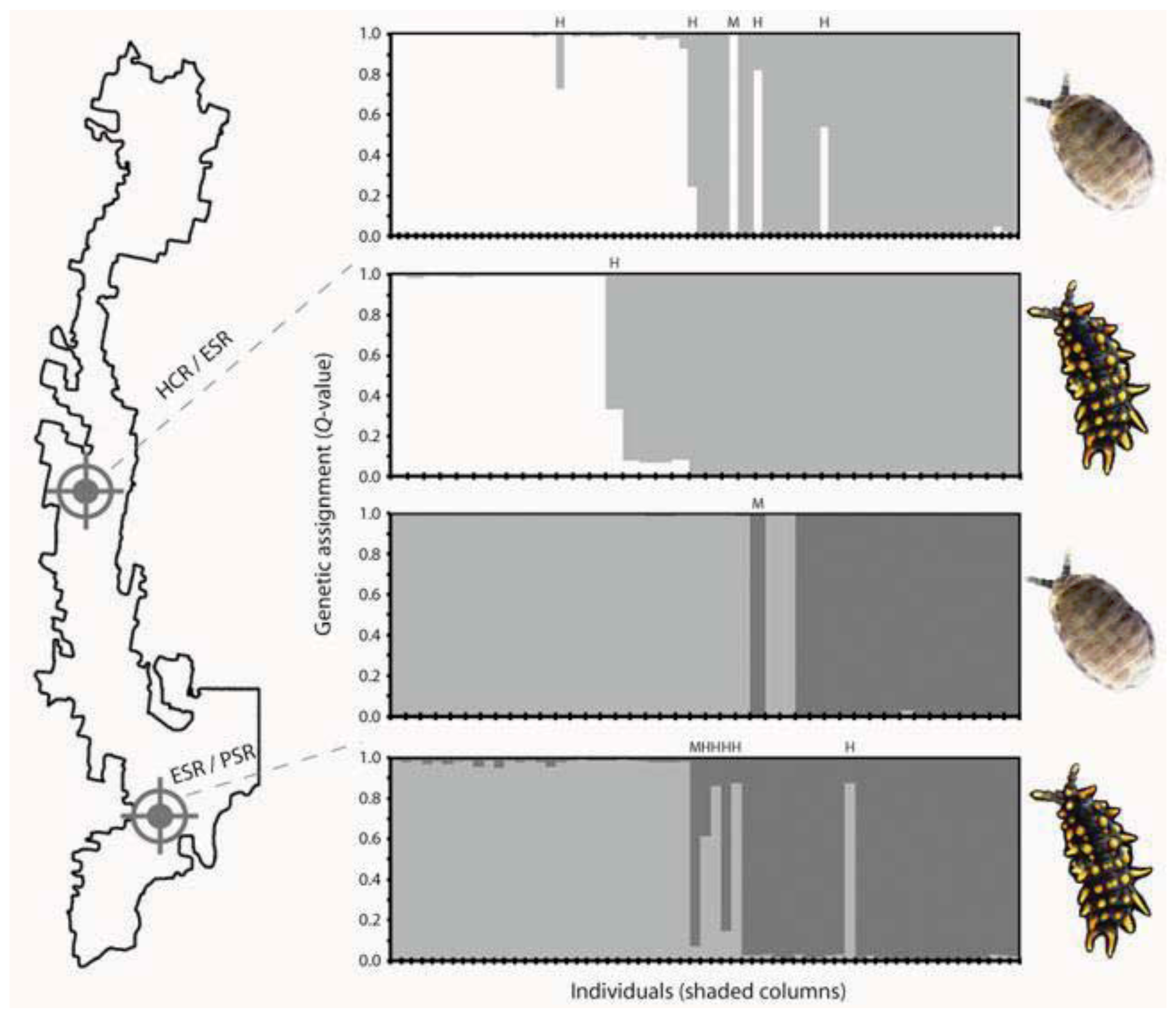
5. Contemporary Gene Flow Dynamics that Maintain Genetic Contact Zones

6. Conservation Considerations
6.1. The Role of Phylogeography in Protecting Saproxylic Invertebrate Biodiversity
6.2. The Current Reserve System at Tallaganda
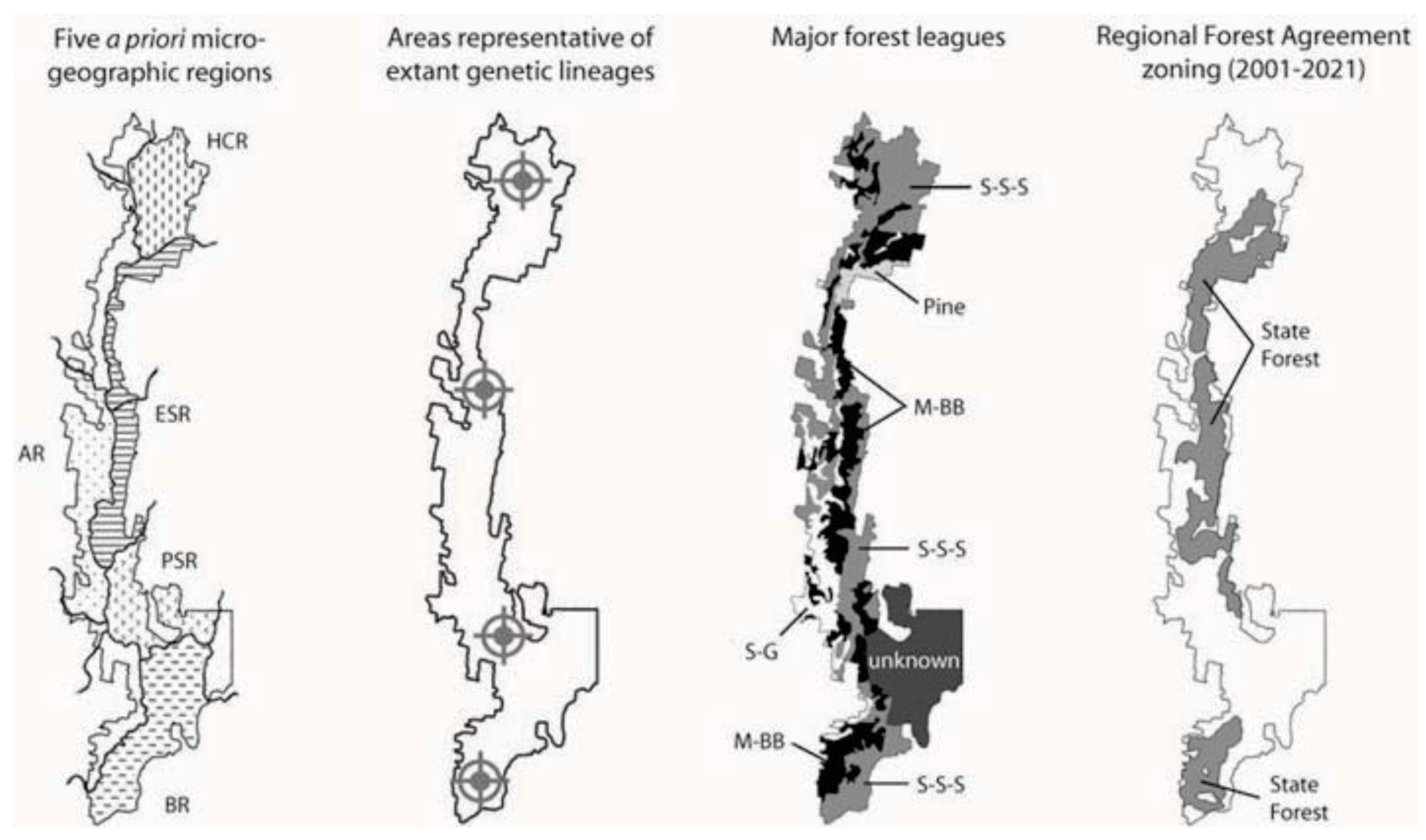
6.3. Ameliorating the Potential Negative Impacts of Commercial Forestry
7. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
References
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Knowles, L.L.; Carstens, B.C. Estimating a geographically explicit model of population divergence. Evolution 2007, 61, 477–493. [Google Scholar] [CrossRef]
- Moritz, C. Defining ‘evolutionarily significant units’ for conservation. Trends Ecol. Evol. 1994, 9, 373–375. [Google Scholar] [CrossRef]
- Moritz, C. Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst. Biol. 2002, 51, 238–254. [Google Scholar] [CrossRef]
- Crandall, K.A.; Bininda-Emonds, O.R.P.; Mace, G.M.; Wayne, R.K. Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 2000, 15, 290–295. [Google Scholar] [CrossRef]
- Garrick, R.C.; Sands, C.J.; Sunnucks, P. The Use and Application of Phylogeography for Invertebrate Conservation Research and Planning. In Insect Biodiversity and Dead Wood: Proceedings of a Symposium for the 22nd International Congress of Entomology, Asheville, NC, USA, 2006; Grove, S.J., Hanula, J.L., Eds.; pp. 15–22, U.S. Department of Agriculture Forest Service Southern Research Station General Technical Report SRS-93.
