Clues to tRNA Evolution from the Distribution of Class II tRNAs and Serine Codons in the Genetic Code
Abstract
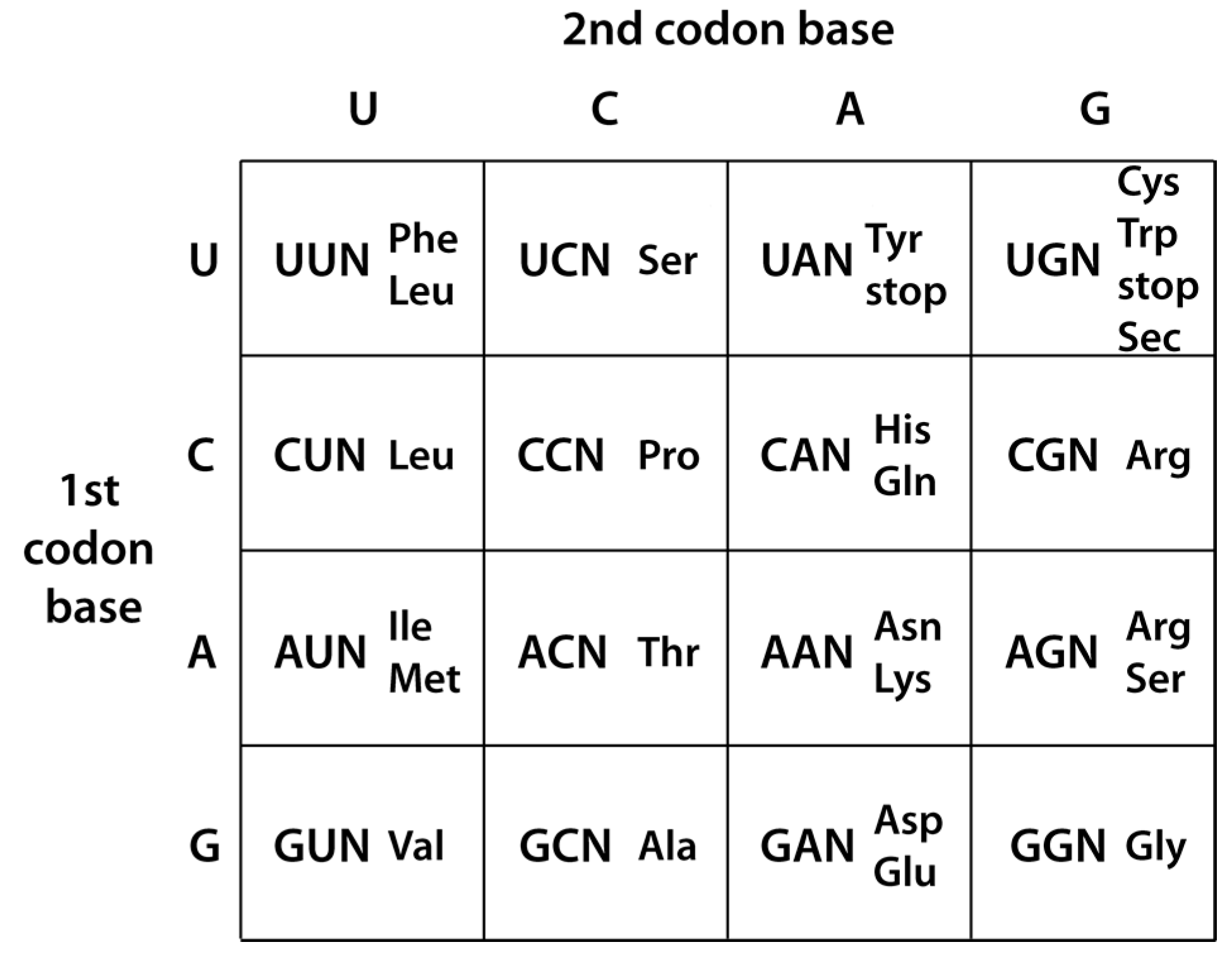
:1. Introduction
2. Hypothesis
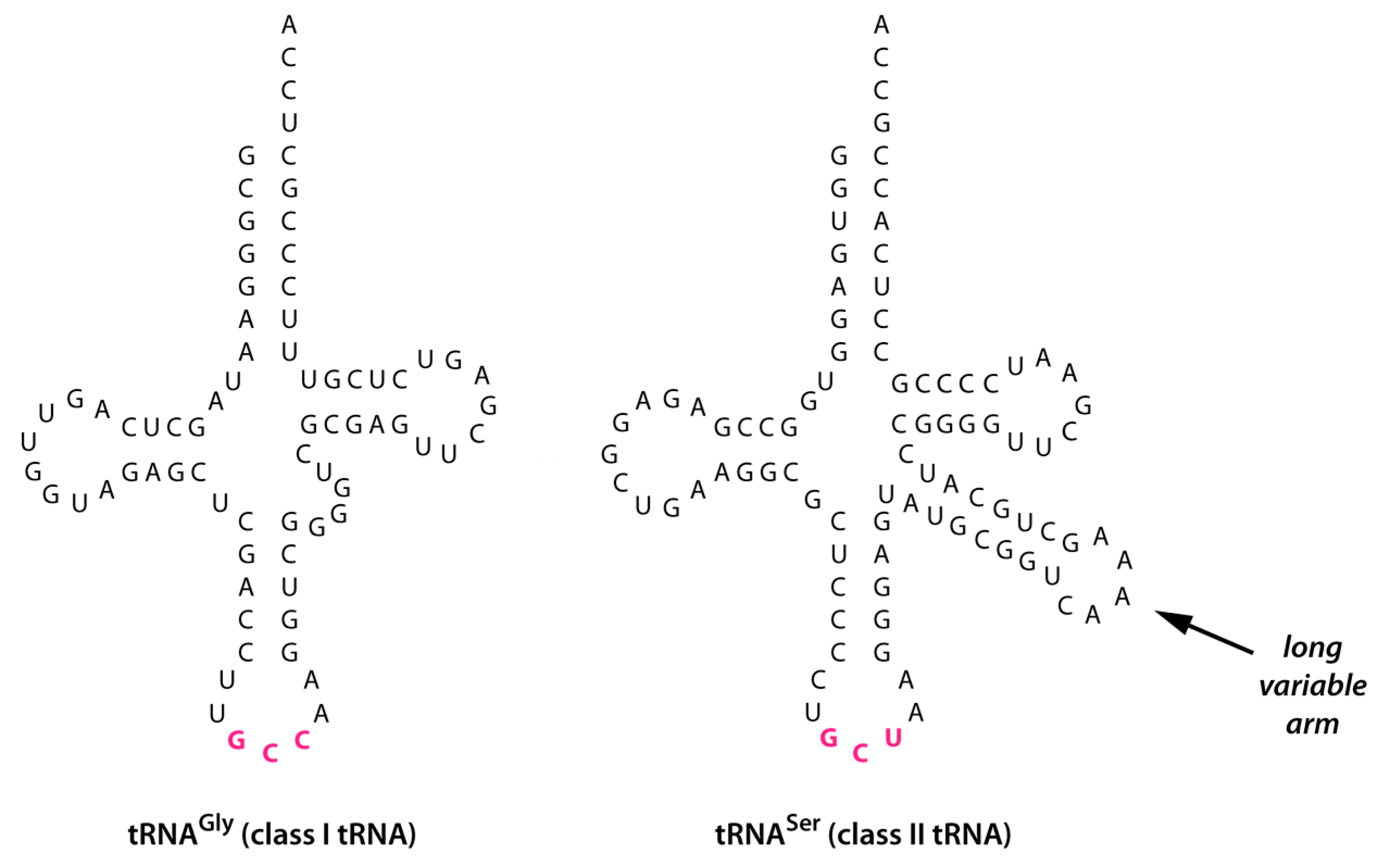
- Evolution of tRNASer(NCU). During the initial duplication and anticodon mutation of tRNAGly(NCC) to form tRNASer(NCU), an enlargement of the variable arm (V arm) occurred—possibly through error-prone replication or retention of an intron located in the V arm region [10]—which produced the long V arm characteristic of a class II tRNA structure (Figure 2).
- Evolution of tRNASer(NGA). Some time following its appearance—and possibly much later—the NCU anticodon of tRNASer(NCU) underwent two transversion mutations to produce tRNASer(NGA) (Figure 3, green arrows). Apart from having a different anticodon, tRNASer(NGA) retained a long V arm and the identity determinants that enabled aminoacylation with serine.
- Evolution of other class II tRNAs. Duplication of tRNASer(NGA) followed by various mutations of its anticodon gave rise to the other class II tRNAs which have codons in the codon boxes surrounding those of tRNASer(NGA): tRNALeu, tRNASec, and bacterial tRNATyr (Figure 3, blue arrows). We have previously proposed that the direction of amino acid incorporation into the genetic code was from the small hydrophilic to the larger hydrophobic amino acids [5]. According to this scenario, this third stage could have occurred relatively late in genetic code evolution, when the synthesis of larger peptides/proteins drove the incorporation of large hydrophobic amino acids that afforded stabilization to the synthesized proteins through formation of a hydrophobic core [11,12].
