DNA Protection Protein, a Novel Mechanism of Radiation Tolerance: Lessons from Tardigrades
Abstract
:1. Tardigrades as Model Animals Tolerant to Various Extreme Environments
2. Radiotolerant Organisms
3. Extraordinary Tolerance to Irradiation in Tardigrades
4. Tardigrade-Unique DNA-Associated Protein, Dsup, Improves Radiotolerance
5. DNA-Association is Necessary for DNA Protection Activity of Dsup Protein
6. What is the Origin of Dsup, a DNA Protective Protein?
7. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Rothschild, L.J.; Mancinelli, R.C. Life in extreme environments. Nature 2001, 409, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G.; Kauschke, S.; Rüdiger, J.; Stevenson, P.A. Neural markers reveal a one-segmented head in tardigrades (water bears). PLoS ONE 2013, 8, e59090. [Google Scholar] [CrossRef] [PubMed]
- Rebecchi, L.; Altiero, T.; Guidetti, R. Anhydrobiosis: The extreme limit of desiccation tolerance. Invertebr. Surviv. J. 2007, 4, 65–81. [Google Scholar]
- Actual checklist of Tardigrada Species. 2015. Available online: http://www.tardigrada.modena.unimo.it/miscellanea/ActualchecklistofTardigrada.pdf (accessed on 29 March 2017).
- Vicente, F.; Bertolani, R. Considerations on the taxonomy of the Phylum Tardigrada. Zootaxa 2013, 3626, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, M.; Komori, O. Effect of lifespan and age on reproductive performance of the tardigrade Acutuncus antarcticus: Minimal reproductive senescence. Hydrobiologia 2016, 772, 93–102. [Google Scholar] [CrossRef]
- Tsujimoto, M.; Imura, S.; Kanda, H. Recovery and reproduction of an Antarctic tardigrade retrieved from a moss sample frozen for over 30 years. Cryobiology 2016, 72, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Saigo, T.; Abe, W.; Kubo, T.; Kunieda, T. Establishment of an isogenic strain of the desiccation-sensitive tardigrade Isohypsibius myrops (Parachela, Eutardigrada) and its life history traits. Zool. J. Linn. Soc. 2016, 178, 863–870. [Google Scholar] [CrossRef]
- Keilin, D. The problem of anabiosis or latent life: History and current concepts. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1959, 150, 149–191. [Google Scholar] [CrossRef]
- Becquerel, P. La suspension de la vie au dessous de 1/20 K absolu par démagnétisation adiabatique de l’alun de fer dans le vide le plus élevé. Comptes Rendus de l'Académie des Sciences 1950, 231, 261–263. [Google Scholar]
