Generation of Nutrients and Detoxification: Possible Roles of Yeasts in Leaf-Cutting Ant Nests
Abstract
:1. Introduction
2. Experimental Section
2.1. Yeast Isolates and Identification
2.2. Assaying Yeasts for Hydrolytic Enzymes
2.3. Assimilation of the Hydrolysis Products
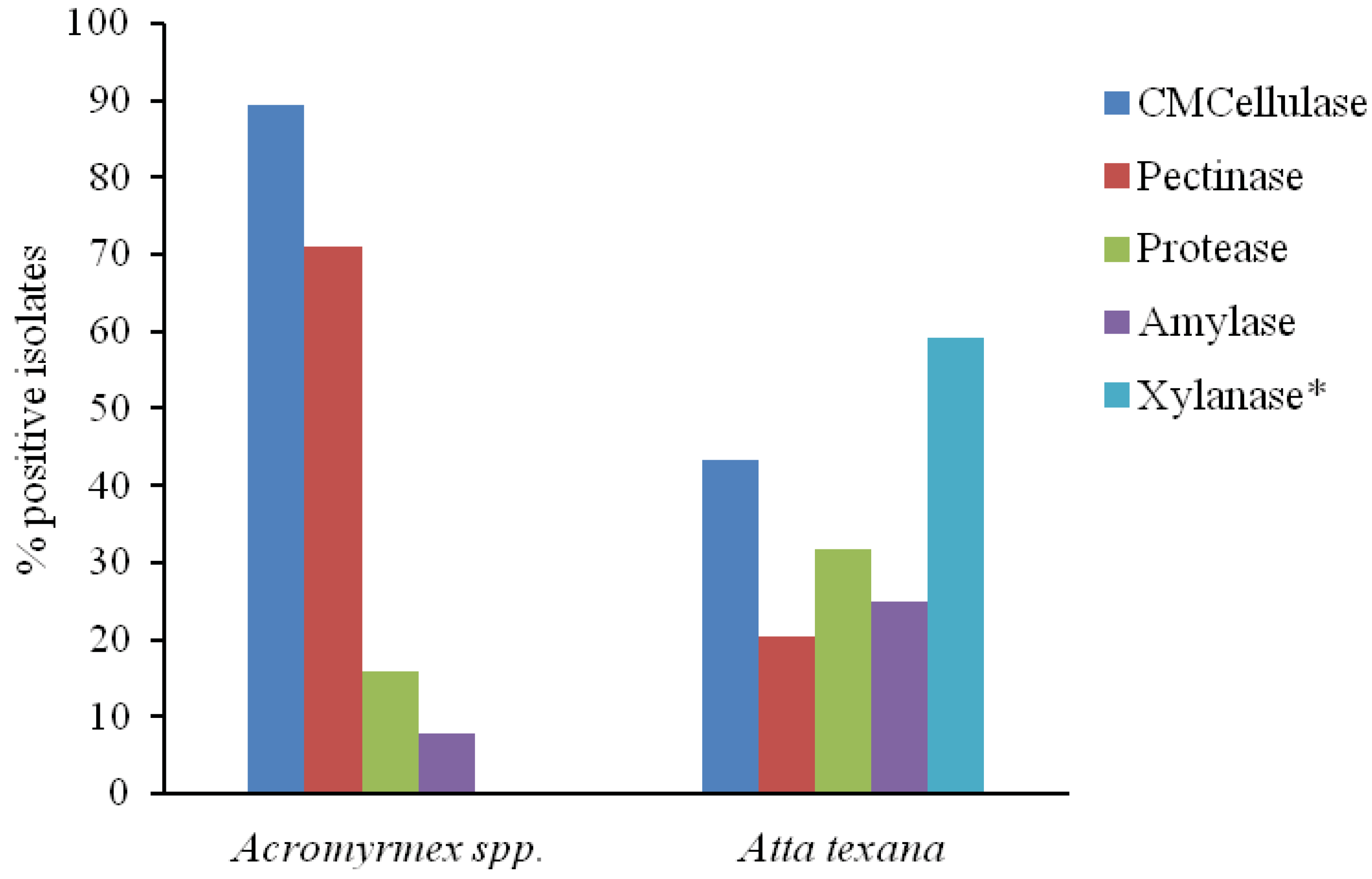
3. Results and Discussion
| Yeast Species 3 | N 4 | Hydrolytic Enzymes 1 | Assimilation 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AM | CEL | P | XYL | PRT | M | C | A | X | |||
| Aureobasidium pullulans | 3 | - | 3 5 | - | 3 | 2 | 3 | 3 | 2 | 3 | |
| Bullera sinensis | 1 | 1 | 1 | - | 1 | - | 1 | 1 | 1 | 1 | |
| Bulleromyces albus | 1 | - | 1 | - | 1 | - | 1 | 1 | 1 | 1 | |
| Candida membranifaciens | 3 | - | - | - | - | - | 3 | 3 | - | 3 | |
| Candida melibiosica | 1 | - | - | - | - | - | 1 | 1 | - | 1 | |
| Cryptococcus laurentii | 3 | 3 | 2 | 1 | 2 | - | 3 | 3 | 3 | 3 | |
| Cryptococcus flavus | 3 | 3 | 3 | 1 | 3 | 2 | 3 | 3 | 3 | 3 | |
| Cryptococcus magnus | 3 | - | 3 | - | 2 | 2 | 3 | 3 | - | 3 | |
| Cryptococcus luteolus | 1 | - | - | - | - | 1 | 1 | 1 | 1 | 1 | |
| Cryptococcus podzolicus | 1 | 1 | - | - | 1 | - | 1 | 1 | 1 | 1 | |
| Cryptococcus flavescens | 1 | - | - | - | 1 | - | 1 | 1 | 1 | 1 | |
| Cryptococcus cf. cellulolyticus | 2 | - | 2 | - | 2 | - | 2 | 2 | 2 | 2 | |
| Cryptococcus sp. (ATT178) | 1 | - | - | - | 1 | - | 1 | 1 | 1 | 1 | |
| Cryptococcus sp. 1 (ATT079) | 1 | - | 1 | - | 1 | - | 1 | 1 | - | 1 | |
| Cryptococcus sp. 2 (ATT080) | 1 | - | - | - | - | - | 1 | 1 | - | 1 | |
| Cryptococcus sp. 3 (ATT123) | 1 | - | - | - | - | - | 1 | 1 | 1 | 1 | |
| Cryptococcus sp. 4 (ATT176) | 1 | 1 | - | 1 | 1 | - | 1 | 1 | 1 | 1 | |
| Farysizyma sp. 6 | 1 | - | - | - | 1 | 1 | 1 | 1 | 1 | 1 | |
| Hannaella kunmingensis 6 | 1 | - | - | - | - | - | 1 | 1 | 1 | 1 | |
| Kodamaea ohmeri | 1 | - | - | - | - | - | 1 | 1 | - | 1 | |
| Pseudozyma sp. | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Rhodotorula nothofagi | 1 | 1 | - | 1 | 1 | - | 1 | 1 | - | 1 | |
| Rhodotorula mucilaginosa | 1 | - | - | 1 | - | - | 1 | 1 | 1 | 1 | |
| Rhodotorula lactosa | 1 | - | - | - | - | - | 1 | 1 | 1 | 1 | |
| Rhodotorula sp. HB 1211 | 1 | - | - | - | - | - | 1 | 1 | 1 | 1 | |
| Sporidiobolus ruineniae | 2 | - | - | 2 | - | - | 2 | 2 | - | 2 | |
| Sporisorium penniseti (yeast-like) | 2 | - | - | 1 | 1 | 2 | 2 | 2 | 2 | 2 | |
| Sympodiomycopsis paphiopedili | 2 | - | 1 | - | 2 | 1 | 2 | 2 | - | 2 | |
| unidentified yeast-like fungus | 2 | - | 1 | - | 1 | 2 | 2 | 2 | 2 | 2 | |
| Total | 44 | 11 | 19 | 9 | 26 | 14 | 44 | 44 | 28 | 44 | |
| Yeast Species | N 4 | Closest Relative 1 | Hydrolytic Enzymes 2 | Assimilation 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | Accession | AM | CEL | P | PRT | M | C | A | X | ||||
| Cryptococcus laurentii | 1 | 100 | JQ317168 | - | - | - | 1 5 | 1 | 1 | 1 | 1 | ||
| Galactomyces candidus | 15 | 100 | JQ317161 | - | 15 | 12 | 2 | 0 | 0 | 14 | 15 | ||
| Meyerozyma caribbica | 1 | 100 | JQ317162 | - | 1 | - | - | 1 | 1 | 1 | 1 | ||
| Meyerozyma guilliermondii | 6 | 100 | JQ317164 | - | 6 | 6 | 1 | 6 | 6 | 3 | 6 | ||
| Trichosporon chiarellii | 6 | 100 | JQ317165 | - | 3 | 4 | 1 | 6 | 6 | 6 | 6 | ||
| Trichosporon montevideense | 6 | 100 | JQ317167 | 1 | 6 | 2 | 1 | 6 | 6 | 3 | 6 | ||
| Trichosporon multisporum | 3 | 100 | JQ317166 | 2 | 3 | 3 | - | 3 | 3 | 2 | 3 | ||
| Total | 38 | 3 | 34 | 27 | 6 | 23 | 23 | 30 | 38 | ||||
4. Conclusion
Acknowledgments
References