3. Discussion
Acknowledgments
Conflicts of Interest
Abbreviations
| tRNA | Transfer RNA |
| Gly | Glycine |
| Ser | Serine |
| Leu | Leucine |
| Sec | Selenocysteine |
| Tyr | Tyrosine |
References
- Bernhardt, H.S.; Tate, W.P. Evidence from glycine transfer RNA of a frozen accident at the dawn of the genetic code. Biol. Direct. 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, H.S.; Tate, W.P. The transition from noncoded to coded protein synthesis: Did coding mRNAs arise from stability-enhancing binding partners to tRNA? Biol. Direct. 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.P.; Wos, J.D.; Nooner, D.W.; Oró, J. Chemical evolution. XXI. The amino acids released on hydrolysis of HCN oligomers. J. Mol. Evol. 1974, 3, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Cleaves, H.J.; Chalmers, J.H.; Lazcano, A.; Miller, S.L.; Bada, J.L. A reassessment of prebiotic organic synthesis in neutral planetary atmospheres. Orig. Life Evol. Biosph. 2008, 38, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, H.S.; Patrick, W.M. Genetic code evolution started with the incorporation of glycine, followed by other small hydrophilic amino acids. J. Mol. Evol. 2014, 78, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, P.; Giegé, R.; Moras, D.; Yokoyama, S. An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl. Acad. Sci. USA 1993, 90, 8763–8768. [Google Scholar] [CrossRef] [PubMed]
- Bada, J.L. New insights into prebiotic chemistry from Stanley Miller's spark discharge experiments. Chem. Soc. Rev. 2013, 42, 2186–2196. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. On the evolution of the genetic code. Proc. Natl. Acad. Sci. USA 1965, 54, 1546–1552. [Google Scholar] [CrossRef] [PubMed]
- Turk, R.M.; Chumachenko, N.V.; Yarus, M. Multiple translational products from a five-nucleotide ribozyme. Proc. Natl. Acad. Sci. USA 2010, 107, 4585–4589. [Google Scholar] [CrossRef] [PubMed]
- Kjems, J.; Leffers, H.; Olesen, T.; Garrett, R.A. A unique tRNA intron in the variable loop of the extreme thermophile Thermofilum endens and its possible evolutionary implications. J. Biol. Chem. 1989, 264, 17834–17837. [Google Scholar] [PubMed]
- Fournier, G.P.; Gogarten, J.P. Signature of a primitive genetic code in ancient protein lineages. J. Mol. Evol. 2007, 65, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Cleaves, H.J. The origin of the biologically coded amino acids. J. Theor. Biol. 2010, 263, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Mörl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Pütz, J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009, 37, D159–D162. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.J.; Caetano-Anollés, G. The evolutionary significance of the long variable arm in transfer RNA. Complexity 2008, 14, 26–39. [Google Scholar] [CrossRef]
- Sun, F.J.; Caetano-Anollés, G. Transfer RNA and the origins of diversified life. Sci. Prog. 2008, 91, 265–284. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. On the origin of the transfer RNA molecule. J. Theor. Biol. 1992, 159, 199–214. [Google Scholar] [CrossRef]
- Dick, T.P.; Schamel, W.A. Molecular evolution of transfer RNA from two precursor hairpins: Implications for the origin of protein synthesis. J. Mol. Evol. 1995, 41. [Google Scholar] [CrossRef]
- Marck, C.; Grosjean, H. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 2002, 8, 1189–1232. [Google Scholar] [CrossRef] [PubMed]
- Hamashima, K.; Fujishima, K.; Masuda, T.; Sugahara, J.; Tomita, M.; Kanai, A. Nematode-specific tRNAs that decode an alternative genetic code for leucine. Nucleic Acids Res. 2012, 40, 3653–3662. [Google Scholar] [CrossRef] [PubMed]
- Hamashima, K.; Mori, M.; Andachi, Y.; Tomita, M.; Kohara, Y.; Kanai, A. Analysis of genetic code ambiguity arising from nematode-specific misacylated tRNAs. PLoS ONE 2015, 10, e0116981. [Google Scholar] [CrossRef] [PubMed]
- Staves, M.P.; Bloch, D.P.; Lacey, J.C., Jr. Evolution of E. coli tRNA(Ile): Evidence of derivation from other tRNAs. Z. Naturforsch. C 1987, 42, 129–133. [Google Scholar] [PubMed]
- McClain, W.H. Rules that govern tRNA identity in protein synthesis. J. Mol. Biol. 1993, 234, 257–280. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.Q.; Gross, H.J. The long extra arms of human tRNA(Ser)Sec and tRNASer function as major identity elements for serylation in an orientation-dependent, but not sequence-specific manner. Nucleic Acids Res. 1993, 21, 5589–5594. [Google Scholar] [CrossRef] [PubMed]