- Horikawa, D.D.; Kunieda, T.; Abe, W.; Watanabe, M.; Nakahara, Y.; Yukuhiro, F.; Sakashita, T.; Hamada, N.; Wada, S.; Funayama, T.; et al. Establishment of a rearing system of the extremotolerant tardigrade Ramazzottius varieornatus: A new model animal for astrobiology. Astrobiology 2008, 8, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Hengherr, S.; Worland, M.R.; Reuner, A.; Brümmer, F.; Schill, R.O. High-temperature tolerance in anhydrobiotic tardigrades is limited by glass transition. Physiol. Biochem. Zool. 2009, 82, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Ono, F.; Saigusa, M.; Uozumi, T.; Matsushima, Y; Ikeda, H.; Saini, N.L.; Yamashita, M. Effect of high hydrostatic pressure on to life of the tiny animal tardigrade. J. Phys. Chem. Solids 2008, 69, 2297–2300. [Google Scholar] [CrossRef]
- Ramløv, H.; Westh, P. Cryptobiosis in the eutardigrade Adorybiotus (Richtersius) coronifer: Tolerance to alcohols, temperature and de novo protein synthesis. Zool. Anz. J. Comp. Zool. 2001, 240, 517–523. [Google Scholar] [CrossRef]
- Horikawa, D.D.; Sakashita, T.; Katagiri, C.; Watanabe, M.; Kikawada, T.; Nakahara, Y.; Hamada, N.; Wada, S.; Funayama, T.; Higashi, S.; et al. Radiation tolerance in the tardigrade Milnesium tardigradum. Int. J. Radiat. Biol. 2006, 82, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, K.I.; Harms-Ringdahl, M.; Torudd, J. Radiation tolerance in the eutardigrade Richtersius coronifer. Int. J. Radiat. Biol. 2005, 81, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, K.I.; Rabbow, E.; Schill, R.O.; Harms-Ringdahl, M.; Rettberg, P. Tardigrades survive exposure to space in low Earth orbit. Curr. Biol. 2008, 18, R729–R731. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Horikawa, D.D.; Saito, Y.; Kuwahara, H.; Kozuka-Hata, H.; Shin-I, T.; Minakuchi, Y.; Ohishi, K.; Motoyama, A.; Aizu, T.; et al. Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nat. Commun. 2016, 7, 12808. [Google Scholar] [CrossRef] [PubMed]
- Koutsovoulos, G.; Kumar, S.; Laetsch, D.R.; Stevens, L.; Daub, J.; Conlon, C.; Maroon, H.; Thomas, F.; Aboobaker, A.A.; Blaxter, M. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc. Natl. Acad. Sci. USA 2016, 113, 5053–5058. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Koutsovoulos, G.; Laetsch, D.R.; Stevens, L.; Kumar, S.; Horikawa, D.D.; Ishino, K.; Komine, S.; Kunieda, T.; Tomita, M.; et al. Comparative genomics of the tardigrades Hypsibius dujardini and Ramazzottius varieornatus. Available online: https://doi.org/10.1101/112664 (accessed on 8 June 2017).
