Pharmacogenomics Guided-Personalization of Warfarin and Tamoxifen
Abstract
:1. Introduction
2. Personalizing Warfarin Therapy
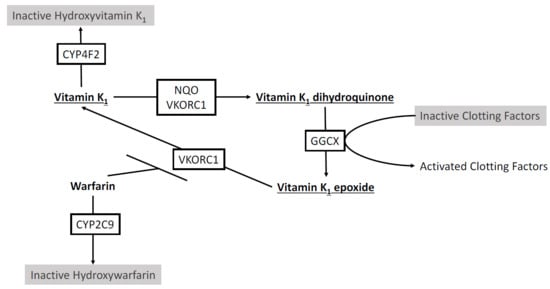
2.1. Warfarin Metabolism by Cytochrome P450s
2.2. The Warfarin Clinical Trial Debate
2.3. Tailoring Pharmacogenomics-Based Warfarin Dosing Algorithms
2.4. Insights from Clinical Implementation of Pharmacogenomics-Guided Warfarin Therapy
3. Personalizing Tamoxifen Therapy
3.1. Tamoxifen Metabolism by Cytochrome P450s
3.2. CYP2D6 and Tamoxifen Clinical Outcomes
3.3. Tamoxifen Metabolism by Other CYP P450 Enzymes
3.4. Therapeutic Drug Monitoring of Tamoxifen
3.5. Clinical Perspectives Based on Experience from a Personalized Tamoxifen Clinic
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nelson, W.W.; Wang, L.; Baser, O.; Damaraju, C.V.; Schein, J.R. Out-of-range INR values and outcomes among new warfarin patients with non-valvular atrial fibrillation. Int. J. Clin. Pharm. 2015, 37, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Budnitz, D.S.; Pollock, D.A.; Weidenbach, K.N.; Mendelsohn, A.B.; Schroeder, T.J.; Annest, J.L. National surveillance of emergency department visits for outpatient adverse drug events. JAMA 2006, 296, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Budnitz, D.S.; Shehab, N.; Kegler, S.R.; Richards, C.L. Medication use leading to emergency department visits for adverse drug events in older adults. Ann. Intern. Med. 2007, 147, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Choonara, I.A.; Cholerton, S.; Haynes, B.P.; Breckenridge, A.M.; Park, B.K. Stereoselective interaction between the R enantiomer of warfarin and cimetidine. Br. J. Clin. Pharmacol. 1986, 21, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Kaminsky, L.S.; Zhang, Z.Y. Human P450 metabolism of warfarin. Pharmacol. Ther. 1997, 73, 67–74. [Google Scholar] [CrossRef]
- Rettie, A.E.; Korzekwa, K.R.; Kunze, K.L.; Lawrence, R.F.; Eddy, A.C.; Aoyama, T.; Gelboin, H.V.; Gonzalez, F.J.; Trager, W.F. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: A role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem. Res. Toxicol. 1992, 5, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Furuya, H.; Fernandez-Salguero, P.; Gregory, W.; Taber, H.; Steward, A.; Gonzalez, F.J.; Idle, J.R. Genetic polymorphism of CYP2C9 and its effect on warfarin maintenance dose requirement in patients undergoing anticoagulation therapy. Pharmacogenet. Genom. 1995, 5, 389–392. [Google Scholar] [CrossRef]
- Stubbins, M.J.; Harries, L.W.; Smith, G.; Tarbit, M.H.; Wolf, C.R. Genetic analysis of the human cytochrome P450 CYP2C9 locus. Pharmacogenet. Genom. 1996, 6, 429–439. [Google Scholar] [CrossRef]
- Aithal, G.P.; Day, C.P.; Kesteven, P.J.; Daly, A.K. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet 1999, 353, 717–719. [Google Scholar] [CrossRef]
- Higashi, M.K.; Veenstra, D.L.; Kondo, L.M.; Wittkowsky, A.K.; Srinouanprachanh, S.L.; Farin, F.M.; Rettie, A.E. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 2002, 287, 1690–1698. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.M.; Johnson, J.A.; Langaee, T.Y.; Feng, H.; Stanaway, I.B.; Schwarz, U.I.; Ritchie, M.D.; Stein, C.M.; Roden, D.M.; Smith, J.D.; et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 2008, 112, 1022–1027. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, F.; McGinnis, R.; Bourgeois, S.; Barnes, C.; Eriksson, N.; Soranzo, N.; Whittaker, P.; Ranganath, V.; Kumanduri, V.; McLaren, W.; et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009, 5, e1000433. [Google Scholar] [CrossRef] [PubMed]
- Lindh, J.D.; Holm, L.; Andersson, M.L.; Rane, A. Influence of CYP2C9 genotype on warfarin dose requirements—A systematic review and meta-analysis. Eur. J. Clin. Pharmacol. 2009, 65, 365–375. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.G.; Rieder, M.J.; Nakano, M.; Hsia, C.K.; Rettie, A.E. CYP4F2 is a vitamin K1 oxidase: An explanation for altered warfarin dose in carriers of the V433M variant. Mol. Pharmacol. 2009, 75, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, M.D.; Awad, T.; Johnson, J.A.; Gage, B.F.; Falkowski, M.; Gardina, P.; Hubbard, J.; Turpaz, Y.; Langaee, T.Y.; Eby, C.; et al. CYP4F2 genetic variant alters required warfarin dose. Blood 2008, 111, 4106–4112. [Google Scholar] [CrossRef] [PubMed]
- Bejarano-Achache, I.; Levy, L.; Mlynarsky, L.; Bialer, M.; Muszkat, M.; Caraco, Y. Effects of CYP4F2 polymorphism on response to warfarin during induction phase: A prospective, open-label, observational cohort study. Clin. Ther. 2012, 34, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Tatarunas, V.; Lesauskaite, V.; Veikutiene, A.; Grybauskas, P.; Jakuska, P.; Jankauskiene, L.; Bartuseviciute, R.; Benetis, R. The effect of CYP2C9, VKORC1 and CYP4F2 polymorphism and of clinical factors on warfarin dosage during initiation and long-term treatment after heart valve surgery. J. Thromb. Thrombolysis 2014, 37, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Suttie, J.W. The biochemical basis of warfarin therapy. Adv. Exp. Med. Biol. 1987, 214, 3–16. [Google Scholar] [PubMed]
