Testing the Domino Theory of Gene Loss in Buchnera aphidicola: The Relevance of Epistatic Interactions
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Ortholog Identification between Buchnera and E. coli
3.2. Essential Genes in E. coli Tend to be Retained in the Genomes of Buchnera
3.3. Correlated Gene Losses along Buchnera Phylogeny
3.4. Is Correlated Gene Loss in Buchnera Explained by Epistasis Data in E. coli?
4. Discussion
4.1. Conservation of Epistatic Relationships between E. coli and Buchnera
4.2. The Role of Epistasis in Evolution of Genome Reduction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lo, W.-S.; Huang, Y.-Y.; Kuo, C.H. Winding paths to simplicity: Genome evolution in facultative insect symbionts. FEMS Microbiol. Rev. 2016, 40, 855–874. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cano, D.J.; Reyes-Prieto, M.; Martínez-Romero, E.; Partida-Martínez, L.P.; Latorre, A.; Moya, A.; Delaye, L. Evolution of small prokaryotic genomes. Front. Microbiol. 2015, 5, 742. [Google Scholar] [CrossRef] [PubMed]
- Shigenobu, S.; Wilson, A.C.C. Genomic revelations of a mutualism: The pea aphid and its obligate bacterial symbiont. Cell. Mol. Life Sci. 2011, 68, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Sapp, J. Paul Buchner (1886–1978) and hereditary symbiosis in insects. Int. Microbiol. 2002, 5, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Shigenobu, S.; Watanabe, H.; Hattori, M.; Sakaki, Y.; Ishikawa, H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 2000, 407, 81–86. [Google Scholar] [PubMed]
- Tamas, I.; Klasson, L.; Canbäck, B.; Näslund, A.K.; Eriksson, A.S.; Wernegreen, J.J.; Sandström, J.P.; Moran, N.A.; Andersson, S.G. 50 million years of genomic stasis in endosymbiotic bacteria. Science 2002, 296, 2376–2379. [Google Scholar] [CrossRef] [PubMed]
- Van Ham, R.C.; Kamerbeek, J.; Palacios, C.; Rausell, C.; Abascal, F.; Bastolla, U.; Fernández, J.M.; Jiménez, L.; Postigo, M.; Silva, F.J.; et al. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 2003, 100, 581–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Aphid Genomics Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010, 8, e1000313. [Google Scholar] [CrossRef] [Green Version]