- Hölldobler, E.; Wilson, E.O. Journey to the Ants: A Story of Scientific Exploration, 3rd ed.; Harvard University Press: Cambridge, London, UK, 1995. [Google Scholar]
- Currie, C.R. A community of ants, fungi and bacteria: A multilateral approach to study symbiosis. Annu. Rev. Microbiol. 2001, 55, 357–380. [Google Scholar] [CrossRef]
- Siqueira, C.G.; Bacci, M., Jr.; Pagnocca, F.C.; Bueno, O.C.; Hebling, M.J.A. Metabolism of plant polysaccharides by Leucoagaricus gongylophorus, the symbiotic fungus of the leaf-cutting ant Atta sexdens L. Appl. Environ. Microbiol. 1998, 64, 4820–4822. [Google Scholar]
- Silva, A.; Bacci, M., Jr.; Pagnocca, F.C.; Bueno, O.C.; Hebling, M.J.A. Production of polysaccharidases in different carbon sources by Leucoagaricus gongylophorus Möller (Singer), the symbiotic fungus of the leaf-cutting ant Atta sexdens Linnaeus. Curr. Microbiol. 2006, 53, 68–71. [Google Scholar]
- Schiott, M.; Rogowska-Wrzesinka, A.; Roepstorff, P.; Boomsma, J.J. Leaf-cutting ant fungi produce cell wall degrading pectinase complexes reminiscent of phytopathogenic fungi. BMC Biol. 2010, 8, 1–12. [Google Scholar] [CrossRef]
- Kooij, P.W.; Schiott, M.; Boomsma, J.J.; de Fine Licht, H.H. Rapid shifts in Atta cephalotes fungus-garden enzyme activity after a change in fungal substrate (Attini, Formicidae). Insectes Soc. 2011, 58, 145–151. [Google Scholar] [CrossRef]
- Silva, A.; Bacci, M., Jr.; Siqueira, G.C.; Bueno, O.C.; Pagnocca, F.C.; Hebling, M.J.A. Survival of Atta sexdens workers on different food sources. J. Insect Physiol. 2003, 49, 307–313. [Google Scholar] [CrossRef]
- Weber, N.A. Gardening Ants: The Attines, Memoirs of American Philosophical Society; American Philosophical Society: Philadelphia, PA, USA, 1972; Volume 92, pp. 1–146. [Google Scholar]
- Currie, C.R.; Stuart, A.E. Weeding and grooming of pathogens in agriculture by ants. Proc. R. Soc. Lond. B Biol. Sci. 2001, 268, 1033–1039. [Google Scholar] [CrossRef]
- Beattie, A.J.; Turnbull, C.L.; Hough, T.; Knox, R.B. Antibiotic production: A possible function for the metapleural glands of ants (Hymenoptera: Formicidae). Ann. Entomol. Soc. Am. 1986, 79, 448–450. [Google Scholar]
- Nascimento, R.R.; Schoeters, E.; Morgan, E.D.; Billen, J.; Stradling, D.J. Chemistry of metapleural gland secretions of three attine ants, Atta sexdens rubropilosa, Atta cephalotes, and Acromyrmex octospinosus (Hymenoptera: Formicidae). J. Chem. Ecol. 1996, 22, 987–1000. [Google Scholar] [CrossRef]
- Poulsen, M.; Hughes, W.O.H.; Boomsma, J.J. Differential resistance and the importance of antibiotic production in Acromyrmex echinatior leaf-cutting ant castes towards the entomopathogenic fungus Aspergillus nomius. Insectes Soc. 2006, 53, 349–355. [Google Scholar] [CrossRef]
- Marsaro, A.L., Jr.; Della Lucia, T.M.C.; Barbosa, L.C.A.; Mafia, L.A.; Morandi, M.A.B. Efeito de secreções da glândula mandibular de Atta sexdens rubropilosa Forel (Hymenoptera: Formicidae) sobre a germinação de conídios de Botrytis cinerea Pers. Fr. Neotrop. Entomol. 2001, 30, 403–406. [Google Scholar] [CrossRef]
- Rodrigues, A.; Carletti, C.D.; Bueno, O.C.; Pagnocca, F.C. Leaf-cutting ant faecal fluid and mandibular gland secretion: Effects on microfungi spore germination. Braz. J. Microbiol. 2008, 39, 64–67. [Google Scholar] [CrossRef]
- Currie, C.R.; Scott, J.A.; Summerbell, R.C.; Malloch, D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 1999, 398, 701–704. [Google Scholar]
- Currie, C.R.; Scott, J.A.; Summerbell, R.C.; Malloch, D. Corrigendum: Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 2003, 423, 461. [Google Scholar]