- Zimmermann, A.; Matschiner, J.T. Biochemical basis of hereditary resistance to warfarin in the rat. Biochem. Pharmacol. 1974, 23, 1033–1040. [Google Scholar] [CrossRef]
- Li, T.; Chang, C.Y.; Jin, D.Y.; Lin, P.J.; Khvorova, A.; Stafford, D.W. Identification of the gene for vitamin K epoxide reductase. Nature 2004, 427, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Rost, S.; Fregin, A.; Ivaskevicius, V.; Conzelmann, E.; Hortnagel, K.; Pelz, H.J.; Lappegard, K.; Seifried, E.; Scharrer, I.; Tuddenham, E.G.; et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 2004, 427, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Rieder, M.J.; Reiner, A.P.; Gage, B.F.; Nickerson, D.A.; Eby, C.S.; McLeod, H.L.; Blough, D.K.; Thummel, K.E.; Veenstra, D.L.; Rettie, A.E. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N. Engl. J. Med. 2005, 352, 2285–2293. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, M.D.; Berg, R.L.; Zhang, K.Q.; Glurich, I.; Schmelzer, J.R.; Yale, S.H.; Vidaillet, H.J.; Burmester, J.K. Evaluation of genetic factors for warfarin dose prediction. Clin. Med. Res. 2007, 5, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.Y.; Chen, J.J.; Lee, M.T.; Wung, J.C.; Chen, Y.F.; Charng, M.J.; Lu, M.J.; Hung, C.R.; Wei, C.Y.; Chen, C.H.; et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum. Mol. Genet. 2005, 14, 1745–1751. [Google Scholar] [CrossRef] [PubMed]
- Bodin, L.; Verstuyft, C.; Tregouet, D.A.; Robert, A.; Dubert, L.; Funck-Brentano, C.; Jaillon, P.; Beaune, P.; Laurent-Puig, P.; Becquemont, L.; et al. Cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase (VKORC1) genotypes as determinants of acenocoumarol sensitivity. Blood 2005, 106, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Limdi, N.A.; Wadelius, M.; Cavallari, L.; Eriksson, N.; Crawford, D.C.; Lee, M.T.; Chen, C.H.; Motsinger-Reif, A.; Sagreiya, H.; Liu, N.; et al. Warfarin pharmacogenetics: A single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood 2010, 115, 3827–3834. [Google Scholar] [CrossRef] [PubMed]
- Hillman, M.A.; Wilke, R.A.; Caldwell, M.D.; Berg, R.L.; Glurich, I.; Burmester, J.K. Relative impact of covariates in prescribing warfarin according to CYP2C9 genotype. Pharmacogenetics 2004, 14, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Gage, B.F.; Eby, C.; Johnson, J.A.; Deych, E.; Rieder, M.J.; Ridker, P.M.; Milligan, P.E.; Grice, G.; Lenzini, P.; Rettie, A.E.; et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin. Pharmacol. Ther. 2008, 84, 326–331. [Google Scholar] [CrossRef] [PubMed]
- International Warfarin Pharmacogenetics Consortium; Klein, T.E.; Altman, R.B.; Eriksson, N.; Gage, B.F.; Kimmel, S.E.; Lee, M.T.; Limdi, N.A.; Page, D.; Roden, D.M.; et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med. 2009, 360, 753–764. [Google Scholar] [PubMed]
- Wu, A.H.; Wang, P.; Smith, A.; Haller, C.; Drake, K.; Linder, M.; Valdes, R., Jr. Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: Comparison with other equations. Pharmacogenomics 2008, 9, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Gong, I.Y.; Tirona, R.G.; Schwarz, U.I.; Crown, N.; Dresser, G.K.; Larue, S.; Langlois, N.; Lazo-Langner, A.; Zou, G.; Roden, D.M.; et al. Prospective evaluation of a pharmacogenetics-guided warfarin loading and maintenance dose regimen for initiation of therapy. Blood 2011, 118, 3163–3171. [Google Scholar] [CrossRef] [PubMed]
- Pirmohamed, M.; Burnside, G.; Eriksson, N.; Jorgensen, A.L.; Toh, C.H.; Nicholson, T.; Kesteven, P.; Christersson, C.; Wahlstrom, B.; Stafberg, C.; et al. A randomized trial of genotype-guided dosing of warfarin. N. Engl. J. Med. 2013, 369, 2294–2303. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, S.E.; French, B.; Kasner, S.E.; Johnson, J.A.; Anderson, J.L.; Gage, B.F.; Rosenberg, Y.D.; Eby, C.S.; Madigan, R.A.; McBane, R.B.; et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N. Engl. J. Med. 2013, 369, 2283–2293. [Google Scholar] [CrossRef] [PubMed]
- Pengo, V.; Zambon, C.F.; Fogar, P.; Padoan, A.; Nante, G.; Pelloso, M.; Moz, S.; Frigo, A.C.; Groppa, F.; Bozzato, D.; et al. A randomized trial of pharmacogenetic warfarin dosing in naive patients with non-valvular atrial fibrillation. PLoS ONE 2015, 10, e0145318. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, S.E.; French, B.; Geller, N.L.; Investigators, C. Genotype-guided dosing of vitamin K antagonists. N. Engl. J. Med. 2014, 370, 1763–1764. [Google Scholar] [PubMed]
- Gong, I.Y.; Schwarz, U.I.; Crown, N.; Dresser, G.K.; Lazo-Langner, A.; Zou, G.; Roden, D.M.; Stein, C.M.; Rodger, M.; Wells, P.S.; et al. Clinical and genetic determinants of warfarin pharmacokinetics and pharmacodynamics during treatment initiation. PLoS ONE 2011, 6, e27808. [Google Scholar] [CrossRef] [PubMed]