- Parmesan, C.; Root, T.L.; Willig, M.R. Impacts of extreme weather and climate on terrestrial biota. Bull. Am. Meteorol. Soc. 2000, 81, 443–450. [Google Scholar] [CrossRef]
- McLachlan, J.S.; Clark, J.S.; Manos, P.S. Molecular indicators of tree migration capacity under rapid climate change. Ecology 2005, 86, 2088–2098. [Google Scholar] [CrossRef]
- Cordellier, M.; Pfenninger, M. Inferring the past to predict the future: Climate modelling predictions and phylogeography for the freshwater gastropod Radix balthica (Pulmonata, Basommatophora). Mol. Ecol. 2009, 18, 534–544. [Google Scholar] [CrossRef]
- Norgate, M.; Chamings, J.; Pavlova, A.; Bull, J.K.; Murray, N.D.; Sunnucks, P. Mitochondrial DNA indicates Late Pleistocene divergence of populations of Heteronympha merope, an emerging model in environmental change biology. PLoS One 2009, 4, e7950. [Google Scholar]
- Eckert, A.J. Seeing the forest for the trees: Statistical phylogeography in a changing world. New Phytol. 2011, 189, 894–897. [Google Scholar] [CrossRef]
- Smith, C.I.; Farrell, B.D. Range expansions in the flightless longhorn cactus beetles, Moneilema gigas and Moneilema armatum, in response to Pleistocene climate changes. Mol. Ecol. 2005, 14, 1025–1044. [Google Scholar] [CrossRef]
- Carstens, B.C.; Brunsfeld, S.J.; Dembowski, J.R.; Good, J.M.; Sullivan, J. Investigating the evolutionary history of the Pacific Northwest mesic forest ecosystem: Hypothesis-testing within a comparative phylogeographic framework. Evolution 2005, 59, 1639–1652. [Google Scholar]
- Bermingham, E.; Moritz, C. Comparative phylogeography: Concepts and applications. Mol. Ecol. 1998, 7, 367–369. [Google Scholar] [CrossRef]
- Moritz, C.; Faith, D.P. Comparative phylogeography and the identification of genetically divergent areas for conservation. Mol. Ecol. 1998, 7, 419–429. [Google Scholar] [CrossRef]
- Lapointe, F.-.J.; Rissler, L.J. Congruence, consensus, and comparative phylogeography of codistributed species in California. Am. Nat. 2005, 166, 290–299. [Google Scholar] [CrossRef]
- Hickerson, M.J.; Stahl, E.A.; Lessios, H.A. Test for simultaneous divergence using approximate Bayesian computation. Evolution 2006, 60, 2435–2453. [Google Scholar]
- Carnaval, A.C.; Hickerson, M.J.; Haddad, C.F.B.; Rodrigues, M.T.; Moritz, C. Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science 2009, 323, 785–789. [Google Scholar]
- Lemey, P.; Rambaut, A.; Drummond, A.J.; Suchard, M.A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009, 5, e1000520. [Google Scholar] [CrossRef]
- Lemey, P.; Rambaut, A.; Welch, J.J.; Suchard, M.A. Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 2010, 27, 1877–1885. [Google Scholar] [CrossRef]
- Aoki, K.; Kato, M.; Murakami, N. Phylogeography of phytophagous weevils and plant species in broadleaved evergreen forests: A congruent genetic gap between western and eastern parts of Japan. Insects 2011, 2, 128–150. [Google Scholar] [CrossRef]
- Goldberg, J.; Trewick, S.A. Exploring phylogeographic congruence in a continental island system. Insects 2011, 2, 369–399. [Google Scholar] [CrossRef]
- Hughes, J.M.; Huey, J.A.; McLean, A.J.; Baggiano, O. Aquatic insects in eastern Australia: A window on ecology and evolution of dispersal in streams. Insects 2011, 2, 447–461. [Google Scholar] [CrossRef]
- Kawakami, T.; Butlin, R.K.; Cooper, S.J.B. Chromosomal speciation revisited: Modes of diversification in Australian morabine grasshoppers (Vandiemenella, viatica species group). Insects 2011, 2, 49–61. [Google Scholar] [CrossRef]
- Maekawa, K.; Nalepa, C.A. Biogeography and phylogeny of wood-feeding cockroaches in the genus Cryptocercus. Insects 2011, 2, 354–368. [Google Scholar] [CrossRef]