- Sen, R.; Ishak, H.D.; Estrada, D.; Dowd, S.E.; Hong, E.; Mueller, U.G. Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc. Natl. Acad. Sci. USA 2009, 106, 17805–17810. [Google Scholar]
- Mueller, U.G.; Dash, D.; Rabeling, C.; Rodrigues, A. Coevolution between attine ants and actinomycete bacteria: A reevaluation. Evolution 2008, 62, 2894–2912. [Google Scholar] [CrossRef]
- Zucchi, T.D.; Guidolin, A.S.; Cônsoli, F.L. Isolation and characterization of actinobacteria ectosymbionts from Acromyrmex subterraneus brunneus (Hymenoptera, Formicidae). Microbiol. Res. 2011, 166, 68–76. [Google Scholar] [CrossRef]
- Reynolds, H.T.; Currie, C.R. Pathogenicity of Escovopsis weberi: The parasite of the attine ant-microbe symbiosis directly consumes the ant-cultivated fungus. Mycologia 2004, 96, 955–959. [Google Scholar] [CrossRef]
- Fisher, P.J.; Stradling, D.J.; Sutton, B.C.; Petrini, L.E. Microfungi in the fungus gardens of the leaf-cutting ant Atta cephalotes: A preliminary study. Mycol. Res. 1996, 100, 541–546. [Google Scholar] [CrossRef]
- Craven, S.E.; Dix, M.W.; Michaelis, G.E. Attine fungus garden contain yeasts. Science 1970, 169, 184–189. [Google Scholar]
- Angelis, C.; Serzedello, A.; de Angelis, D.F. Yeasts found in gardens of Atta sexdens rubropilosa and Atta laevigata. Naturalia 1983, 8, 149–151. [Google Scholar]
- Carreiro, S.C.; Pagnocca, F.C.; Bueno, O.C.; Bacci, M., Jr.; Hebling, M.J. A.; da Silva, O.A. Yeasts associated with nests of the leaf-cutting ant Atta sexdens rubropilosa Forel, 1908. Antonie van Leeuwenhoek 1997, 71, 243–248. [Google Scholar] [CrossRef]
- Pagnocca, F.C.; Rodrigues, A.; Nagamoto, N.S.; Bacci, M., Jr. Yeasts and filamentous fungi carried by the gynes of leaf-cutting ants. Antonie van Leeuwenhoek 2008, 94, 517–526. [Google Scholar] [CrossRef]
- Guedes, A.F.A.; Attili-Angelis, D.; Pagnocca, F.C. Selective isolation of dematiaceous fungi from the workers of Atta laevigata (Formicidae: Attini). Folia Microbiol. 2011. [Google Scholar]
- Little, A.E.F.; Currie, C.R. Symbiotic complexity: Discovery of a fifth symbiont in the attine ant-microbe symbiosis. Biol. Lett. 2007, 3, 501–504. [Google Scholar] [CrossRef]
- Rodrigues, A.; Cable, R.N.; Mueller, U.G.; Bacci, M., Jr.; Pagnocca, F.C. Antagonistic interactions between gardens yeasts and microfungal garden pathogens of leaf-cutting ants. Antonie van Leeuwenhoek 2009, 96, 331–342. [Google Scholar] [CrossRef]
- Ribeiro, S.B. Caracterização de espécies bacterianas encontradas em ninhos de Atta sexdens L. e isolamento de Streptomyces de formigas da tribo Attini. Ph.D. Thesis, Instituto de Biociências, Universidade Estadual Paulista, Rio Claro, Brazil, 2000. [Google Scholar]
- Bacci, M., Jr.; Ribeiro, S.B.; Casarotto, M.E.F.; Pagnocca, F.C. Biopolymer-degrading bacteria from nests of the leaf-cutting ant Atta sexdens rubropilosa. Braz. J. Med. Biol. Res. 1995, 28, 79–82. [Google Scholar]
- Rodrigues, A.; Bacci, M., Jr.; Mueller, U.G.; Ortiz, A.; Pagnocca, F.C. Microfungal “weeds” in the leafcutter ant symbiosis. Microb. Ecol. 2008, 56, 604–614. [Google Scholar] [CrossRef]
- Carreiro, S.C.; Pagnocca, F.C.; Bacci, M., Jr.; Lachance, M.A.; Bueno, O.C.; Hebling, M.J.A.; Ruivo, C.C.; Rosa, C.A. Simpodiomyces attinorum sp. nov., a yeast species associated with nests of the leaf-cutting ant Atta sexdens. Int. J. Syst. Evol. Microbiol. 2004, 54, 1891–1894. [Google Scholar]