- Hansen, A.K.; Moran, N.A. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. USA 2011, 108, 2849–2854. [Google Scholar] [CrossRef] [PubMed]
- McCutcheon, J.P.; Moran, N.A. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 2012, 10, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Wernegreen, J.J. Endosymbiont evolution: Predictions from theory and surprises from genomes. Ann. N. Y. Acad. Sci. 2015, 1360, 16–35. [Google Scholar] [CrossRef] [PubMed]
- Wernegreen, J.J. Ancient bacterial endosymbionts of insects: Genomes as sources of insight and springboards for inquiry. Exp. Cell. Res. 2017, 358, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Wernegreen, J.J. In it for the long haul: evolutionary consequences of persistent endosymbiosis. Curr. Opin. Genet. Dev. 2017, 47, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Fares, M.A.; Moya, A.; Barrio, E. Adaptive evolution in GroEL from distantly related endosymbiotic bacteria of insects. J. Evol. Biol. 2005, 18, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Delmotte, F.; Rispe, C.; Schaber, J.; Silva, F.J.; Moya, A. Tempo and mode of early gene loss in endosymbiotic bacteria from insects. BMC Evol. Biol. 2006, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Toft, C.; Fares, M.A. Selection for translational robustness in Buchnera aphidicola, endosymbiotic bacteria of aphids. Mol. Biol. Evol. 2009, 26, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Sabater-Muñoz, B.; Toft, C.; Alvarez-Ponce, D.; Fares, M.A. Chance and necessity in the genome evolution of endosymbiotic bacteria of insects. ISME J. 2017, 11, 1291–1304. [Google Scholar] [CrossRef] [PubMed]