- Gage, B.F.; Bass, A.R.; Lin, H.; Woller, S.C.; Stevens, S.M.; Al-Hammadi, N.; Li, J.; Rodriguez, T., Jr.; Miller, J.P.; McMillin, G.A.; et al. Effect of genotype-guided warfarin dosing on clinical events and anticoagulation control among patients undergoing hip or knee arthroplasty: The GIFT randomized clinical trial. JAMA 2017, 318, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Caraco, Y.; Blotnick, S.; Muszkat, M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: A prospective randomized controlled study. Clin. Pharmacol. Ther. 2008, 83, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Verhoef, T.I.; Ragia, G.; de Boer, A.; Barallon, R.; Kolovou, G.; Kolovou, V.; Konstantinides, S.; Le Cessie, S.; Maltezos, E.; van der Meer, F.J.; et al. A randomized trial of genotype-guided dosing of acenocoumarol and phenprocoumon. N. Engl. J. Med. 2013, 369, 2304–2312. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.L.; Horne, B.D.; Stevens, S.M.; Grove, A.S.; Barton, S.; Nicholas, Z.P.; Kahn, S.F.; May, H.T.; Samuelson, K.M.; Muhlestein, J.B.; et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation 2007, 116, 2563–2570. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Ng, S.S.; Oldenburg, J.; Chong, P.Y.; Rost, S.; Guo, J.Y.; Yap, H.L.; Rankin, S.C.; Khor, H.B.; Yeo, T.C.; et al. Interethnic variability of warfarin maintenance requirement is explained by VKORC1 genotype in an Asian population. Clin. Pharmacol. Ther. 2006, 79, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, T.; Ghosh, K.; Shetty, S. VKORC1 and CYP2C9 genotype distribution in Asian countries. Thromb. Res. 2014, 134, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Perera, M.A.; Gamazon, E.; Cavallari, L.H.; Patel, S.R.; Poindexter, S.; Kittles, R.A.; Nicolae, D.; Cox, N.J. The missing association: Sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin. Pharmacol. Ther. 2011, 89, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, W.; Gamazon, E.R.; Aquino-Michaels, K.; Patel, S.; O’Brien, T.J.; Harralson, A.F.; Kittles, R.A.; Barbour, A.; Tuck, M.; McIntosh, S.D.; et al. Ethnicity-specific pharmacogenetics: The case of warfarin in African Americans. Pharmacogenom. J. 2014, 14, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Mega, J.L.; Walker, J.R.; Ruff, C.T.; Vandell, A.G.; Nordio, F.; Deenadayalu, N.; Murphy, S.A.; Lee, J.; Mercuri, M.F.; Giugliano, R.P.; et al. Genetics and the clinical response to warfarin and edoxaban: Findings from the randomised, double-blind ENGAGE AF-TIMI 48 trial. Lancet 2015, 385, 2280–2287. [Google Scholar] [CrossRef]
- Higgins, M.J.; Stearns, V. CYP2D6 polymorphisms and tamoxifen metabolism: Clinical relevance. Curr. Oncol. Rep. 2010, 12, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Zembutsu, H. Pharmacogenomics toward personalized tamoxifen therapy for breast cancer. Pharmacogenomics 2015, 16, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Jameera Begam, A.; Jubie, S.; Nanjan, M.J. Estrogen receptor agonists/antagonists in breast cancer therapy: A critical review. Bioorg. Chem. 2017, 71, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Bhave, M.A.; Henry, N.L. Extended Endocrine Therapy: Is 5 Years Enough? Curr. Oncol. Rep. 2017, 19, 16. [Google Scholar] [CrossRef] [PubMed]
- Davies, C.; Pan, H.; Godwin, J.; Gray, R.; Arriagada, R.; Raina, V.; Abraham, M.; Medeiros Alencar, V.H.; Badran, A.; Bonfill, X.; et al. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet 2013, 381, 805–816. [Google Scholar] [CrossRef]
- Gray, R.G.; Rea, D.; Handley, K.; Bowden, S.J.; Perry, P.; Earl, H.M.; Poole, C.J.; Bates, T.; Chetiyawardana, S.; Dewar, J.A.; et al. aTTom: Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years in 6953 women with early breast cancer. J. Clin. Oncol. 2013, 31. [Google Scholar] [CrossRef]
- Del Re, M.; Citi, V.; Crucitta, S.; Rofi, E.; Belcari, F.; van Schaik, R.H.; Danesi, R. Pharmacogenetics of CYP2D6 and tamoxifen therapy: Light at the end of the tunnel? Pharmacol. Res. 2016, 107, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Hawse, J.R.; Subramaniam, M.; Cicek, M.; Wu, X.; Gingery, A.; Grygo, S.B.; Sun, Z.; Pitel, K.S.; Lingle, W.L.; Goetz, M.P.; et al. Endoxifen’s molecular mechanisms of action are concentration dependent and different than that of other anti-estrogens. PLoS ONE 2013, 8, e54613. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.C.; Desta, Z.; Flockhart, D.A.; Skaar, T.C. Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother. Pharmacol. 2005, 55, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Madlensky, L.; Natarajan, L.; Tchu, S.; Pu, M.; Mortimer, J.; Flatt, S.W.; Nikoloff, D.M.; Hillman, G.; Fontecha, M.R.; Lawrence, H.J.; et al. Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clin. Pharmacol. Ther. 2011, 89, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Gaedigk, A. Complexities of CYP2D6 gene analysis and interpretation. Int. Rev. Psychiatry 2013, 25, 534–553. [Google Scholar] [CrossRef] [PubMed]