- Middelhoven, W.J.; Fonseca, A.; Carreiro, S.C.; Pagnocca, F.C.; Bueno, O.C. Cryptococcus haglerorum sp. nov., an anamorphic basidiomycetous yeast isolated from nests of the leaf-cutting ant Atta sexdens. Antonie van Leeuwenhoek 2003, 83, 167–174. [Google Scholar] [CrossRef]
- Pagnocca, F.C.; Legaspe, M.F.C.; Rodrigues, A.; Ruivo, C.C.C.; Nagamoto, N.S.; Bacci, M., Jr.; Forti, L.C. Yeast isolated from a fungus-growing ant nest including the description of Trichosporon chiarellii sp. nov., an anamorphic basidiomycetous yeast. Int. J. Syst. Evol. Microbiol. 2010, 60, 1454–1459. [Google Scholar] [CrossRef]
- Suen, G.; Scott, J.J.; Aylward, F.O.; Adams, S.M.; Tringe, S.G.; Pinto-Tomás, A.A.; Foster, C.E.; Pauly, M.; Weimer, P.J.; Barry, K.W.; et al. An insect herbivore microbiome with high plant biomass-degrading capacity. PloS Genet. 2011, 6, 1–14. [Google Scholar]
- Pinto-Tomás, A.A.; Anderson, M.A.; Suen, G.; Stevenson, D.M.; Chu, F.S.T.; Cleland, W.W.; Weimer, P.J.; Currie, C.R. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 2009, 326, 1120–1123. [Google Scholar]
- de Almeida, J.M.G.C.F. Yeast community survey in the Tagus estuary. FEMS Microbiol. Lett. 2005, 52, 295–303. [Google Scholar]
- Sampaio, J.P.; Gadanho, M.; Santos, S.; Duarte, F.L.; Pais, C.; Fonseca, A.; Fell, J.W. Polyphasic taxonomy of basidiomycetous yeasts genus Rhodosporidium: Rhodosporidium kratochvilovae and related anamorphic species. Int. J. Syst. Evol. Microbiol. 2001, 51, 687–697. [Google Scholar]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar]
- Looder, J. The Yeast,a Taxonomic Study, 2nd ed.; North-Holland Pub. Co.: Amsterdam, The Netherlands, 1970. [Google Scholar]
- Maijala, P.; Fagerstedt, K.V.; Raudaskoski, M. Detection of extracellular cellulolytic and proteolytic activity in ectomycorrhyzal fungi and Hetererobasidion annosum (Fr.) Bref. New Phytol. 1991, 117, 643–648. [Google Scholar] [CrossRef]
- Mckay, A.M. A plate assay method for the detection of fungal polygalacturonase secretion. FEMS Microbiol. Lett. 1988, 56, 355–358. [Google Scholar] [CrossRef]
- Williams, A.G. Staining reactions for the detection of hemicellulose-degrading bacteria. FEMS Microbiol. Lett. 1983, 20, 253–358. [Google Scholar] [CrossRef]
- Carreiro, S.C.; Pagnocca, F.C.; Bacci, M., Jr.; Bueno, O.C.; Hebling, M.J.A.; Middelhoven, W.J. Occurrence of killer yeasts in leaf-cutting ant nests. Folia Microbiol. 2002, 47, 259–262. [Google Scholar] [CrossRef]
- Inacio, J.; Pereira, P.; de Carvalho, M.; Fonseca, A.; Amaral-Collaço, M.T.; Spencer-Martins, I. Estimation and diversity of phylloplane mycobiota on selected plants in a Mediterranean-type ecosystem in Portugal. Microb. Ecol. 2002, 44, 344–353. [Google Scholar] [CrossRef]
- Glushakova, A.M.; Chernov, I.Y. Seasonal dynamics of the structure of epiphytic yeasts communities. Microbiology 2010, 79, 830–839. [Google Scholar] [CrossRef]
- Spencer, J.F.T.; Spencer, D.M. Yeasts in Natural and Artificial Habitats; Springer: Berlin, Germany, 1997. [Google Scholar]
- Sosa-Calvo, J.; Schultz, T.R. Three remarkable new fungus-growing ant species of the genus Myrmicocrypta (Hymenoptera: Formicidae), with a reassessment of the characters that define the genus and its position within the Attini. Ann. Entomol. Soc. Am. 2010, 103, 181–195. [Google Scholar] [CrossRef]
- Phaff, H.J.; Starmer, W.T. Yeasts associated with plants, insects and soil. In The Yeasts, 2nd ed.; Rose, A.H., Harrison, J.S., Eds.; Academic Press: London, UK, 1987; pp. 123–180. [Google Scholar]
