Amino Acid Transporters and Release of Hydrophobic Amino Acids in the Heterocyst-Forming Cyanobacterium Anabaena sp. Strain PCC 7120
Abstract
:1. Introduction
| Transporter | TCDB # | Transported amino acids | ORF | Gene | Predicted gene product | Ref. |
|---|---|---|---|---|---|---|
| N-I | 3.A.1.4 | Pro, Phe, Leu, | all1046 | natA | ATPase | [27] |
| Gly, Thr, Ala, | alr1834 | natB | PSB | |||
| Ser, Met, Asn, | all1047 | natC | TM | |||
| His, Orn, Gln, | all1248 | natD | TM | |||
| Glu | all2912 | natE | ATPase | |||
| N-II | 3.A.1.3 | Asp, Glu, Asn, | alr4164 | natF | PSB | [28] |
| Gln, Met, Thr, | alr4165 | natG | TM | |||
| Ala, Ser, Gly, | alr4166 | natH | TM | |||
| His | alr4167 | bgtA | ATPase | |||
| Bgt | 3.A.1.3 | Lys, Arg, Orn, | alr4167 | bgtA | ATPase | [28] |
| His, Gln | alr3187 | bgtB | PSB and TM | |||
| N-III | 3.A.1.4 | Gly, Pro, Glu, | alr2535 | natI | PSB | This work |
| Phe, Leu, Ala, | alr2536 | natJ | TM | |||
| Gln | alr2538 | natK | TM | |||
| alr2539 | natL | ATPase | ||||
| alr2541 | natM | ATPase |
2. Experimental Section
2.1. Bacterial Strains and Growth Conditions
2.2. Plasmid Construction and Genetic Procedures
| Primer | Sequence (5'→3') |
|---|---|
| alr2536-7120-1 | GGA TCC GCT AAC GCT ACT TTG CCG |
| alr2536-7120-2 | GGA TCC GCA ACC CAA AGC CAA TC |
| all0342-7120-1 | GGA TCC GTT GAC CAA TAC CCT CAT GGC |
| all0342-7120-2 | GGA TCC GCT TGG AAG GTT ACA GGC |
| alr3429-7120-1 | GGA TCC GGG GTT TAA AGA TGC TGA CGG |
| alr3429-7120-2 | GGA TCC GAG GAT GTT CTC TCA CCC |
| all1189-1 | GGA TCC GGA AAC TCA CAG |