- Dagan, T.; Blekhman, R.; Graur, D. The “domino theory” of gene death: Gradual and mass gene extinction events in three lineages of obligate symbiotic bacterial pathogens. Mol. Biol. Evol. 2006, 23, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Prieto, A.; Yoon, H.S.; Moustafa, A.; Yang, E.C.; Andersen, R.A.; Boo, S.M.; Nakayama, T.; Ishida, K.; Bhattacharya, D. Differential gene retention in plastids of common recent origin. Mol. Biol. Evol. 2010, 27, 1530–1537. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.W.; Chin, J.P.; Quinn, J.P. Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules. Nat. Rev. Microbiol. 2013, 11, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, J.S.; Saini, D.K. Did the loss of two-component systems initiate pseudogene accumulation in Mycobacterium leprae? Microbiology 2004, 150, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Lamelas, A.; Gosalbes, M.J.; Moya, A.; Latorre, A. New clues about the evolutionary history of metabolic losses in bacterial endosymbionts, provided by the genome of Buchnera aphidicola from the aphid Cinara tujafilina. Appl. Environ. Microbiol. 2011, 77, 4446–4454. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Valero, L.; Latorre, A.; Silva, F.J. The evolutionary fate of nonfunctional DNA in the bacterial endosymbiont Buchnera aphidicola. Mol. Biol. Evol. 2004, 21, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Phillips, P.C. Epistasis—The essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008, 9, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.; Díaz-Mejía, J.J.; Vlasblom, J.; Gagarinova, A.; Phanse, S.; Graham, C.; Yousif, F.; Ding, H.; Xiong, X.; Nazarians-Armavil, A.; et al. Genetic interaction maps in Escherichia coli reveal functional crosstalk among cell envelope biogenesis pathways. PLoS Genet. 2011, 7, e1002377. [Google Scholar] [CrossRef] [PubMed]
- Tamas, I.; Klasson, L.M.; Sandström, J.P.; Andersson, S.G. Mutualists and parasites: how to paint yourself into a (metabolic) corner. FEBS Lett. 2001, 498, 135–139. [Google Scholar] [CrossRef]
- Contreras-Moreira, B.; Vinuesa, P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 2013, 79, 7696–7701. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis and visualization of phylogenomic data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Pagel, M. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc. R. Soc. Lond. B. 1994, 255, 37–45. [Google Scholar] [CrossRef]