- McGaughran, A.; Stevens, M.I.; Hogg, I.D.; Carapelli, A. Extreme glacial legacies: A synthesis of the Antarctic springtail phylogeographic record. Insects 2011, 2, 62–82. [Google Scholar] [CrossRef]
- Miraldo, A.; Wirta, H.; Hanski, I. Origin and diversification of dung beetles in Madagascar. Insects 2011, 2, 112–127. [Google Scholar] [CrossRef]
- Pfeiler, E.; Markow, T.A. Phylogeography of the cactophilic Drosophila and other arthropods associated with cactus necroses in the Sonoran Desert. Insects 2011, 2, 218–231. [Google Scholar] [CrossRef]
- Trewick, S.A.; Wallis, G.P.; Morgan-Richards, M. The invertebrate life of New Zealand: A phylogeographic approach. Insects 2011, 2, 297–325. [Google Scholar] [CrossRef]
- Garrick, R.C. Montane refuges and topographic complexity generate and maintain invertebrate biodiversity: Recurring themes across space and time. J. Insect Conserv. 2011, 15, 469–478. [Google Scholar] [CrossRef]
- Beheregaray, L.B. Twenty years of phylogeography: The state of the field and the challenges for the southern hemisphere. Mol. Ecol. 2008, 17, 3754–3774. [Google Scholar]
- Templeton, A.R. Nested clade analyses of phylogeographic data: Testing hypotheses about gene flow and population history. Mol. Ecol. 1998, 7, 381–397. [Google Scholar] [CrossRef]
- Cruzan, M.B.; Templeton, A.R. Paleoecology and coalescence: Phylogeographic analysis of hypotheses from the fossil record. Trends Ecol. Evol. 2000, 15, 491–496. [Google Scholar] [CrossRef]
- Hugall, A.; Moritz, C.; Moussalli, A.; Stanisic, J. Reconciling paleodistribution models and comparative phylogeography in the Wet Tropics rainforest land snail Gnarosophia bellendenkerensis (Brazier 1875). Proc. Natl. Acad. Sci. USA 2002, 99, 6112–6117. [Google Scholar]
- Graham, C.H.; Moritz, C.; Williams, S.E. Habitat history improves prediction of biodiversity in rainforest fauna. Proc. Natl. Acad. Sci. USA 2006, 103, 632–636. [Google Scholar]
- Heatwole, H. Major Components and Distributions of the Terrestrial Fauna. In The Fauna of Australia; Dyne, G.R., Walton, D.W., Eds.; Australian Government Publishing Service: Canberra, Australia, 1987; Volume 1, pp. 69–100. [Google Scholar]
- Moritz, C.; Richardson, K.S.; Ferrier, S.; Monteith, G.B.; Stanisic, J.; Williams, S.E.; Whiffin, T. Biogeographical concordance and efficiency of taxon indicators for establishing conservation priority in a tropical rainforest biota. Proc. R. Soc. Lond. B 2001, 268, 1875–1881. [Google Scholar] [CrossRef]
- Yeates, D.K.; Bouchard, P.; Monteith, G.B. Patterns and levels of endemism in the Australian Wet Tropics rainforest: Evidence from flightless insects. Invertebr. Syst. 2002, 16, 605–619. [Google Scholar] [CrossRef]
- Barclay, S.; Ash, J.E.; Rowell, D.M. Environmental factors influencing the presence and abundance of a log-dwelling invertebrate, Euperipatoides rowelli (Onychophora: Peripatopsidae). J. Zool. 2000, 250, 425–436. [Google Scholar] [CrossRef]
- Bond, J.E.; Beamer, D.A.; Lamb, T.; Hedin, M. Combining genetic and geospatial analyses to infer population extinction in mygalomorph spiders endemic to the Los Angeles region. Anim. Conserv. 2006, 9, 145–157. [Google Scholar] [CrossRef]
- Grove, S.; Meggs, J.; Goodwin, A. A Review of Biodiversity Conservation Issues Relating to Coarse Woody Debris Management in the Wet Eucalypt Production Forests of Tasmania. Division of Forest Management, Forestry Tasmania Technical Report 22: Hobart, Tasmania, Australia, 2002; pp. 1–72. [Google Scholar]
- Speight, M.C.D. Saproxylic Invertebrates and Their Conservation. Nature and Environment Series No. 42; Council of Europe: Strasbourg, France, 1989.