- Papa, P.J.; Papa, F. Inhibition des bactéries dans les nids d’Acromyrmex octospinosus Reich. Bull. Soc. Pathol. Exot. 1982, 75, 415–425. [Google Scholar]
- Sakay, T.; Sakamoto, T.; Hallaert, J.; Vandamme, E.J. Pectin, pectinase, and protopectinase: Production, properties, and applications. Adv. Appl. Microbiol. 1993, 39, 213–293. [Google Scholar] [CrossRef]
- Carreiro, S.C. Pesquisa de fator killer e análise de degradação de polissacarídeos vegetais por leveduras associadas aos ninhos de Atta sexdens. Ph.D. Thesis, Instituto de Biociências de Rio Claro, Universidade Estadual Paulista, Rio Claro, Brazil, 1999. [Google Scholar]
- Stolp, H. Microbial Ecology: Organisms, Habitats, Activities, 1st ed.; Cambridge University Press: London, UK, 1988. [Google Scholar]
- Martin, M.M.; Weber, N.A. The celullose-utilizing capability of the fungus cultured by the attine ant Atta colombica tonsipes. Ann. Entomol. Soc. Am. 1969, 62, 1386–1387. [Google Scholar]
- Bacci, M., Jr.; Anversa, M.M.; Pagnocca, F.C. Cellulose degradation by Leucocoprinus gongylophorus, the fungus cultured by the leaf-cutting ant Atta sexdens rubropilosa. Antonie van Leeuwenhoek 1995, 67, 385–386. [Google Scholar] [CrossRef]
- Abril, A.B.; Bucher, E.H. Evidence that the fungus cultured by leaf-cutting ants does not metabolize cellulose. Ecol. Lett. 2002, 5, 325–328. [Google Scholar] [CrossRef]
- Silva, A.; Bacci, M., Jr.; Pagnocca, F.C.; Bueno, O.C.; Hebling, M.J.A. Starch metabolism in Leucoagaricus gongylophorus, the symbiotic fungus of leaf-cutting ants. Microbiol. Res. 2005, 161, 299–303. [Google Scholar]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study, 5th ed.; Elsevier: San Diego, CA, USA, 2011. [Google Scholar]
- Abril, A.B.; Bucher, E.H. Nutritional sources of the fungus cultured by leaf-cutting ants. Appl. Soil Ecol. 2003, 26, 243–247. [Google Scholar]
Supplemental Material

| Strain Code | Yeast Species | Nest ID | Ant Species | City/State/Country 1 | Nest Location 2 |
|---|---|---|---|---|---|
| 8a | Galactomyces candidus | AOMB100904-04 | Acromyrmex heyeri | Sentinela do Sul/RS/Brazil | S 30º37'09''; W 51º33'18'' |
| 10a | Meyerozyma guilliermondii | AOMB120904-05 | Acromyrmex subterraneus | Santana da Boa Vista/RS/Brazil | S 31º19'35''; W 52º44'40'' |
| 15a | Galactomyces candidus | ||||
| 16a1 | Meyerozyma guilliermondii | AOMB130904-02 | Acromyrmex coronatus | Vacaria/RS/Brazil | S 28º27'51''; W 50º53'07'' |
| 16a2 | Meyerozyma guilliermondii | ||||
| 24a | Galactomyces candidus | AOMB040904-02 | Acromyrmex coronatus | Piraquara/PR/Brazil | S 25º25'50''; W 49º04'56'' |
| 27a | Galactomyces candidus | AOMB110904-04 | Acromyrmex lundi | Chuvisca/RS/Brazil | S 30º50'10''; W 51º55'10'' |
| 27a1 | Galactomyces candidus | ||||
| 27b1 | Trichosporon chiarellii | ||||
| 27c1 | Trichosporon chiarellii | ||||
| 27c2’ | Cryptococcus laurentii | ||||
| 28b2 | Galactomyces candidus | AOMB110904-05 | Acromyrmex lundi | Chuvisca/RS/Brazil | S 30º50'10''; W 51º55'10'' |
| 30b | Trichosporon chiarellii | AOMB110904-03 | Acromyrmex heyeri | Chuvisca/RS/Brazil | S 30º50'10''; W 51º55'10'' |
| 30c | Trichosporon chiarellii | ||||
| 31a | Galactomyces candidus | AOMB120904-09 | Acromyrmex sp. | Santana da Boa Vista/RS/Brazil | S 30º56'40''; W 53º05'10'' |
| 31b | Trichosporon multisporum | ||||
| 31b2 | Trichosporon montevideense | ||||
| 31c | Galactomyces candidus | ||||
| 32a | Trichosporon multisporum | AOMB120904-03 | Acromyrmex ambiguus | Santana da Boa Vista/RS/Brazil | S 31º19'35''; W 52º44'40'' |
| 32c | Trichosporon multisporum | ||||