- Asahara, H.; Himeno, H.; Tamura, K.; Nameki, N.; Hasegawa, T.; Shimizu, M. Escherichia coli seryl-tRNA synthetase recognizes tRNA(Ser) by its characteristic tertiary structure. J. Mol. Biol. 1994, 236, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Chumachenko, N.V.; Novikov, Y.; Yarus, M. Rapid and simple ribozymic aminoacylation using three conserved nucleotides. J. Am. Chem. Soc. 2009, 131, 5257–5263. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. The genetic code did not originate from an mRNA codifying polyglycine because the proto-mRNAs already codified for an amino acid number greater than one. J. Theor. Biol. 2014, 361, 204–205. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Boniecki, M.T.; Jaffe, J.D.; Imai, B.S.; Yau, P.M.; Luthey-Schulten, Z.A.; Martinis, S.A. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl. Acad. Sci. USA 2011, 108, 9378–9383. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.A.; Tuite, M.F. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995, 23, 1481–1486. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.C.; Miranda, I.; Silva, R.M.; Moura, G.R.; Thomas, B.; Akoulitchev, A.; Santos, M.A. A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol. 2007, 8, R206. [Google Scholar] [CrossRef] [PubMed]
- Antonczak, A.K.; Simova, Z.; Yonemoto, I.T.; Bochtler, M.; Piasecka, A.; Czapinska, H.; Brancale, A.; Tippmann, E.M. Importance of single molecular determinants in the fidelity of expanded genetic codes. Proc. Natl. Acad. Sci. USA 2011, 108, 1320–1325. [Google Scholar] [CrossRef] [PubMed]
- Holman, K.M.; Wu, J.; Ling, J.; Simonović, M. The crystal structure of yeast mitochondrial ThrRS in complex with the canonical threonine tRNA. Nucleic Acids Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Rogers, H.H.; Griffiths-Jones, S. tRNA anticodon shifts in eukaryotic genomes. RNA 2014, 20, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.J.; de Henau, S.; Crothers, D.M. On the physical basis for ambiguity in genetic coding interactions. Proc. Natl. Acad. Sci. USA 1978, 75, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Rodin, S.N.; Rodin, A.S. On the origin of the genetic code: Signatures of its primordial complementarity in tRNAs and aminoacyl-tRNA synthetases. Heredity (Edinb.) 2008, 100, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Achsel, T.; Gross, H.J. Identity determinants of human tRNA(Ser): Sequence elements necessary for serylation and maturation of a tRNA with a long extra arm. EMBO J. 1993, 12, 3333–3338. [Google Scholar] [PubMed]
- Ling, J.; O'Donoghue, P.; Söll, D. Genetic code flexibility in microorganisms: Novel mechanisms and impact on physiology. Nat. Rev. Microbiol. 2015, 13, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Plankensteiner, K.; Righi, A.; Rode, B.M. Glycine and diglycine as possible catalytic factors in the prebiotic evolution of peptides. Orig. Life Evol. Biosph. 2002, 32, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Milner-White, E.J.; Russell, M.J. Predicting the conformations of peptides and proteins in early evolution. Biol. Direct. 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Van der Gulik, P.; Massar, S.; Gilis, D.; Buhrman, H.; Rooman, M. The first peptides: The evolutionary transition between prebiotic amino acids and early proteins. J. Theor. Biol. 2009, 261, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Francis, B.R. Evolution of the genetic code by incorporation of amino acids that improved or changed protein function. J. Mol. Evol. 2013, 77, 134–158. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.R. The structure and function of the aspartic proteinases. Annu. Rev. Biophys. Biophys. Chem. 1990, 19, 189–215. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.M.; Jeffares, D.C.; Hoeppner, M.P.; Penny, D. Does the ribosome challenge our understanding of the RNA world? J. Mol. Evol. 2016, 82. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernhardt, H.S. Clues to tRNA Evolution from the Distribution of Class II tRNAs and Serine Codons in the Genetic Code. Life 2016, 6, 10. https://doi.org/10.3390/life6010010
Bernhardt HS. Clues to tRNA Evolution from the Distribution of Class II tRNAs and Serine Codons in the Genetic Code. Life. 2016; 6(1):10. https://doi.org/10.3390/life6010010
Chicago/Turabian StyleBernhardt, Harold S. 2016. "Clues to tRNA Evolution from the Distribution of Class II tRNAs and Serine Codons in the Genetic Code" Life 6, no. 1: 10. https://doi.org/10.3390/life6010010