- Yamaguchi, A.; Tanaka, S.; Yamaguchi, S.; Kuwahara, H.; Takamura, C.; Imajoh-Ohmi, S.; Horikawa, D.D.; Toyoda, A.; Katayama, T.; Arakawa, K.; et al. Two novel heat-soluble protein families abundantly expressed in an anhydrobiotic tardigrade. PLoS ONE 2012, 7, e44209. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Tanaka, J.; Miwa, Y.; Horikawa, D.D.; Katayama, T.; Arakawa, K.; Toyoda, A.; Kubo, T.; Kunieda, T. Novel Mitochondria-Targeted Heat-Soluble Proteins Identified in the Anhydrobiotic Tardigrade Improve Osmotic Tolerance of Human Cells. PLoS ONE 2015, 10, e0118272. [Google Scholar] [CrossRef] [PubMed]
- Boothby, T.C.; Tapia, H.; Brozena, A.H.; Piszkiewicz, S.; Smith, A.E.; Giovannini, I.; Rebecchi, L.; Pielak, G.J.; Koshland, D.; Goldstein, B. Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol. Cell 2017, 65, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P. Sensing and repairing DNA double-strand breaks. Carcinogenesis 2002, 23, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.F. DNA damage as the cause of ionizing radiation-induced gene activation. Radiat. Res. 1994, 138, S85–S88. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, F.A.; Kim, M.Y.; Chappell, L.J.; Huff, J.L. How safe is safe enough? Radiation risk for a human mission to Mars. PLoS ONE 2013, 8, e74988. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.J.; Giaccia, A.J. Radiobiology for the Radiologist, 7th ed.; Lippincott Williams & Wilins: Philadelpha, PA, USA, 2012; pp. 120–121. [Google Scholar]
- Byrne, R.T.; Chen, S.H.; Wood, E.A.; Cabot, E.L.; Cox, M.M. Escherichia coli Genes and Pathways Involved in Surviving Extreme Exposure to Ionizing Radiation. J. Bacteriol. 2014, 196, 3534–3545. [Google Scholar] [CrossRef] [PubMed]
- Battista, J.R. Against all odds: The survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 1997, 51, 203–224. [Google Scholar] [CrossRef] [PubMed]
- Narumi, I. Unlocking radiation resistance mechanisms: Still a long way to go. Trends Microbiol. 2003, 11, 422–425. [Google Scholar] [CrossRef]
- Zahradka, K.; Slade, D.; Bailone, A.; Sommer, S.; Averbeck, D.; Petranovic, M.; Lindner, A.B.; Radman, M. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 2006, 443, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Kiang, J.G.; Fukumoto, R.; Lee, D.Y.; Wehr, N.B.; Viteri, G.A.; Berlett, B.S.; Levine, R.L. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS ONE 2010, 5, e12570. [Google Scholar] [CrossRef] [PubMed]