| 39a | Meyerozyma caribbica | AOMB140904-03 | Acromyrmex disciger | Blumenau/SC/Brazil | S 26º54'04''; W 49º10'51'' |
| 39b | Galactomyces candidus | ||||
| 56b | Galactomyces candidus | AOMB140904-05 | Acromyrmex laticeps | Blumenau/SC/Brazil | S 26º54'04''; W 49º10'51'' |
| 59a | Galactomyces candidus | AOMB100904-07 | Acromyrmex heyeri | Sentinela do Sul/RS/Brazil | S 30º37'09''; W 51º33'18'' |
| 59b | Galactomyces candidus | ||||
| 63a | Trichosporon chiarellii | AOMB110904-11 | Acromyrmex lundi | Chuvisca/RS/Brazil | S 30º50'10''; W 51º55'10'' |
| 63c | Trichosporon chiarellii | ||||
| 77b | Trichosporon montevideense | AOMB110904-20 | Acromyrmex laticeps | Chuvisca/RS/Brazil | S 30º50'10''; W 51º55'10'' |
| 78a | Meyerozyma guilliermondii | AOMB130904-10 | Acromyrmex laticeps | Lages/SC/Brazil | - |
| 78b | Meyerozyma guilliermondii | ||||
| 78b2 | Meyerozyma guilliermondii | ||||
| 79b | Galactomyces candidus | AOMB120904-01 | Acromyrmex ambiguus | Santana da Boa Vista/RS/Brazil | S 31º19'35''; W 52º44'40'' |
| 87a | Galactomyces candidus | AOMB110904-10 | Acromyrmex laticeps | Chuvisca/RS/Brazil | S 30º50'10''; W 51º55'10'' |
| 88a1 | Trichosporon montevideense | AOMB060904-03 | Acromyrmex ambiguus | Nova Petrópolis/RS/Brazil | S 29º23'18''; W 50º54'40'' |
| 88a2 | Trichosporon montevideense | ||||
| 88b | Trichosporon montevideense | ||||
| 88b2 | Galactomyces candidus | ||||
| 88c1 | Trichosporon montevideense | ||||
| ATT001 | Candida membranifaciens | UGM051218-02 | Atta texana | Bastrop County/TX/USA | N 30°05'49''; W 97°13'29'' |
| ATT002 | Candida membranifaciens | ||||
| ATT004 | Candida membranifaciens | ||||
| ATT003 | Candida melibiosica | ||||
| ATT175 | unidentified yeast-like fungus | ||||
| ATT176 | Cryptococcus sp. 4 | ||||
| ATT177 | Rhodotorula nothofagi | ||||
| ATT178 | Cryptococcus sp. | ||||
| ATT203 | unidentified yeast-like fungus | ||||
| ATT204 | Cryptococcus podzolicus | ||||
| ATT205 | Kodamaea ohmeri | ||||
| ATT252 | Rhodotorula mucilaginosa | ||||
| ATT253 | Cryptococcus laurentii | ||||
| ATT254 | Sporidiobolus ruineniae | ||||
| ATT256 | Sporidiobolus ruineniae | ||||
| ATT255 | Sporisorium penniseti (yeast-like) | ||||
| ATT257 | Sporisorium penniseti (yeast-like) | ||||
| ATT258 | Rhodotorula lactosa | ||||
| ATT259 | Cryptococcus flavus | ||||
| ATT064 | Cryptococcus cf. cellulolyticus | UGM060121-02 | Atta texana | Austin/TX/USA | N 30°13'56.40"; W 97°39'10.80" |
| ATT067 | Cryptococcus cf. cellulolyticus | ||||
| ATT068 | Pseudozyma sp. | ||||
| ATT069 | Cryptococcus magnus | ||||
| ATT070 | Farysizyma sp. 3 | ||||
| ATT121 | Cryptococcus magnus | ||||
| ATT122 | Cryptococcus luteolus | ||||
| ATT123 | Cryptococcus sp. 3 | ||||
| ATT147 | Rhodotorula sp. HB 1211 | ||||
| ATT148 | Cryptococcus magnus | ||||
| ATT260 | Cryptococcus flavus | ||||
| ATT262 | Aureobasidium pullulans | ||||
| ATT263 | Aureobasidium pullulans | ||||
| ATT269 | Aureobasidium pullulans | ||||
| ATT073 | Cryptococcus laurentii | UGM060121-01 | Atta texana | Austin/TX/USA | N 30°13'58.38"; W 97°39'06.06" |
| ATT075 | Cryptococcus laurentii | ||||
| ATT074 | Bullera sinensis | ||||
| ATT078 | Bulleromyces albus | ||||
| ATT079 | Cryptococcus sp. 1 | ||||
| ATT080 | Cryptococcus sp. 2 | ||||
| ATT082 | Hannaella kunmingensis 3 | ||||
| ATT120 | Cryptococcus flavescens | ||||
| ATT264 | Sympodiomycopsis paphiopedili | ||||
| ATT268 | Cryptococcus flavus | ||||
| ATT271 | Sympodiomycopsis paphiopedili |