- Barker, D.; Pagel, M. Predicting Functional Gene Links from Phylogenetic-Statistical Analyses of Whole Genomes. PLoS Comput. Biol. 2005, 1, 24–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Guy, L.; Kultima, J.R.; Andersson, S.G.E. genoPlotR: Comparative gene and genome visualization in R. Bioinformatics 2010, 26, 2334–2335. [Google Scholar] [CrossRef] [PubMed]
- Decottignies, A.; Sanchez-Perez, I.; Nurse, P. Schizosaccharomyces pombe essential genes: A pilot study. Genome Res. 2003, 13, 399–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, G.; Rocha, E.; Danchin, A. How essential are nonessential genes? Mol. Biol. Evol. 2005, 22, 2147–2156. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.C.; Lin, C.C.; Chang, J.Y.; Mori, H.; Juan, H.F.; Huang, H.C. Predicting essential genes based on network and sequence analysis. Mol. Biosyst. 2009, 5, 1672–1678. [Google Scholar] [CrossRef] [PubMed]
- Jordan, I.K.; Rogozin, I.B.; Wolf, Y.I.; Koonin, E.V. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002, 12, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Gao, F.; Lin, Y. Evolutionary conservation analysis between the essential and nonessential genes in bacterial genomes. Sci. Rep. 2015, 5, 13210. [Google Scholar] [CrossRef] [PubMed]
- Rancati, G.; Moffat, J.; Typas, A.; Pavelka, N. Emerging and evolving concepts in gene essentiality. Nat. Rev. Genet. 2017, 19, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Roguev, A.; Bandyopadhyay, S.; Zofall, M.; Zhang, K.; Fischer, T.; Collins, S.R.; Qu, H.; Shales, M.; Park, H.O.; Hayles, J.; et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 2008, 322, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Fedyshyn, Y.; Koh, J.L.; Prasad, T.S.; Chahwan, C.; Chua, G.; Toufighi, K.; Baryshnikova, A.; Hayles, J.; Hoe, K.L.; et al. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proc. Natl. Acad. Sci. USA 2008, 105, 16653–16658. [Google Scholar] [CrossRef] [PubMed]