- Bessman, M.J.; Frick, D.N.; O’Handley, S.F. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 1996, 271, 25059–25062. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Shen, J.; Dunn, C.A.; Desai, S.; Bessman, M.J. The Nudix hydrolases of Deinococcus radiodurans. Mol. Microbiol. 2001, 39, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Driedger, A.A. The DNA content of single cells of Micrococcus radiodurans. Can. J. Microbiol. 1970, 16, 1136–1137. [Google Scholar] [CrossRef] [PubMed]
- Slade, D.; Radman, M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, E.; Meselson, M. Extreme resistance of bdelloid rotifers to ionizing radiation. Proc. Natl. Acad. Sci. USA 2008, 105, 5139–5144. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Sakashita, T.; Fujita, A.; Kikawada, T.; Horikawa, D.D.; Nakahara, Y.; Wada, S.; Funayama, T.; Hamada, N.; Kobayashi, Y.; et al. Biological effects of anhydrobiosis in an African chironomid, Polypedilum vanderplanki on radiation tolerance. Int. J. Radiat. Biol. 2006, 82, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Krisko, A.; Leroy, M.; Radman, M.; Meselson, M. Extreme anti-oxidant protection against ionizing radiation in bdelloid rotifers. Proc. Natl. Acad. Sci. USA 2012, 109, 2354–2357. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, P.A. Effects of ionizing radiation on mammalian cells. Naturwissenschaften 1974, 61, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.J.; Giaccia, A.J. Radiobiology for the Radiologist, 7th ed.; Lippincott Williams & Wilins: Philadelpha, PA, USA, 2012; pp. 9–11. [Google Scholar]
- França, M.B.; Panek, A.D.; Eleutherio, E.C.A. Oxidative stress and its effects during dehydration. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 146, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, C.; Foray, N. Chromatin structure and radiation-induced DNA damage: From structural biology to radiobiology. Int. J. Biochem. Cell Biol. 2014, 49, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Levin-Zaidman, S. Ringlike Structure of the Deinococcus radiodurans Genome: A Key to Radioresistance? Science 2003, 299, 254–256. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, J.M.; Battista, J.R. A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol. 2005, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Beltrán-Pardo, E.; Jönsson, K.I.; Harms-Ringdahl, M.; Haghdoost, S.; Wojcik, A. Tolerance to Gamma Radiation in the Tardigrade Hypsibius dujardini from Embryo to Adult Correlate Inversely with Cellular Proliferation. PLoS ONE 2015, 10, e0133658. [Google Scholar]
- Andrews, H.L. Species Differences in Response to High Radiation Doses. Radiat. Res. 2012, 9, 469–477. [Google Scholar] [CrossRef]