- Caudle, K.E.; Dunnenberger, H.M.; Freimuth, R.R.; Peterson, J.F.; Burlison, J.D.; Whirl-Carrillo, M.; Scott, S.A.; Rehm, H.L.; Williams, M.S.; Klein, T.E.; et al. Standardizing terms for clinical pharmacogenetic test results: Consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet. Med. 2017, 19, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Desta, Z.; Stearns, V.; Ward, B.; Ho, H.; Lee, K.H.; Skaar, T.; Storniolo, A.M.; Li, L.; Araba, A.; et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J. Natl. Cancer Inst. 2005, 97, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Borges, S.; Desta, Z.; Li, L.; Skaar, T.C.; Ward, B.A.; Nguyen, A.; Jin, Y.; Storniolo, A.M.; Nikoloff, D.M.; Wu, L.; et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: Implication for optimization of breast cancer treatment. Clin. Pharmacol. Ther. 2006, 80, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Teft, W.A.; Gong, I.Y.; Dingle, B.; Potvin, K.; Younus, J.; Vandenberg, T.A.; Brackstone, M.; Perera, F.E.; Choi, Y.H.; Zou, G.; et al. CYP3A4 and seasonal variation in vitamin D status in addition to CYP2D6 contribute to therapeutic endoxifen level during tamoxifen therapy. Breast Cancer Res. Treat. 2013, 139, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Murdter, T.E.; Schroth, W.; Bacchus-Gerybadze, L.; Winter, S.; Heinkele, G.; Simon, W.; Fasching, P.A.; Fehm, T.; German, T.; Group, A.I.C.; et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin. Pharmacol. Ther. 2011, 89, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Gaedigk, A.; Simon, S.D.; Pearce, R.E.; Bradford, L.D.; Kennedy, M.J.; Leeder, J.S. The CYP2D6 activity score: Translating genotype information into a qualitative measure of phenotype. Clin. Pharmacol. Ther. 2008, 83, 234–242. [Google Scholar] [CrossRef] [PubMed]
- LLerena, A.; Naranjo, M.E.; Rodrigues-Soares, F.; Penas, L.E.M.; Farinas, H.; Tarazona-Santos, E. Interethnic variability of CYP2D6 alleles and of predicted and measured metabolic phenotypes across world populations. Expert Opin. Drug Metab. Toxicol. 2014, 10, 1569–1583. [Google Scholar] [CrossRef] [PubMed]
- Bradford, L.D. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 2002, 3, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Sideras, K.; Ingle, J.N.; Ames, M.M.; Loprinzi, C.L.; Mrazek, D.P.; Black, J.L.; Weinshilboum, R.M.; Hawse, J.R.; Spelsberg, T.C.; Goetz, M.P. Coprescription of tamoxifen and medications that inhibit CYP2D6. J. Clin. Oncol. 2010, 28, 2768–2776. [Google Scholar] [CrossRef] [PubMed]
- Binkhorst, L.; Mathijssen, R.H.; van Herk-Sukel, M.P.; Bannink, M.; Jager, A.; Wiemer, E.A.; van Gelder, T. Unjustified prescribing of CYP2D6 inhibiting SSRIs in women treated with tamoxifen. Breast Cancer Res. Treat. 2013, 139, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.M.; Juurlink, D.N.; Gomes, T.; Duong-Hua, M.; Pritchard, K.I.; Austin, P.C.; Paszat, L.F. Selective serotonin reuptake inhibitors and breast cancer mortality in women receiving tamoxifen: A population based cohort study. BMJ 2010, 340, c693. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Rae, J.M.; Suman, V.J.; Safgren, S.L.; Ames, M.M.; Visscher, D.W.; Reynolds, C.; Couch, F.J.; Lingle, W.L.; Flockhart, D.A.; et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J. Clin. Oncol. 2005, 23, 9312–9318. [Google Scholar] [CrossRef] [PubMed]
- Schroth, W.; Antoniadou, L.; Fritz, P.; Schwab, M.; Muerdter, T.; Zanger, U.M.; Simon, W.; Eichelbaum, M.; Brauch, H. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J. Clin. Oncol. 2007, 25, 5187–5193. [Google Scholar] [CrossRef] [PubMed]
- Ramon y Cajal, T.; Altes, A.; Pare, L.; del Rio, E.; Alonso, C.; Barnadas, A.; Baiget, M. Impact of CYP2D6 polymorphisms in tamoxifen adjuvant breast cancer treatment. Breast Cancer Res. Treat. 2010, 119, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Lammers, L.A.; Mathijssen, R.H.; van Gelder, T.; Bijl, M.J.; de Graan, A.J.; Seynaeve, C.; van Fessem, M.A.; Berns, E.M.; Vulto, A.G.; van Schaik, R.H. The impact of CYP2D6-predicted phenotype on tamoxifen treatment outcome in patients with metastatic breast cancer. Br. J. Cancer 2010, 103, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Schroth, W.; Goetz, M.P.; Hamann, U.; Fasching, P.A.; Schmidt, M.; Winter, S.; Fritz, P.; Simon, W.; Suman, V.J.; Ames, M.M.; et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA 2009, 302, 1429–1436. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Suman, V.J.; Hoskin, T.L.; Gnant, M.; Filipits, M.; Safgren, S.L.; Kuffel, M.; Jakesz, R.; Rudas, M.; Greil, R.; et al. CYP2D6 metabolism and patient outcome in the Austrian Breast and Colorectal Cancer Study Group trial (ABCSG) 8. Clin. Cancer Res. 2013, 19, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Damodaran, S.E.; Pradhan, S.C.; Umamaheswaran, G.; Kadambari, D.; Reddy, K.S.; Adithan, C. Genetic polymorphisms of CYP2D6 increase the risk for recurrence of breast cancer in patients receiving tamoxifen as an adjuvant therapy. Cancer Chemother. Pharmacol. 2012, 70, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Saladores, P.; Murdter, T.; Eccles, D.; Chowbay, B.; Zgheib, N.K.; Winter, S.; Ganchev, B.; Eccles, B.; Gerty, S.; Tfayli, A.; et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharmacogenom. J. 2015, 15, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sun, Y.; Yao, L.; Shi, L.; Wu, Y.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Fan, T.; et al. Association between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann. Oncol. 2008, 19, 1423–1429. [Google Scholar] [CrossRef] [PubMed]