- Beltrao, P.; Ryan, C.; Krogan, N.J. Comparative interaction networks: Bridging genotype to phenotype. Adv. Exp. Med. Biol. 2012, 751, 139–156. [Google Scholar] [PubMed]
- Sipiczki, M. Where does fission yeast sit on the tree of life? Genome Biol. 2000, 1, REVIEWS1011. [Google Scholar] [CrossRef] [PubMed]
- Hedges, S.B. The origin and evolution of model organisms. Nat. Rev. Genet. 2002, 3, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; Munson, M.A.; Baumann, P.; Ishikawa, H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. R. Soc. B 1993, 253, 167–171. [Google Scholar] [CrossRef]
- Kuo, C.-H.; Ochman, H. Inferring clocks when lacking rocks: The variable rates of molecular evolution in bacteria. Biol. Direct. 2009, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Beyer, A.; Bandyopadhyay, S.; Ideker, T. Integrating physical and genetic maps: from genomes to interaction networks. Nat. Rev. Genet. 2007, 8, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Houk, E.J.; Griffiths, G.W.; Hadjokas, N.E.; Beck, S.D. Peptidoglycan in the cell wall of the primary intracellular symbiote of the pea aphid. Science 1977, 198, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Simonet, P.; Duport, G.; Gaget, K.; Weiss-Gayet, M.; Colella, S.; Febvay, G.; Charles, H.; Viñuelas, J.; Heddi, A.; Calevro, F. Direct flow cytometry measurements reveal a fine-tuning of symbiotic cell dynamics according to the host developmental needs in aphid symbiosis. Sci. Rep. 2016, 6, 19967. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.J.; Price, N.D. Genetic co-occurrence network across sequenced microbes. PLoS Comput. Biol. 2011, 7, e1002340. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.; Arnold, R.; Bundalovic-Torma, C.; Gagarinova, A.; Wong, K.S.; Kumar, A.; Stewart, G.; Samanfar, B.; Aoki, H.; Wagih, O.; et al. Quantitative genome-wide genetic interaction screens reveal global epistatic relationships of protein complexes in Escherichia coli. PLoS Genet. 