- Grove, S.J. Tree Basal area and dead wood as surrogate indicators of saproxylic insect faunal integrity: A case study from the Australian lowland tropics. Ecol. Indic. 2002, 1, 171–188. [Google Scholar] [CrossRef]
- Stork, N.E.; Hammond, P.M. Sampling Arthropods from Tree-crowns by Fogging with Knock-down Insecticides: Lessons from Studies of Oak Tree Beetle Assemblages in Richmond Park (UK). In Canopy Arthropods; Stork, N.E., Adis, J., Didham, R.K., Eds.; Chapman and Hall: London, UK, 1997; pp. 5–26. [Google Scholar]
- Hewitt, G.M. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Barrows, T.T.; Stone, J.O.; Fifield, L.K.; Cresswell, R.G. Late Pleistocene glaciation of the Kosciuszko massif, snowy mountains, Australia. Quat. Res. 2001, 55, 179–189. [Google Scholar] [CrossRef]
- Hewitt, G.M. Biodiversity: A climate for colonization. Heredity 2004, 92, 1–2. [Google Scholar] [CrossRef]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. Lond. B 2004, 359, 183–195. [Google Scholar] [CrossRef]
- Keast, A. Distributional Patterns, Regional Biotas, and Adaptations in the Australian Biota: A Synthesis. In Ecological Biogeography of Australia; Keast, A., Ed.; Junk: The Hague: South Holland, The Netherlands, 1981; Volume 3, pp. 1891–1997. [Google Scholar]
- Chapple, D.G.; Keogh, J.S. Parallel adaptive radiations in arid and temperate Australia: Molecular phylogeography and systematics of the Egernia whitii (Lacertilia: Scincidae) species group. Biol. J. Linn. Soc. Lond. 2004, 83, 157–173. [Google Scholar] [CrossRef]
- Chapple, D.G.; Keogh, J.S.; Hutchinson, M.N. Substantial genetic substructuring in southeastern and alpine Australia revealed by molecular phylogeography of the Egernia whitii (Lacertilia: Scincidae) species group. Mol. Ecol. 2005, 14, 1279–1292. [Google Scholar] [CrossRef]
- Hodges, K.M.; Rowell, D.M.; Keogh, J.S. Remarkably different phylogeographic structure in two closely related lizard species in a zone of sympatry in south-eastern Australia. J. Zool. 2007, 272, 64–72. [Google Scholar] [CrossRef]
- Symula, R.; Keogh, J.S.; Cannatella, D.C. Ancient phylogeographic divergence in southeastern Australia among populations of the widespread common froglet, Crinia signifera. Mol. Phylogenet. Evol. 2008, 47, 569–580. [Google Scholar] [CrossRef]
- Byrne, M. Evidence for multiple refugia at different time scales during Pleistocene climatic oscillations in southern Australia inferred from phylogeography. Quat. Sci. Rev. 2008, 27, 2576–2585. [Google Scholar] [CrossRef]
- Byrne, M.; Steane, D.A.; Joseph, L.; Yeates, D.K.; Jordan, G.J.; Crayn, D.; Aplin, K.; Cantrill, D.J.; Cook, L.G.; Crisp, M.D.; et al. Decline of a biome: Evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. J. Biogeogr. 2011, 38, 1635–1656. [Google Scholar] [CrossRef]
- Kershaw, A.P.; D’Costa, D.M.; McEwen Mason, J.R.C.; Wagstaff, B.E. Palynological evidence for Quaternary vegetation and environments of mainland southeastern Australia. Quat. Sci. Rev. 1991, 10, 391–404. [Google Scholar] [CrossRef]
- Hope, G.S. Quaternary Vegetation. In History of the Australian Vegetation: Cretaceous to Recent; Hill, R.S., Ed.; Cambridge University Press: Cambridge, UK, 1994; pp. 368–389. [Google Scholar]
- Bowler, J.M. Aridity in the Late Tertiary and Quaternary of Australia. In Evolution of the Flora and Fauna of Arid Australia; Barker, W.R., Greenslade, P.J.M., Eds.; Peacock Publications: Frewville, Australia, 1982; pp. 35–45. [Google Scholar]
- Frakes, L.A.; McGowran, B.; Bowler, J.M. Evolution of Australian Environments. In The Fauna of Australia; Dyne, G.R., Walton, D.W., Eds.; Australian Government Publishing Service: Canberra, Australia, 1987; Volume 1, pp. 1–16. [Google Scholar]
- Garrick, R.C.; Sands, C.J.; Rowell, D.M.; Tait, N.N.; Greenslade, P.; Sunnucks, P. Phylogeography recapitulates topography: Very fine-scale local endemism of a saproxylic ‘giant’ springtail at Tallaganda in the Great Dividing Range of south-east Australia. Mol. Ecol. 2004, 13, 3329–3344. [Google Scholar]
- Greenslade, P.; Moore, S.; Farrow, R. Observations on the feeding behaviour of Uchidanurinae (Collembola: Neanuridae) in Australia. Vic. Nat. 2002, 119, 221–223. [Google Scholar]