- Kiyotani, K.; Mushiroda, T.; Sasa, M.; Bando, Y.; Sumitomo, I.; Hosono, N.; Kubo, M.; Nakamura, Y.; Zembutsu, H. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 2008, 99, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Kiyotani, K.; Mushiroda, T.; Imamura, C.K.; Hosono, N.; Tsunoda, T.; Kubo, M.; Tanigawara, Y.; Flockhart, D.A.; Desta, Z.; Skaar, T.C.; et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J. Clin. Oncol. 2010, 28, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.H.; Gomes, M.; Menezes, F.; Afonso, N.; Abreu, P.H.; Medeiros, R.; Pereira, D.; Lopes, C. CYP2D6*4 polymorphism: A new marker of response to hormonotherapy in male breast cancer? Breast 2015, 24, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Rae, J.M.; Drury, S.; Hayes, D.F.; Stearns, V.; Thibert, J.N.; Haynes, B.P.; Salter, J.; Sestak, I.; Cuzick, J.; Dowsett, M.; et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J. Natl. Cancer Inst. 2012, 104, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Regan, M.M.; Leyland-Jones, B.; Bouzyk, M.; Pagani, O.; Tang, W.; Kammler, R.; Dell’orto, P.; Biasi, M.O.; Thurlimann, B.; Lyng, M.B.; et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: The breast international group 1–98 trial. J. Natl. Cancer Inst. 2012, 104, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.E.; Maranian, M.J.; Driver, K.E.; Platte, R.; Kalmyrzaev, B.; Baynes, C.; Luccarini, C.; Shah, M.; Ingle, S.; Greenberg, D.; et al. CYP2D6 gene variants: Association with breast cancer specific survival in a cohort of breast cancer patients from the United Kingdom treated with adjuvant tamoxifen. Breast Cancer Res. 2010, 12, R64. [Google Scholar] [CrossRef] [PubMed]
- Nowell, S.A.; Ahn, J.; Rae, J.M.; Scheys, J.O.; Trovato, A.; Sweeney, C.; MacLeod, S.L.; Kadlubar, F.F.; Ambrosone, C.B. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res. Treat. 2005, 91, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Ro, J.; Park, S.; Lim, H.S.; Lee, K.S.; Kang, H.S.; Jung, S.Y.; Lee, S. Lack of any association between functionally significant CYP2D6 polymorphisms and clinical outcomes in early breast cancer patients receiving adjuvant tamoxifen treatment. Breast Cancer Res. Treat. 2012, 131, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Ahern, T.P.; Hertz, D.L.; Damkier, P.; Ejlertsen, B.; Hamilton-Dutoit, S.J.; Rae, J.M.; Regan, M.M.; Thompson, A.M.; Lash, T.L.; Cronin-Fenton, D.P. Cytochrome P-450 2D6 (CYP2D6) Genotype and Breast Cancer Recurrence in Tamoxifen-Treated Patients: Evaluating the Importance of Loss of Heterozygosity. Am. J. Epidemiol. 2017, 185, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Hertz, D.L.; Kidwell, K.M.; Hilsenbeck, S.G.; Oesterreich, S.; Osborne, C.K.; Philips, S.; Chenault, C.; Hartmaier, R.J.; Skaar, T.C.; Sikora, M.J.; et al. CYP2D6 genotype is not associated with survival in breast cancer patients treated with tamoxifen: Results from a population-based study. Breast Cancer Res. Treat. 2017, 166, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Kiyotani, K.; Mushiroda, T.; Hosono, N.; Tsunoda, T.; Kubo, M.; Aki, F.; Okazaki, Y.; Hirata, K.; Takatsuka, Y.; Okazaki, M.; et al. Lessons for pharmacogenomics studies: Association study between CYP2D6 genotype and tamoxifen response. Pharmacogenet. Genom. 2010, 20, 565–568. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Liu, Y.; Liu, Z.; You, J.; Chen, Z.; Wang, J.; Peng, Q.; Xie, L.; Li, R.; Li, S.; et al. CYP2D6 polymorphisms influence tamoxifen treatment outcomes in breast cancer patients: A meta-analysis. Cancer Chemother. Pharmacol. 2013, 72, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Lum, D.W.; Perel, P.; Hingorani, A.D.; Holmes, M.V. CYP2D6 genotype and tamoxifen response for breast cancer: A systematic review and meta-analysis. PLoS ONE 2013, 8, e76648. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.A.; Lim, H.S. Association between CYP2D6 genotypes and the clinical outcomes of adjuvant tamoxifen for breast cancer: A meta-analysis. Pharmacogenomics 2014, 15, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Province, M.A.; Goetz, M.P.; Brauch, H.; Flockhart, D.A.; Hebert, J.M.; Whaley, R.; Suman, V.J.; Schroth, W.; Winter, S.; Zembutsu, H.; et al. CYP2D6 genotype and adjuvant tamoxifen: Meta-analysis of heterogeneous study populations. Clin. Pharmacol. Ther. 2014, 95, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.S.; Bhat, R.; Crutchley, R.D.; Trivedi, M.V. Impact of CYP2D6 polymorphisms on endoxifen concentrations and breast cancer outcomes. Pharmacogenom. J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Binkhorst, L.; Mathijssen, R.H.; Jager, A.; van Gelder, T. Individualization of tamoxifen therapy: Much more than just CYP2D6 genotyping. Cancer Treat. Rev. 2015, 41, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.L.; Buys, S.S.; Fletcher, D.; Melis, R.; Johnson-Davis, K.L.; Lyon, E.; Malmberg, E.M.; McMillin, G.A. Multigene and drug interaction approach for tamoxifen metabolite patterns reveals possible involvement of CYP2C9, CYP2C19, and ABCB1. J. Clin. Pharmacol. 2016, 56, 1570–1581. [Google Scholar] [CrossRef] [PubMed]
- Marcath, L.A.; Deal, A.M.; Van Wieren, E.; Danko, W.; Walko, C.M.; Ibrahim, J.G.; Weck, K.E.; Jones, D.R.; Desta, Z.; McLeod, H.L.; et al. Comprehensive assessment of cytochromes P450 and transporter genetics with endoxifen concentration during tamoxifen treatment. Pharmacogenet. Genom. 2017, 27, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Lamba, J.K.; Lin, Y.S.; Schuetz, E.G.; Thummel, K.E. Genetic contribution to variable human CYP3A-mediated metabolism. Adv. Drug Deliv. Rev. 2002, 54, 1271–1294. [Google Scholar] [CrossRef]
- Binkhorst, L.; van Gelder, T.; Loos, W.J.; de Jongh, F.E.; Hamberg, P.; Moghaddam-Helmantel, I.M.; de Jonge, E.; Jager, A.; Seynaeve, C.; van Schaik, R.H.; et al. Effects of CYP induction by rifampicin on tamoxifen exposure. Clin. Pharmacol. Ther. 2012, 92, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Henderson, S.L.; Teft, W.A.; Kim, R.B. Profound reduction in tamoxifen active metabolite endoxifen in a breast cancer patient treated with rifampin prior to initiation of an anti-TNFα biologic for ulcerative colitis: A case report. BMC Cancer 2016, 16, 304. [Google Scholar] [CrossRef] [PubMed]
- Antunes, M.V.; de Oliveira, V.; Raymundo, S.; Staudt, D.E.; Gossling, G.; Biazus, J.V.; Cavalheiro, J.A.; Rosa, D.D.; Mathy, G.; Wallemacq, P.; et al. CYP3A4*22 is related to increased plasma levels of 4-hydroxytamoxifen and partially compensates for reduced CYP2D6 activation of tamoxifen. Pharmacogenomics 2015, 16, 601–617. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.S.; Chen, X.A.; Singh, O.; Yap, Y.S.; Ng, R.C.; Wong, N.S.; Wong, M.; Lee, E.J.; Chowbay, B. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br. J. Clin. Pharmacol. 2011, 71, 737–750. [Google Scholar] [CrossRef] [PubMed]
- van Schaik, R.H.; Kok, M.; Sweep, F.C.; van Vliet, M.; van Fessem, M.; Meijer-van Gelder, M.E.; Seynaeve, C.; Lindemans, J.; Wesseling, J.; Van’t Veer, L.J.; et al. The CYP2C19*2 genotype predicts tamoxifen treatment outcome in advanced breast cancer patients. Pharmacogenomics 2011, 12, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; He, J.; He, G.H.; He, J.C.; Xu, F.; Xu, G.L. Association of CYP2C19 polymorphisms with survival of breast cancer patients using tamoxifen: Results of a meta-analysis. Asian Pac. J. Cancer Prev. 2014, 15, 8331–8335. [Google Scholar] [CrossRef] [PubMed]
- Moyer, A.M.; Suman, V.J.; Weinshilboum, R.M.; Avula, R.; Black, J.L.; Safgren, S.L.; Kuffel, M.J.; Ames, M.M.; Ingle, J.N.; Goetz, M.P. SULT1A1, CYP2C19 and disease-free survival in early breast cancer patients receiving tamoxifen. Pharmacogenomics 2011, 12, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Damkier, P.; Kjaersgaard, A.; Barker, K.A.; Cronin-Fenton, D.; Crawford, A.; Hellberg, Y.; Janssen, E.A.M.; Langefeld, C.; Ahern, T.P.; Lash, T.L. CYP2C19*2 and CYP2C19*17 variants and effect of tamoxifen on breast cancer recurrence: Analysis of the International Tamoxifen Pharmacogenomics Consortium dataset. Sci. Rep. 2017, 7, 7727. [Google Scholar] [CrossRef] [PubMed]
- Kiyotani, K.; Mushiroda, T.; Nakamura, Y.; Zembutsu, H. Pharmacogenomics of tamoxifen: Roles of drug metabolizing enzymes and transporters. Drug Metab. Pharmacokinet. 2012, 27, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Santander, A.; Gaibar, M.; Novillo, A.; Romero-Lorca, A.; Rubio, M.; Chicharro, L.M.; Tejerina, A.; Bandres, F. Relationship between genotypes Sult1a2 and Cyp2d6 and tamoxifen metabolism in breast cancer patients. PLoS ONE 2013, 8, e70183. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lorca, A.; Novillo, A.; Gaibar, M.; Bandres, F.; Fernandez-Santander, A. Impacts of the Glucuronidase Genotypes UGT1A4, UGT2B7, UGT2B15 and UGT2B17 on Tamoxifen Metabolism in Breast Cancer Patients. PLoS ONE 2015, 10, e0132269. [Google Scholar] [CrossRef] [PubMed]