2014, 10, e1004120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Beloglazova, N.; Bundalovic-Torma, C.; Phanse, S.; Deineko, V.; Gagarinova, A.; Musso, G.; Vlasblom, J.; Lemak, S.; Hooshyar, M.; et al. Conditional Epistatic Interaction Maps Reveal Global Functional Rewiring of Genome Integrity Pathways in Escherichia coli. Cell. Rep. 2016, 14, 648–661. [Google Scholar] [PubMed]
- Rajagopala, S.V.; Sikorski, P.; Kumar, A.; Mosca, R.; Vlasblom, J.; Arnold, R.; Franca-Koh, J.; Pakala, S.B.; Phanse, S.; Ceol, A.; et al. The binary protein-protein interaction landscape of Escherichia coli. Nat. Biotechnol. 2014, 32, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Yizhak, K.; Tuller, T.; Papp, B.; Ruppin, E. Metabolic modeling of endosymbiont genome reduction on a temporal scale. Mol. Syst. Biol. 2011, 7, 479. [Google Scholar] [CrossRef] [PubMed]


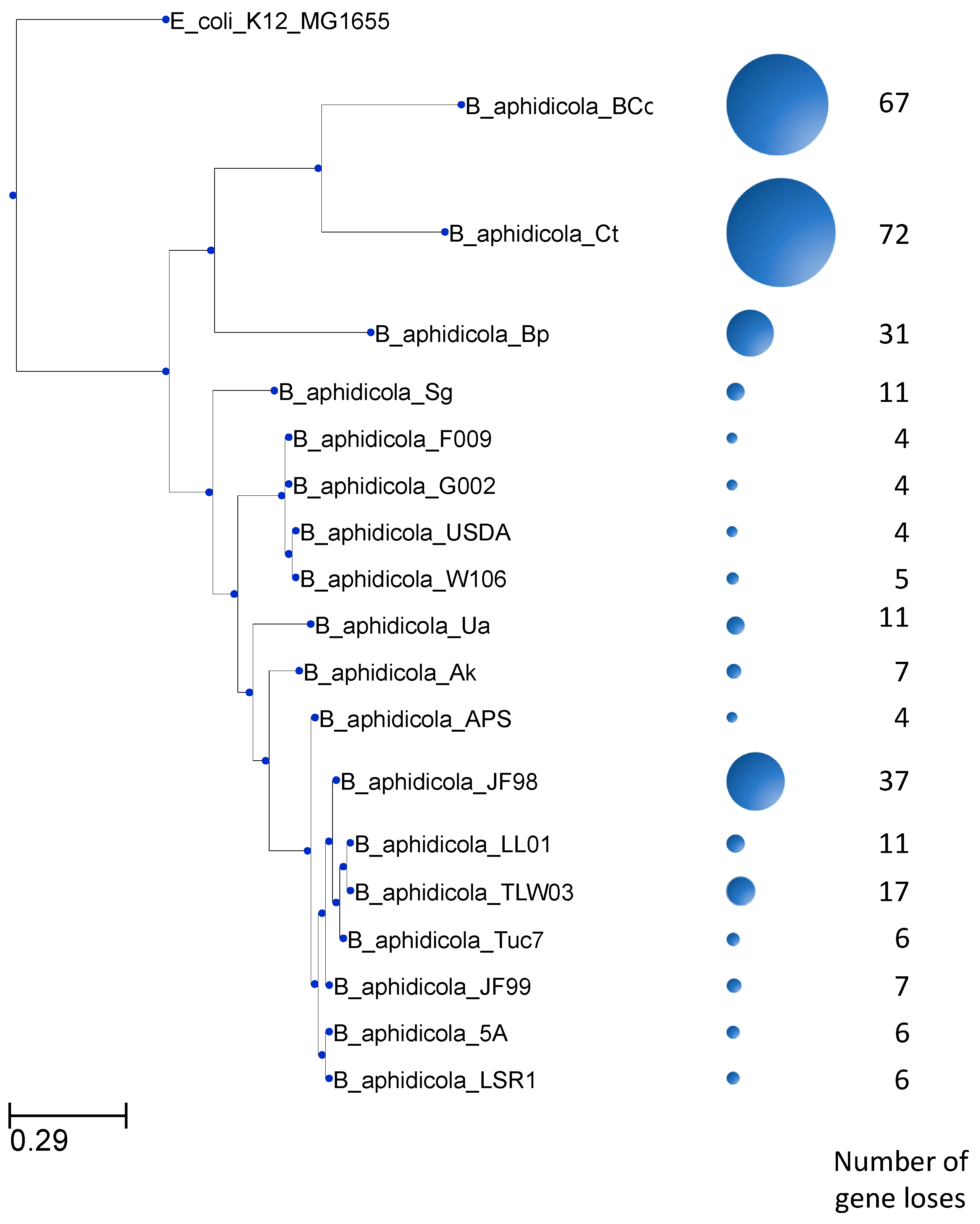
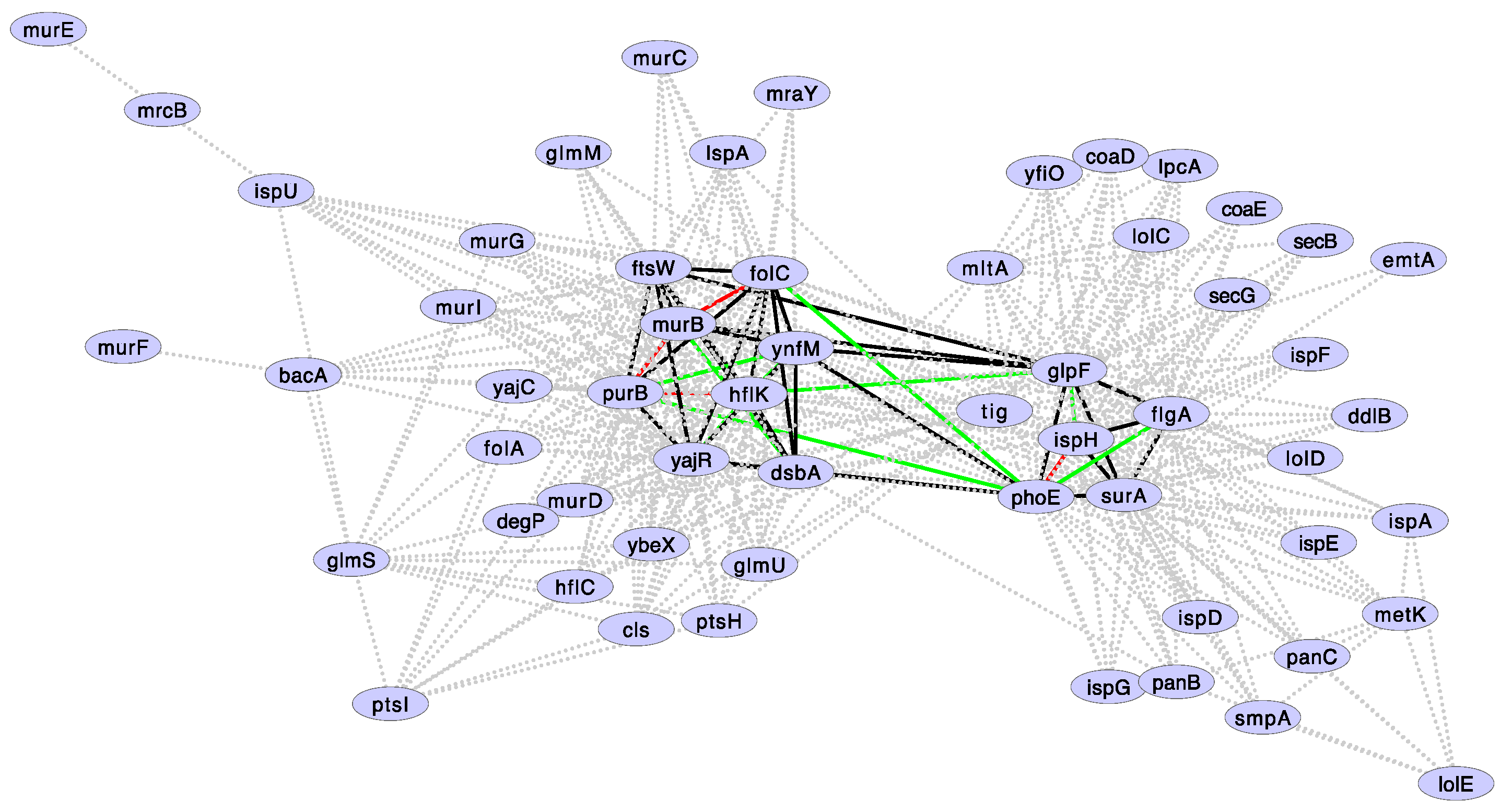
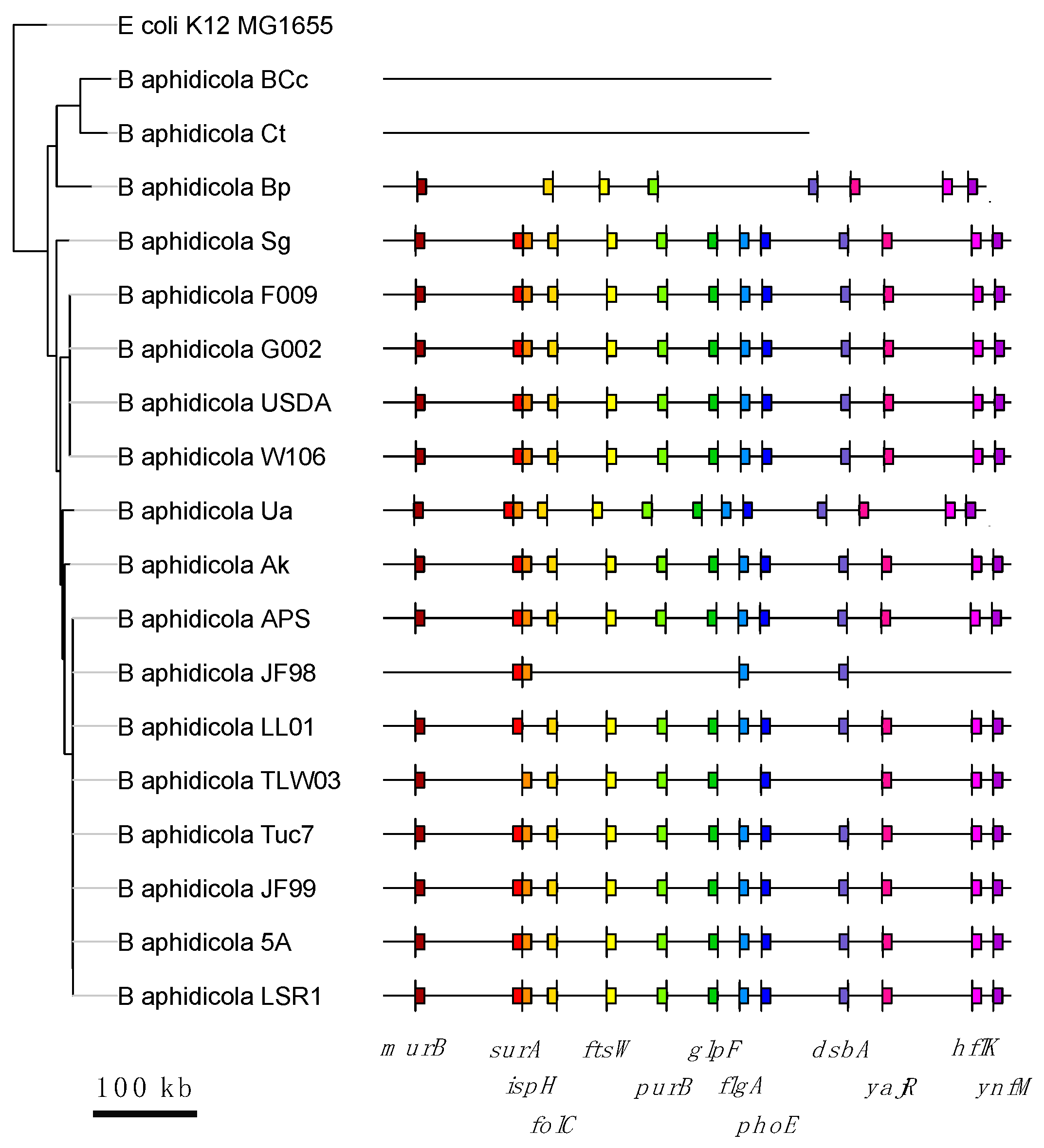


| Correlated Losses Gene Set Defined by | Media | p-Value | 95% CI | Odds Ratio |
|---|---|---|---|---|
| FDR < 0.1 | RM | 0.31 | 0.29 to ∞ | Inf. |
| (p-value < 0.01) | MM | 0.02 | 1.09 to 17.7 | 3.93 |
| FDR < 0.45 | RM | 0.28 | 0.75 to 3.08 | 1.48 |
| (p-value < 0.05) | MM | 0.01 | 1.11 to 2.78 | 1.76 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Cano, D.J.; Bor, G.; Moya, A.; Delaye, L. Testing the Domino Theory of Gene Loss in Buchnera aphidicola: The Relevance of Epistatic Interactions. Life 2018, 8, 17. https://doi.org/10.3390/life8020017
Martínez-Cano DJ, Bor G, Moya A, Delaye L. Testing the Domino Theory of Gene Loss in Buchnera aphidicola: The Relevance of Epistatic Interactions. Life. 2018; 8(2):17. https://doi.org/10.3390/life8020017
Chicago/Turabian StyleMartínez-Cano, David J., Gil Bor, Andrés Moya, and Luis Delaye. 2018. "Testing the Domino Theory of Gene Loss in Buchnera aphidicola: The Relevance of Epistatic Interactions" Life 8, no. 2: 17. https://doi.org/10.3390/life8020017