- Winsor, L. Aspects of taxonomy and functional histology in terrestrial flatworms (Tricladida: Terricola). Pedobiologia 1998, 42, 412–432. [Google Scholar]
- Winsor, L. The Australian terrestrial flatworm fauna (Tricladida: Terricola). Pedobiologia 1998, 42, 457–463. [Google Scholar]
- Sluys, R. Global diversity of land planarians (Platyhelminthes, Tricladida, Terricola): A new indicator-taxon in biodiversity and conservation studies. Biodivers. Conserv. 1999, 8, 1663–1681. [Google Scholar] [CrossRef]
- Dendy, A. On the Victorian land planarians. Trans. Roy. Soc. Vic. 1891, 2, 65–80. [Google Scholar]
- Moseley, H.N. Notes on the structure of several forms of land planarian with a description of two new genera and several new species, and a list of all species at present known. Q. J. Microsc. Sci. 1877, 17, 273–292. [Google Scholar]
- Trewick, S.A.; Wallis, G.P. Bridging the “beech-gap”: New Zealand invertebrate phylogeography implicates Pleistocene glaciation and Pliocene Isolation. Evolution 2001, 55, 2170–2180. [Google Scholar]
- Nalepa, C.A.; Luykx, P.; Klass, K-.D.; Deitz, L.L. Distribution of karyotypes of the Cryptocercus punctulatus species complex (Dictyoptera: Cryptoceridae) in the southern Appalachians: Relation to habitat and history. Ann. Entomol. Soc. Am. 2002, 95, 276–287. [Google Scholar] [CrossRef]
- Schmuki, C.; Vorburger, C.; Runciman, D.; MacEachern, S.; Sunnucks, P. When log-dwellers meet loggers: Impacts of forest fragmentation on endemic log-dwelling beetles in southeastern Australia. Mol. Ecol. 2006, 15, 1481–1492. [Google Scholar] [CrossRef]
- Leschen, R.A.B.; Buckley, T.R.; Harman, H.M.; Shulmeister, J. Determining the origin and age of the westland beech (Nothofagus) gap, New Zealand, using fungus beetle genetics. Mol. Ecol. 2008, 17, 1256–1276. [Google Scholar] [CrossRef]
- Thomas, S.M.; Hedin, M. Multigenic phylogeographic divergence in the paleoendemic southern Appalachian opilionid Fumontana deprehendor Shear (Opiliones, Laniatores, Triaenonychidae). Mol. Phylogenet. Evol. 2008, 46, 645–658. [Google Scholar] [CrossRef]
- Marske, K.A.; Leschen, R.A.B.; Barker, G.M.; Buckley, T.R. Phylogeography and ecological niche modelling implicate coastal refugia and trans-alpine dispersal of a New Zealand fungus beetle. Mol. Ecol. 2009, 18, 5126–5142. [Google Scholar] [CrossRef]
- Walker, M.J.; Stockman, A.K.; Marek, P.E.; Bond, J.E. Pleistocene glacial refugia across the Appalachian Mountains and coastal plain in the millipede genus Narceus: Evidence from population genetic, phylogeographic, and paleoclimatic data. BMC Evol. Biol. 2009, 9, 25. [Google Scholar]
- Garrick, R.C.; Rowell, D.M.; Simmons, C.S.; Hillis, D.M.; Sunnucks, P. Fine-scale phylogeographic congruence despite demographic incongruence in two low-mobility saproxylic springtails. Evolution 2008, 62, 1103–1118. [Google Scholar] [CrossRef]
- Garrick, R.C.; Sunnucks, P. Development and application of three-tiered nuclear genetic markers for basal hexapods using single-stranded conformation polymorphism coupled with targeted DNA sequencing. BMC Genet. 2006, 7, 11. [Google Scholar] [CrossRef]
- Garrick, R.C.; Sands, C.J.; Rowell, D.M.; Hillis, D.M.; Sunnucks, P. Catchments catch all: Long-term population history of a giant springtail from the southeast Australian highlands—A multigene approach. Mol. Ecol. 2007, 16, 1865–1882. [Google Scholar] [CrossRef]
- Sunnucks, P.; Blacket, M.J.; Taylor, J.M.; Sands, C.J.; Ciavaglia, S.A.; Garrick, R.C.; Tait, N.N.; Rowell, D.M.; Pavlova, A. A tale of two flatties: Different responses of two terrestrial flatworms to past environmental climatic fluctuations at Tallaganda in montane southeastern Australia. Mol. Ecol. 2006, 15, 4513–4531. [Google Scholar]
- Edwards, S.V.; Beerli, P. Perspective: Gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution 2000, 54, 1839–1854. [Google Scholar]
- Arbogast, B.S.; Edwards, S.V.; Wakeley, J.; Beerli, P.; Slowinski, J.B. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu. Rev. Ecol. Syst. 2002, 33, 707–740. [Google Scholar] [CrossRef]