- Horikawa, D.D.; Yamaguchi, A.; Sakashita, T.; Tanaka, D.; Hamada, N.; Yukuhiro, F.; Kuwahara, H.; Kunieda, T.; Watanabe, M.; Nakahara, Y.; et al. Tolerance of anhydrobiotic eggs of the tardigrade Ramazzottius varieornatus to extreme environments. Astrobiology 2012, 12, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Gusev, O.; Nakahara, Y.; Vanyagina, V.; Malutina, L.; Cornette, R.; Sakashita, T.; Hamada, N.; Kikawada, T.; Kobayashi, Y.; Okuda, T. Anhydrobiosis-associated nuclear DNA damage and repair in the sleeping chironomid: Linkage with radioresistance. PLoS ONE 2010, 5, e14008. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Reuner, A.; Brümmer, F.; Schill, R.O. DNA damage in storage cells of anhydrobiotic tardigrades. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2009, 153, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Rebecchi, L.; Cesari, M.; Altiero, T.; Frigieri, A.; Guidetti, R. Survival and DNA degradation in anhydrobiotic tardigrades. J. Exp. Biol. 2009, 212, 4033–4039. [Google Scholar] [CrossRef] [PubMed]
- May, R.M.; Maria, M.; Guimard, J. Actions différentielles des rayons x et ultraviolets sur le tardigrade Macrobiotus areolatus, à l’état actif et desséché. Bull. Biol. Fr. Belg. 1964, 98, 349–367. [Google Scholar]
- Nilsson, E.J.C.; Jönsson, K.I.; Pallon, J. Tolerance to proton irradiation in the eutardigrade Richtersius coronifer—A nuclear microprobe study. Int. J. Radiat. Biol. 2010, 86, 1–8. [Google Scholar]
- Jönsson, K.I.; Wojcik, A. Tolerance to X-rays and Heavy Ions (Fe, He) in the Tardigrade Richtersius coronifer and the Bdelloid Rotifer Mniobia russeola. Astrobiology 2017, 17, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, K.I.; Hygum, T.L.; Andersen, K.N.; Clausen, L.K.B.; Møbjerg, N. Tolerance to Gamma Radiation in the Marine Heterotardigrade, Echiniscoides sigismundi. PLoS ONE 2016, 11, e0168884. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Boon, C.; Redon, C.; Bonner, W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999, 146, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Andrievski, A.; Wilkins, R.C. The response of gamma-H2AX in human lymphocytes and lymphocytes subsets measured in whole blood cultures. Int. J. Radiat. Biol. 2009, 85, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Taneja, N.; Davis, M.; Choy, J.S.; Beckett, M.A.; Singh, R.; Kron, S.J.; Weichselbaum, R.R. Histone H2AX Phosphorylation as a Predictor of Radiosensitivity and Target for Radiotherapy. J. Biol. Chem. 2004, 279, 2273–2280. [Google Scholar] [CrossRef] [PubMed]