- Novillo, A.; Romero-Lorca, A.; Gaibar, M.; Rubio, M.; Fernandez-Santander, A. Tamoxifen metabolism in breast cancer treatment: Taking the focus off the CYP2D6 gene. Pharmacogenom. J. 2017, 17, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.; Balleine, R.L.; Lee, C.; Gao, B.; Balakrishnar, B.; Menzies, A.M.; Yeap, S.H.; Ali, S.S.; Gebski, V.; Provan, P.; et al. Dose Escalation of Tamoxifen in Patients with Low Endoxifen Level: Evidence for Therapeutic Drug Monitoring-The TADE Study. Clin. Cancer Res. 2016, 22, 3164–3171. [Google Scholar] [CrossRef] [PubMed]
- Gong, I.Y.; Teft, W.A.; Ly, J.; Chen, Y.H.; Alicke, B.; Kim, R.B.; Choo, E.F. Determination of clinically therapeutic endoxifen concentrations based on efficacy from human MCF7 breast cancer xenografts. Breast Cancer Res. Treat. 2013, 139, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Irvin, W.J., Jr.; Walko, C.M.; Weck, K.E.; Ibrahim, J.G.; Chiu, W.K.; Dees, E.C.; Moore, S.G.; Olajide, O.A.; Graham, M.L.; Canale, S.T.; et al. Genotype-guided tamoxifen dosing increases active metabolite exposure in women with reduced CYP2D6 metabolism: A multicenter study. J. Clin. Oncol. 2011, 29, 3232–3239. [Google Scholar] [CrossRef] [PubMed]
- Kiyotani, K.; Mushiroda, T.; Imamura, C.K.; Tanigawara, Y.; Hosono, N.; Kubo, M.; Sasa, M.; Nakamura, Y.; Zembutsu, H. Dose-adjustment study of tamoxifen based on CYP2D6 genotypes in Japanese breast cancer patients. Breast Cancer Res. Treat. 2012, 131, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Martinez de Duenas, E.; Ochoa Aranda, E.; Blancas Lopez-Barajas, I.; Ferrer Magdalena, T.; Bandres Moya, F.; Chicharro Garcia, L.M.; Gomez Capilla, J.A.; Zafra Ceres, M.; de Haro, T.; Romero Llorens, R.; et al. Adjusting the dose of tamoxifen in patients with early breast cancer and CYP2D6 poor metabolizer phenotype. Breast 2014, 23, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Dezentje, V.O.; Opdam, F.L.; Gelderblom, H.; Hartigh den, J.; Van der Straaten, T.; Vree, R.; Maartense, E.; Smorenburg, C.H.; Putter, H.; Dieudonne, A.S.; et al. CYP2D6 genotype- and endoxifen-guided tamoxifen dose escalation increases endoxifen serum concentrations without increasing side effects. Breast Cancer Res. Treat. 2015, 153, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Hertz, D.L.; Rae, J.M. Individualized tamoxifen dose escalation: Confirmation of feasibility, question of utility. Clin. Cancer Res. 2016, 22, 3121–3123. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Suman, V.J.; Reid, J.M.; Northfelt, D.W.; Mahr, M.A.; Ralya, A.T.; Kuffel, M.; Buhrow, S.A.; Safgren, S.L.; McGovern, R.M.; et al. First-in-human phase I study of the tamoxifen metabolite Z-endoxifen in women with endocrine-refractory metastatic breast cancer. J. Clin. Oncol. 2017, 35, 3391–3400. [Google Scholar] [CrossRef] [PubMed]
- Gryn, S.E.; Teft, W.A.; Kim, R.B. Profound reduction in the tamoxifen active metabolite endoxifen in a patient on phenytoin for epilepsy compared with a CYP2D6 genotype matched cohort. Pharmacogenet. Genom. 2014, 24, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.A.; Caudle, K.E.; Gong, L.; Whirl-Carrillo, M.; Stein, C.M.; Scott, S.A.; Lee, M.T.; Gage, B.F.; Kimmel, S.E.; Perera, M.A.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin. Pharmacol. Ther. 2017, 102, 397–404. [Google Scholar] [CrossRef] [PubMed]
| Studies | Design | N | Population | Alleles | Outcomes | P |
|---|---|---|---|---|---|---|
| Positive association | ||||||
| Primohamed et al., 2013 [32] | RCT, genotype guided vs. standard dose | 455 | 98% White 1% Black 1% Asian | CYP2C9*2 CYP2C9*3 VKORC1*2 | Improved time within therapeutic INR (67.4% vs. 60.3%); reduction in INR > 4, reduced time to therapeutic INR | <0.001 |
| Gage et al., 2017 [37] | RCT, genotype guided vs. clinical | 1597 | 91% White 6% Black 2% Asian 1% Other | CYP2C9*2 CYP2C9*3 VKORC1*2 CYP4F2*3 | Reduced composite measure of major bleeding, INR > 4, death, and VTE (10.8% vs. 14.7%). In hip and knee arthroplasty patients | <0.02 |
| Caraco et al., 2008 [38] | RCT, Genotype vs. clinical | 191 | Unavailable | CYP2C9*2 CYP2C9*3 | Reduction in time to first therapeutic INR (2.73 days earlier) and reduction in time to stable INR (18.1 days earlier) | <0.001 |
| Gage et al., 2008 [28] | Validation of dosing algorithm | 292 | 93% Caucasian 15% Black 2% Hispanic | CYP2C9*2 CYP2C9*3 VKORC1*2 | Pharmacogenomic dose prediction more accurate than clinical dose prediction (53% vs. 17% of explained variability, respectively) | <0.0001 |
| IWPC, 2009 [29] | Validation of dosing algorithm | 1009 | 55% White 30% Asian 10% Black 5% Other | CYP2C9*2 CYP2C9*3 VKORC1*2 # | Pharmacogenomic dose prediction more accurate than clinical dose prediction (accurately identified 49.4% vs. 33.3% of patients requiring ≤21 mg warfarin per week, respectively) | <0.001 |
| Gong et al., 2011 [31] | Validation of dosing algorithm | 167 | 95% White 2% Black 2%Asian 1% Other | CYP2C9*2 CYP2C9*3 VKORC1*2 CYP4F2*3 | Demonstrated the safe effective prediction of dose limiting variation | N/A |
| Negative association | ||||||