- Ho, S.Y.W.; Phillips, M.J.; Cooper, A.; Drummond, A.J. Time dependency of molecular rate estimates and systematic overestimation of recent divergence dates. Mol. Biol. Evol. 2005, 22, 1561–1568. [Google Scholar] [CrossRef]
- Brower, A.V.Z. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6491–6495. [Google Scholar] [CrossRef]
- Barrows, T.T.; Stone, J.O.; Fifield, L.K. Exposure ages for Pleistocene periglacial deposits in Australia. Quat. Sci. Rev. 2004, 23, 697–708. [Google Scholar] [CrossRef]
- Beavis, A.S.; Rowell, D.M. Phylogeography of Two Australian Species of Funnel Web Spider (Araneae: Mygalomorphae: Hexathelidae) in Tallaganda State Forest, New South Wales. In Insect Biodiversity and Dead Wood: Proceedings of a Symposium for the 22nd International Congress of Entomology, Asheville, NC, USA, 2006; Grove, S.J., Hanula, J.L., Eds.; pp. 23–29, U.S. Department of Agriculture Forest Service Southern Research Station General Technical Report SRS-93.
- Beavis, A.S.; Sunnucks, P.; Rowell, D.M. Microhabitat preferences drive phylogeographic disparities in two Australian funnel web spiders. Biol. J. Linn. Soc. Lond. 2011, 104, 805–819. [Google Scholar] [CrossRef]
- Sunnucks, P.; Wilson, A.C.C. Microsatellite markers for the onychophoran Euperipatoides rowelli. Mol. Ecol. 1999, 8, 895–906. [Google Scholar] [CrossRef]
- Sunnucks, P.; Tait, N. Velvet worms: Tales of the unexpected. Nat. Aust. 2001, 27, 61–69. [Google Scholar]
- Barton, N.H.; Hewitt, G.M. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985, 16, 113–148. [Google Scholar]
- Morgan-Richards, M.; Wallis, G.P. A comparison of five hybrid zones of the weta Hemideina thoracica (Orthoptera: Anostostomatidae): Degree of cytogenetic differentiation fails to predict zone width. Evolution 2003, 54, 849–861. [Google Scholar]
- Moritz, C.; Hoskin, C.H.; MacKenzie, J.B.; Phillips, B.L.; Tonione, M.; Silva, N.; VanDerWal, J.; Williams, S.E.; Graham, C.H. Identification and dynamics of a cryptic suture zone in tropical rainforest. Proc. R. Soc. Lond. B 2009, 276, 1235–1244. [Google Scholar] [CrossRef]
- Mesibov, R. A remarkable case of mosaic parapatry in millipedes. ZooKeys 2011, 156, 71–84. [Google Scholar] [CrossRef]
- Shaw, D.D.; Wilkinson, P. Chromosome differentiation, hybrid breakdown and maintenance of a narrow hybrid zone in Caledia. Chromosoma 1980, 80, 1–31. [Google Scholar] [CrossRef]
- Deharveng, L.; Bedos, A.; Gisclard, C. Environmental factors, microgeographic patterns of endemism and hybrid zones in Monobella grassei (Insecta: Collembola: Neanuridae). Biol. J. Linn. Soc. Lond. 1998, 64, 527–554. [Google Scholar]
- Pritchard, J.K.; Stevens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002; pp. 1–20. [Google Scholar]
- Grove, S.J.; Stork, N.E. An inordinate fondness for beetles. Invertebr. Taxon. 2000, 14, 733–739. [Google Scholar] [CrossRef]
- Grove, S.J. Extent and composition of dead wood in Australian lowland tropical rainforest with management histories. For. Ecol. Manage. 2001, 154, 35–53. [Google Scholar] [CrossRef]
- Yee, M.; Yuan, Z.-.Q.; Mohammed, C. Not just waste wood: Decaying logs as key habitats in Tasmania’s wet sclerophyll Eucalyptus obliqua production forests: The ecology of large and small logs compared. Tasforests 2001, 13, 119–128. [Google Scholar]
- Gibb, H.; Hjältén, J.; Ball, J.P.; Atlegrim, O.; Pettersson, R.B.; Hilszczanski, J.; Johansson, T.; Danell, K. Effects of landscape composition and substrate availability on saproxylic beetles in boreal forests: A study using experimental logs for monitoring assemblages. Ecography 2006, 29, 191–204. [Google Scholar] [CrossRef]
- Yee, M.; Grove, S.J.; Richardson, A.M.M.; Mohammed, C.L. Brown Rot in Inner Heartwood: Why Large Logs Support Characteristic Saproxylic Beetle Assemblages of Conservation Concern. In Insect Biodiversity and Dead Wood: Proceedings of a Symposium for the 22nd International Congress of Entomology, Asheville, NC, USA, 2006; Grove, S.J., Hanula, J.L., Eds.; pp. 42–56, U.S. Department of Agriculture Forest Service Southern Research Station General Technical Report SRS-93.