| Strain Code | Yeast Species | Hydrolytic Enzymes 1 | Assimilation 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AM | CEL | P | XYL | PRT | M | C | A | X | |||
| 27c2’ | Cryptococcus laurentii 4 | - | - | - | ND 3 | + | + | + | + | + | |
| 8a | Galactomyces candidus 4 | - | + | + | ND | - | - | - | + | + | |
| 15a | Galactomyces candidus 4 | - | + | + | ND | + | - | - | + | + | |
| 24a | Galactomyces candidus | - | + | + | ND | - | - | - | + | + | |
| 27a | Galactomyces candidus | - | + | + | ND | - | - | - | + | + | |
| 27a1 | Galactomyces candidus | - | + | + | ND | - | - | - | + | + | |
| 28b2 | Galactomyces candidus | - | + | + | ND | - | - | - | + | + | |
| 31a | Galactomyces candidus | - | + | + | ND | - | - | - | + | + | |
| 31c | Galactomyces candidus | - | + | + | ND | - | - | - | + | + | |
| 39b | Galactomyces candidus | - | + | + | ND | - | - | - | + | + | |
| 56b | Galactomyces candidus | - | + | - | ND | - | - | - | + | + | |
| 59a | Galactomyces candidus | - | + | + | ND | - | - | - | + | + | |
| 59b | Galactomyces candidus | - | + | + | ND | - | - | - | + | + | |
| 79b | Galactomyces candidus | - | + | + | ND | + | - | - | + | + | |
| 87a | Galactomyces candidus | - | + | - | ND | - | - | - | + | + | |
| 88b2 | Galactomyces candidus | - | + | - | ND | - | - | - | - | + | |
| 39a | Meyerozyma caribbica 4 | - | + | - | ND | - | + | + | + | + | |
| 10a | Meyerozyma guilliermondii | - | + | + | ND | - | + | + | + | + | |
| 16a1 | Meyerozyma guilliermondii 4 | - | + | + | ND | - | + | + | - | + | |
| 16a2 | Meyerozyma guilliermondii | - | + | + | ND | - | + | + | - | + | |
| 78a | Meyerozyma guilliermondii | - | + | + | ND | + | + | + | + | + | |
| 78b | Meyerozyma guilliermondii | - | + | + | ND | - | + | + | + | + | |
| 78b2 | Meyerozyma guilliermondii | - | + | + | ND | - | + | + | - | + | |
| 27b1 | Trichosporon chiarellii | - | - | + | ND | - | + | + | + | + | |
| 27c1 | Trichosporon chiarellii | - | + | - | ND | - | + | + | + | + | |
| 30b | Trichosporon chiarellii | - | - | + | ND | - | + | + | + | + | |
| 30c | Trichosporon chiarellii 4 | - | - | + | ND | - | + | + | + | + | |
| 63a | Trichosporon chiarellii | - | + | + | ND | + | + | + | + | + | |
| 63c | Trichosporon chiarellii | - | + | - | ND | - | + | + | + | + | |
| 31b2 | Trichosporon montevideense | - | + | + | ND | + | + | + | + | + | |
| 77b | Trichosporon montevideense | + | + | + | ND | - | + | + | + | + | |
| 88a1 | Trichosporon montevideense | - | + | - | ND | - | + | + | - | + | |
| 88a2 | Trichosporon montevideense | - | + | - | ND | - | + | + | - | + | |
| 88b | Trichosporon montevideense | - | + | - | ND | - | + | + | - | + | |
| 88c1 | Trichosporon montevideense 4 | - | + | - | ND | - | + | + | + | + | |
| 31b | Trichosporon multisporum 4 | + | + | + | ND | - | + | + | + | + | |
| 32a | Trichosporon multisporum | + | + | + | ND | - | + | + | - | + | |
| 32c | Trichosporon multisporum | - | + | + | ND | - | + | + | + | + | |
| ATT262 | Aureobasidium pullulans | - | + | - | + | - | + | + | + | + | |
| ATT263 | Aureobasidium pullulans | - | + | - | + | + | + | + | + | + | |
| ATT269 | Aureobasidium pullulans | - | + | - | + | + | + | + | - | + | |
| ATT003 | Candida melibiosica | - | - | - | - | - | + | + | - | + | |
| ATT001 | Candida membranifaciens | - | - | - | - | - | + | + | - | + | |
| ATT002 | Candida membranifaciens | - | - | - | - | - | + | + | - | + | |
| ATT004 | Candida membranifaciens | - | - | - | - | - | + | + | - | + | |
| ATT074 | Bullera sinensis | + | + | - | + | - | + | + | + | + | |