| Kimmel et al., 2013 [33] | RCT, Genotype guided vs. clinical | 1015 | 66%White 27% Black 7% Hispanic | CYP2C9*2 CYP2C9*3 VKORC1*2 | No difference in time in therapeutic INR (45.2% vs. 45.4%) No difference in anticoagulation control or dose prediction | 0.91 |
| Verhoef et al., 2013 [39] | RCT, Genotype guided vs. clinical | 1597 | 98% White | CYP2C9*2 CYP2C9*3 VKORC1*2 | No difference in time in therapeutic INR range (61.6% vs. 60.2%) | 0.47 |
| Pengo et al., 2015 [34] | RCT, Genotype guided vs. standard | 180 | 100% White | CYP2C9*2 CYP2C9*3 VKORC1*2 CYP4F2*3 | No difference in out of range INRs (45.6% vs. 43.6%) or time in therapeutic INR range (51.9% vs. 53.3%) | 0.79 0.71 |
| Anderson et al., 2007 [40] | RCT, Genotype guided vs. standard | 200 | 94% White | CYP2C9*2 CYP2C9*3 VKORC1*2 | No difference in time in therapeutic INR range (30.7% vs. 33.1%) | 0.47 |
| Studies | N | Alleles | DNA Source | Conclusions | Outcome | HR (95% CI) | P |
|---|---|---|---|---|---|---|---|
| Positive association | |||||||
| Goetz et al., 2005 [68] | 190 | *4 | PE-tissue, buccal swabs | *4/*4 patients had worse RFS and DFS | RFS DFS | 2.71 (1.15–6.41) 2.44 (1.22–4.90) | 0.023 0.012 |
| Schroth et al., 2007 [69] | 206 | *4, *5, *10, *41, CNV | normal breast tissue | Decreased function alleles (*4, *5, *10 and *41) were associated with higher rates of recurrence and shorter relapse free periods | RFS EFS | 2.24 (1.16–4.33) 1.89 (1.10–3.25) | 0.02 0.02 |
| Ramón et al., 2010 [70] | 91 | 33 alleles | blood | Patients with *4/*4, *4/*41, *1/*5 or *2/*5 genotypes had shorter DFS | 0.016 | ||
| Lammers et al., 2010 [71] | 99 | *3, *4, *5, *6, *10, *41 | blood | PMs had worse overall survival compared to NMs | OS | 2.09 (1.06–4.12) | 0.034 |
| Schroth et al., 2009 [72] | 1325 | *3,*4, *5, *10, *41 | blood, fresh frozen or PE-tissue | Decreased activity (NM/IM; PM) had worse EFS and DFS | EFS DFS | 1.35 (1.08–1.68) 1.31 (1.06–1.61) | 0.007 0.02 |
| Goetz et al., 2013 [73] | 453 | *3, *4, *6, *10, *41 | PE- tissue | PM/PM patients had higher risk of disease event compared to NM/NM patients | OR | 2.45 (1.05–5.73 | 0.04 |
| Damodaran et al., 2012 [74] | 132 | *1, *2, *4, *5, *10 | blood | CYP2D6 activity scores <0.5 had worse RFS compared to activity scores >1 | RFS | 7.29 (2.92–18.2) | <0.001 |
| Saladores et al., 2015 [75] | 587 | *3, *4, *5, *6, *9, *10, *41, CNV | blood | Improved DRFS was associated with increased CYP2D6 activity score | DRFS | 0.62 (0.43–0.9) | 0.013 |
| Xu et al., 2008 [76] | 152 | *10 | blood, fresh frozen or PE-tissue | *10/*10 was associated with worse DFS | DFS | 4.7 (1.1–20.0) | 0.04 |
| Kiyotani et al., 2008 [77] | 67 | *4, *5, *6, *10, *14, *18, *21, *41 | blood | *10/*10 genotype had worse RFS | RFS | 10.04 (1.17–86.3) | 0.036 |
| Kiyotani et al., 2010 [78] | 282 | *4, *5, *6, *10, *14B, *18, *21, *36, *41, CNV | blood | Presence of two variant alleles was associated with worse RFS compared to patients with no variants | RFS | 9.52 (2.79–32.45) | <0.0001 |
| Negative association | |||||||
| Rae et al., 2012 [80] | 588 | *2, *3, *4, *6, *10, *41 | PE-tissue | PMs did not have reduced recurrence rates compared to NMs | RFS | 0.99 (0.48–2.08) | 0.99 |
| Regan et al., 2012 [81] | 973 | *2, *3, *4, *5, *6, *7, *10, *17, *41 | PE-tissue | IMs and PMs treated with tamoxifen monotherapy were not associated with BCFI | BCFI | 0.86 (0.6–1.24) | 0.35 |
| Abraham et al., 2010 [82] | 3155 | *4, *5, *6, *9, *10, *41, CNV | blood | PM/IM patients did not have reduced survival outcomes compared to NMs | BCSS | 0.93 (0.55–1.57) | 0.78 |
| Nowell et al., 2005 [83] | 160 | *3, *4, *6 | PE-tissue | *4/*4, *1/*4 were not associated with reduced DFS compared to *1/*1 | DFS | 0.67 (0.33–1.35) | 0.19 |
| Park et al., 2012 [84] | 716 | *2, *5, *10, *41 | blood | Homozygous variant carriers did not have reduced RFS | RFS | 1.14 (0.68–1.92) | 0.61 |
| Hertz et al., 2017 [86] | 476 | *2, *3, *4, *6, *10, *41, CNV | Fresh frozen tumors | CYP2D6 activity score was not associated with RFS | RFS | 1.16 (0.84–1.62) | 0.37 |
| Kiyotani et al., 2010 [87] | 167 | *1, *4, *5, *10, *21, *36, *41 | blood | No association between genotype and RFS in patients on tamoxifen-combined therapy | RFS | 0.64 (0.20–1.99) | 0.44 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wigle, T.J.; Jansen, L.E.; Teft, W.A.; Kim, R.B. Pharmacogenomics Guided-Personalization of Warfarin and Tamoxifen. J. Pers. Med. 2017, 7, 20. https://doi.org/10.3390/jpm7040020
Wigle TJ, Jansen LE, Teft WA, Kim RB. Pharmacogenomics Guided-Personalization of Warfarin and Tamoxifen. Journal of Personalized Medicine. 2017; 7(4):20. https://doi.org/10.3390/jpm7040020
Chicago/Turabian StyleWigle, Theodore J., Laura E. Jansen, Wendy A. Teft, and Richard B. Kim. 2017. "Pharmacogenomics Guided-Personalization of Warfarin and Tamoxifen" Journal of Personalized Medicine 7, no. 4: 20. https://doi.org/10.3390/jpm7040020