- Puck, T.T.; Marcus, P.I. Action of x-rays on mammalian cells. J. Exp. Med. 1956, 103, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Spurio, R.; Dürrenberger, M.; Falconi, M.; La Teana, A.; Pon, C.L.; Gualerzi, C.O. Lethal overproduction of the Escherichia coli nucleoid protein H-NS: Ultramicroscopic and molecular autopsy. Mol. Gen. Genet. 1992, 231, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Setlow, B.; Hand, A.R.; Setlow, P. Synthesis of a Bacillus subtilis small, acid-soluble spore protein in Escherichia coli causes cell DNA to assume some characteristics of spore DNA. J. Bacteriol. 1991, 173, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Wise, M. POPP the question: What do LEA proteins do? Trends Plant Sci. 2004, 9, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [PubMed]
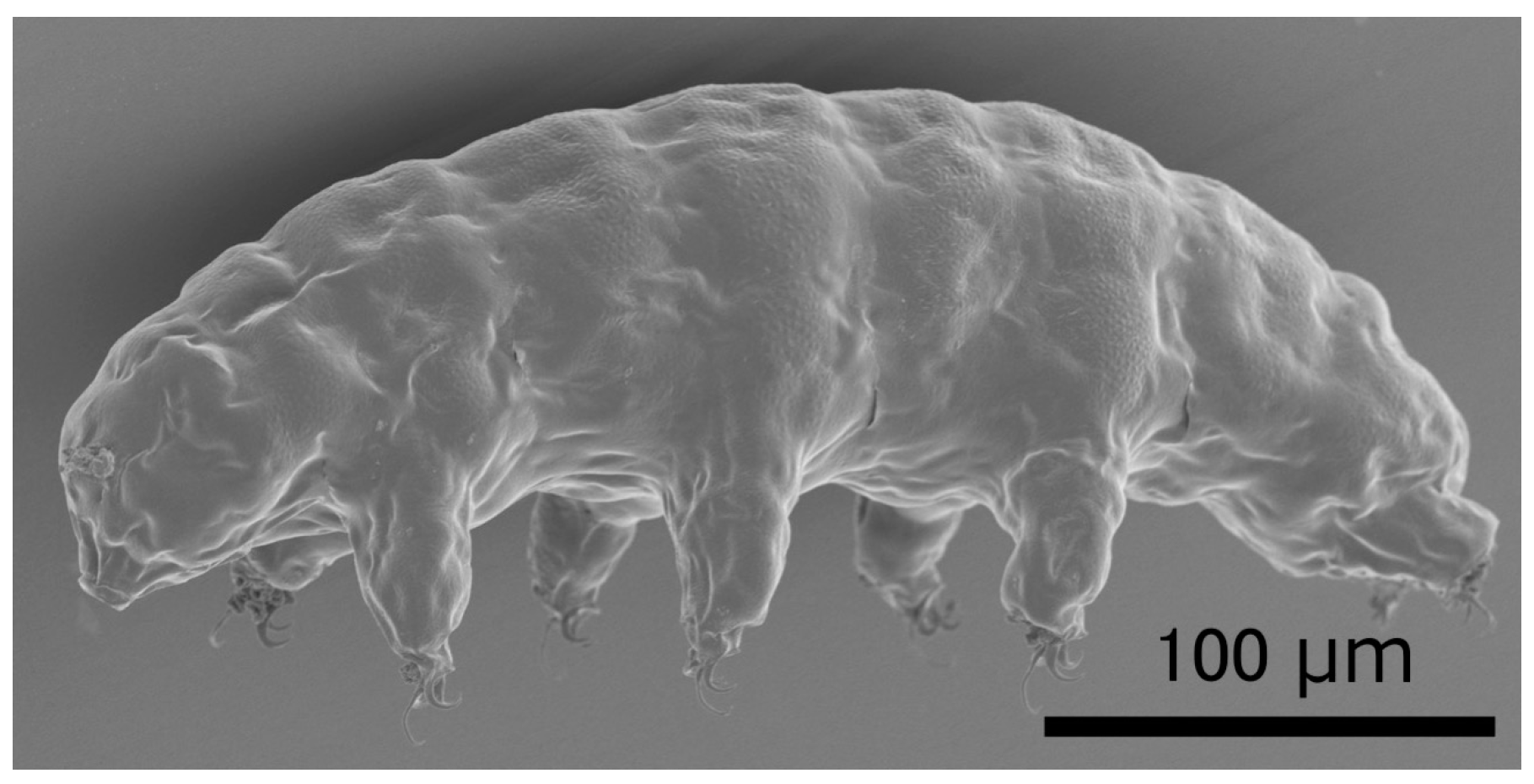



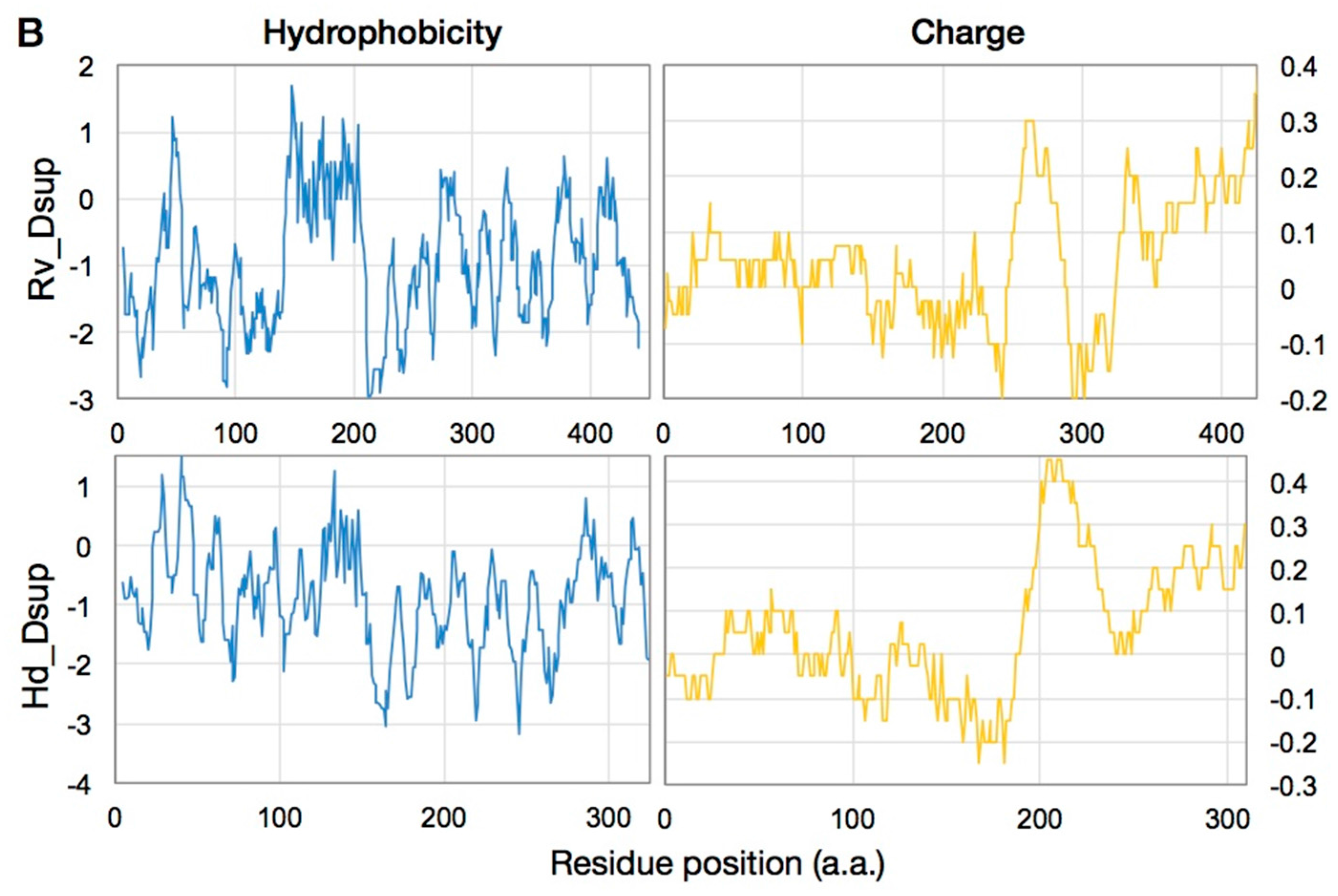

| Tardigrade Species | State 1 | Ionizing Radiation | Radiotolerance | Reference |
|---|---|---|---|---|
| Macrobiotus areolatus | Dehyd. | X-ray | 5700 Gy (LD50/1 day after) | [55] |
| Richtersius coronifer | Hyd. | γ-rays | 4700 Gy (LD50/18 h) | [16] |
| Hyd. | γ-rays | 2500 Gy (LD50/30 days) | [16] | |
| Dehyd. | γ-rays | 3000 Gy (LD50/22 h) | [16] | |
| Dehyd. | proton-beam | 10240 Gy (LD50/24 h) | [56] | |
| Dehyd. | X-ray | 2000 Gy (Few animals revived within 7 days) | [57] | |
| Dehyd. | 4He, 56Fe | 2000 Gy (Most animals revived within 7 days) | [57] | |
| Milnesium tardigradum | Hyd. | γ-rays | 5000 Gy (LD50/48 h) | [15] |
| Dehyd. | γ-rays | 4400 Gy (LD50/48 h) | [15] | |
| Hyd. | 4He | 6200 Gy (LD50/48 h) | [15] | |
| Dehyd. | 4He | 5200 Gy (LD50/48 h) | [15] | |
| Ramazzottius varieornatus | Hyd. | 4He | 4000 Gy (Most animals survived for 48 h) | [11] |
| Dehyd. | 4He | 4000 Gy (Most animals revived within 48 h) | [11] | |
| Hypsibius dujardini | Hyd. | γ-rays | 4180 Gy (LD50/48 h) | [49] |
| Echiniscoides sigismundi 2 | Hyd. | γ-rays | 1270 Gy (LD50/48 h) | [58] |
| Hyd. | γ-rays | 1550 Gy (LD50/7 days) | [58] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashimoto, T.; Kunieda, T. DNA Protection Protein, a Novel Mechanism of Radiation Tolerance: Lessons from Tardigrades. Life 2017, 7, 26. https://doi.org/10.3390/life7020026
Hashimoto T, Kunieda T. DNA Protection Protein, a Novel Mechanism of Radiation Tolerance: Lessons from Tardigrades. Life. 2017; 7(2):26. https://doi.org/10.3390/life7020026
Chicago/Turabian StyleHashimoto, Takuma, and Takekazu Kunieda. 2017. "DNA Protection Protein, a Novel Mechanism of Radiation Tolerance: Lessons from Tardigrades" Life 7, no. 2: 26. https://doi.org/10.3390/life7020026