- Lindenmayer, D.B.; Margules, C.R.; Botkin, D.B. Indicators of biodiversity for ecologically sustainable forest management. Conserv. Biol. 2000, 14, 941–950. [Google Scholar] [CrossRef]
- Lindenmayer, D.B.; Cunningham, R.B.; Donnelly, C.F.; Lesslie, R. On the use of landscape surrogates as ecological indicators in fragmented forests. For. Ecol. Manage. 2002, 159, 203–216. [Google Scholar] [CrossRef]
- Grove, S.J. Saproxylic insect ecology and the sustainable management of forests. Annu. Rev. Ecol. Syst. 2002, 33, 1–23. [Google Scholar] [CrossRef]
- Australian Government. Regional Forest Agreement for Southern New South Wales, April 2001; Australian Government, 2001; pp. 1–110. Available online: http://www.affa.gov.au (accessed on 20 February 2012).
- Grove, S.J.; Bashford, R. Beetle assemblages from the Warra log-decay project: Insights from the first year of sampling. Tasforests 2003, 14, 117–129. [Google Scholar]
- State Forests of New South Wales. Proposed Forestry Operations in the Queanbeyan and Badja Management Area, Environmental Impact Statement; State Forests of New South Wales, Southern Region: Pennant Hills, NSW, Australia, 1995. [Google Scholar]
- Grove, S.J. The influence of forest management history on the integrity of the saproxylic beetle fauna in an Australian tropical rainforest. Biol. Conserv. 2002, 104, 149–171. [Google Scholar] [CrossRef]
- Grove, S.J. A Research Agenda for Insects and Dead Wood. In Insect Biodiversity and Dead Wood: Proceedings of a Symposium for the 22nd International Congress of Entomology, Asheville, NC, USA, 2006; Grove, S.J., Hanula, J.L., Eds.; pp. 98–108, U.S. Department of Agriculture Forest Service Southern Research Station General Technical Report SRS-93.
- Langor, D.W.; Spence, J.R.; Hammond, H.E.J.; Jacobs, J.; Cobb, T.P. Maintaining Saproxylic Insects in Canada’s Extensively Managed Boreal Forests: A Review. In Insect Biodiversity and Dead Wood: Proceedings of a Symposium for the 22nd International Congress of Entomology, Asheville, NC, USA, 2006; Grove, S.J., Hanula, J.L., Eds.; pp. 83–97, U.S. Department of Agriculture Forest Service Southern Research Station General Technical Report SRS-93.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Garrick, R.C.; Rowell, D.M.; Sunnucks, P. Phylogeography of Saproxylic and Forest Floor Invertebrates from Tallaganda, South-eastern Australia. Insects 2012, 3, 270-294. https://doi.org/10.3390/insects3010270
Garrick RC, Rowell DM, Sunnucks P. Phylogeography of Saproxylic and Forest Floor Invertebrates from Tallaganda, South-eastern Australia. Insects. 2012; 3(1):270-294. https://doi.org/10.3390/insects3010270
Chicago/Turabian StyleGarrick, Ryan C., David M. Rowell, and Paul Sunnucks. 2012. "Phylogeography of Saproxylic and Forest Floor Invertebrates from Tallaganda, South-eastern Australia" Insects 3, no. 1: 270-294. https://doi.org/10.3390/insects3010270