| ATT078 | Bulleromyces albus | - | + | - | + | - | + | + | + | + | |
| ATT064 | Cryptococcus cf. cellulolyticus | - | + | - | + | - | + | + | + | + | |
| ATT067 | Cryptococcus cf. cellulolyticus | - | + | - | + | - | + | + | + | + | |
| ATT259 | Cryptococcus flavus | + | + | - | + | + | + | + | + | + | |
| ATT260 | Cryptococcus flavus | + | + | - | + | + | + | + | + | + | |
| ATT268 | Cryptococcus flavus | + | + | + | + | - | + | + | + | + | |
| ATT120 | Cryptococcus flavescens | - | - | - | + | - | + | + | + | + | |
| ATT253 | Cryptococcus laurentii | + | - | + | + | - | + | + | + | + | |
| ATT073 | Cryptococcus laurentii | + | + | - | + | - | + | + | + | + | |
| ATT075 | Cryptococcus laurentii | + | + | - | - | - | + | + | + | + | |
| ATT122 | Cryptococcus luteolus | - | - | - | - | + | + | + | + | + | |
| ATT069 | Cryptococcus magnus | - | + | - | + | + | + | + | - | + | |
| ATT148 | Cryptococcus magnus | - | + | - | - | + | + | + | - | + | |
| ATT121 | Cryptococcus magnus | - | + | - | + | - | + | + | - | + | |
| ATT204 | Cryptococcus podzolicus | + | - | - | + | - | + | + | + | + | |
| ATT178 | Cryptococcus sp. | - | - | - | + | - | + | + | + | + | |
| ATT079 | Cryptococcus sp. 1 | - | + | - | + | - | + | + | - | + | |
| ATT080 | Cryptococcus sp. 2 | - | - | - | - | - | + | + | - | + | |
| ATT123 | Cryptococcus sp. 3 | - | - | - | - | - | + | + | + | + | |
| ATT176 | Cryptococcus sp. 4 | + | - | + | + | - | + | + | + | + | |
| ATT070 | Farysizyma sp. | - | - | - | + | + | + | + | + | + | |
| ATT082 | Hannaella kunmingensis | - | - | - | - | - | + | + | + | + | |
| ATT205 | Kodamaea ohmeri | - | - | - | - | - | + | + | - | + | |
| ATT068 | Pseudozyma sp. | + | + | + | + | + | + | + | + | + | |
| ATT258 | Rhodotorula lactose | - | - | - | - | - | + | + | + | + | |
| ATT252 | Rhodotorula mucilaginosa | - | - | + | - | - | + | + | + | + | |
| ATT177 | Rhodotorula nothofagi | + | - | + | + | - | + | + | - | + | |
| ATT147 | Rhodotorula sp. HB 1211 | - | - | - | - | - | + | + | + | + | |
| ATT254 | Sporidiobolus ruineniae | - | - | + | - | - | + | + | - | + | |
| ATT256 | Sporidiobolus ruineniae | - | - | + | - | - | + | + | - | + | |
| ATT255 | Sporisorium penniseti (yeast-like) | - | - | + | + | + | + | + | + | + | |
| ATT257 | Sporisorium penniseti (yeast-like) | - | - | - | - | + | + | + | + | + | |
| ATT264 | Sympodiomycopsis paphiopedili | - | + | - | + | - | + | + | - | + | |
| ATT271 | Sympodiomycopsis paphiopedili | - | - | - | + | + | + | + | - | + | |
| ATT175 | unidentified yeast-like fungus | - | + | - | + | + | + | + | + | + | |
| ATT203 | unidentified yeast-like fungus | - | - | - | - | + | + | + | + | + | |
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mendes, T.D.; Rodrigues, A.; Dayo-Owoyemi, I.; Marson, F.A.L.; Pagnocca, F.C. Generation of Nutrients and Detoxification: Possible Roles of Yeasts in Leaf-Cutting Ant Nests. Insects 2012, 3, 228-245. https://doi.org/10.3390/insects3010228
Mendes TD, Rodrigues A, Dayo-Owoyemi I, Marson FAL, Pagnocca FC. Generation of Nutrients and Detoxification: Possible Roles of Yeasts in Leaf-Cutting Ant Nests. Insects. 2012; 3(1):228-245. https://doi.org/10.3390/insects3010228
Chicago/Turabian StyleMendes, Thais D., André Rodrigues, Ifeloju Dayo-Owoyemi, Fernando A. L. Marson, and Fernando C. Pagnocca. 2012. "Generation of Nutrients and Detoxification: Possible Roles of Yeasts in Leaf-Cutting Ant Nests" Insects 3, no. 1: 228-245. https://doi.org/10.3390/insects3010228