| all1189-2 | GCG GAT CCA GGA TAA TAG |
| alr1538-1 | GGA TCC TGG CTG TGT ATT TAG |
| alr1538-2 | GGA TCC TTT GGG CAG AAG |
| all3551-1 | GGA TCC AGC CCA ATA GTT G |
| all3551-2 | GGA TCC CTG CCA AAG AC |
| AA-1 | GAG CCA TAC AAG CTC TGA TTC ATG G |
| AA-2 | ACG CGA TCG CTG ACT CCT GCC |
2.3. Growth Tests and Sample Preparation
2.4. Substrate Transport Assays
3. Results
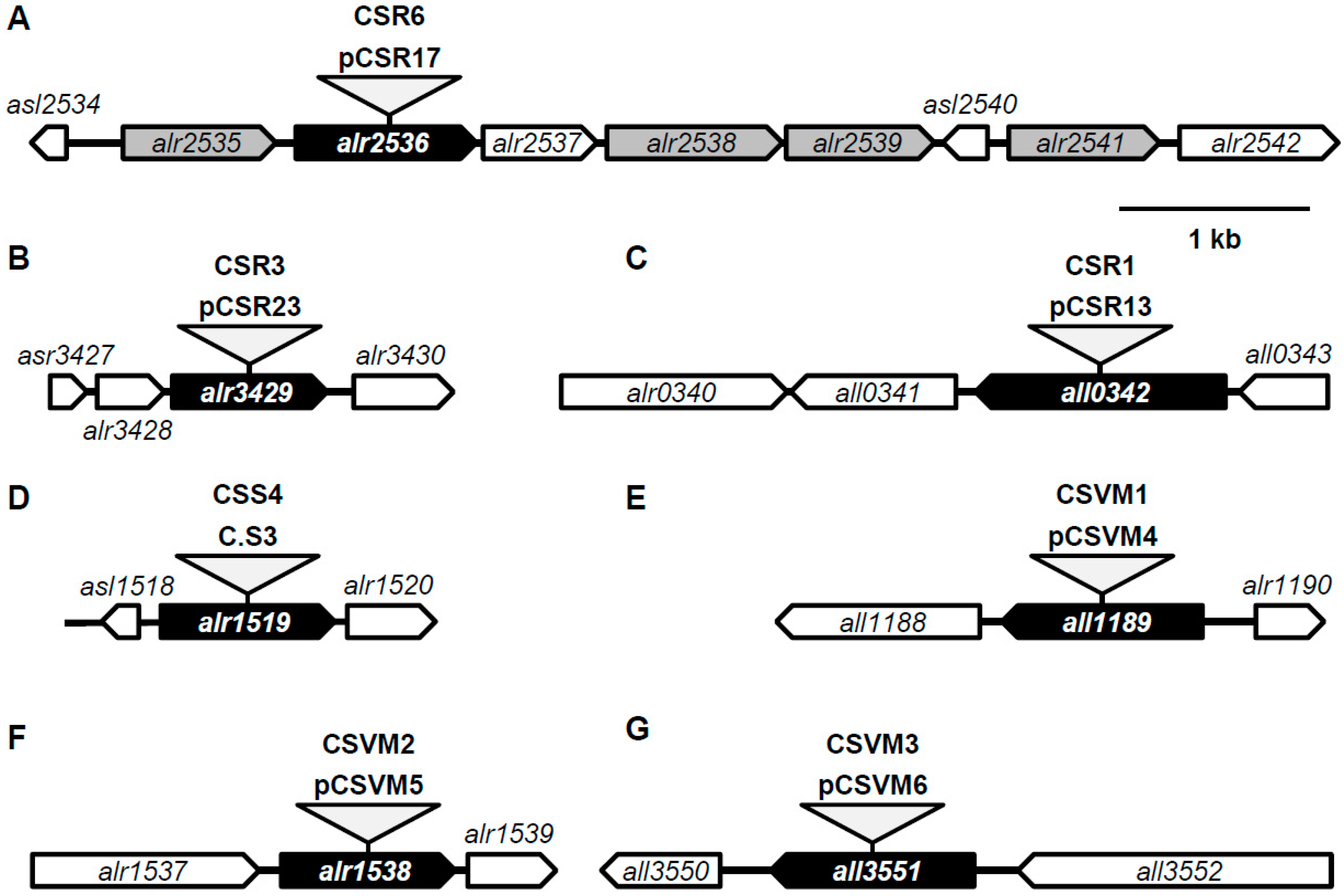
3.1. Predicted ABC-Type Amino Acid Transporters
| Substrate | Transport activity (nmol [mg Chl]−1) | ||||||
|---|---|---|---|---|---|---|---|
| PCC 7120 | Mutant strain (inactivated ORF) | ||||||
| CSR6 (alr2536) | CSR3 (alr3429) | CSR1 (all0342) | |||||
| Mean ± SE (n) | Mean ± SE (n) | % (P) | Mean ± SE (n) | % (P) | Mean ± SE (n) | % (P) | |
| Basic | |||||||
| l-Arg | 125 ± 1.59 (25) | 115 ± 16.4 (3) | 92% (0.077) | 148 ± 6.60 (4) | 119% (0.187) | 195 ± 12.5 (3) | 156% (0.068) |
| l-Lys | 138 ± 1.55 (16) | 151 ± 3.92 (2) | 110% (0.167) | 163 ± 6.15 (3) | 118% (0.119) | 140 ± 8.40 (2) | 102% (0.882) |
| l-His | 88.0 ± 1.10 (16) | 71.7 ± 4.80 (3) | 81% (0.174) | 90.0 ± 4.94 (4) | 102% (0.863) | 132 ± 5.28 (3) | 150% (0.023) |
| Acidic | |||||||
| l-Asp | 37.3 ± 0.39 (27) | 33.5 ± 2.18 (3) | 90% (0.440) | 31.1 ± 1.67 (4) | 83% (0.170) | 38.8 ± 2.19 (4) | 104% (0.768) |
| l-Glu | 10.5 ± 0.12 (25) | 6.62 ± 0.70 (3) | 63% (0.061) | 10.2 ± 0.26 (4) | 97% (0.721) | 16.9 ± 1.56 (4) | 161% (0.130) |
| Neutral polar | |||||||
| l-Gln | 99.4 ± 0.90 (30) | 77.0 ± 6.89 (3) | 77% (0.189) | 99.3 ± 6.31 (4) | 100% (0.994) | 158 ± 6.48 (4) | 158% (0.014) |
| l-Ser | 218 ± 2.50 (10) | 195 ± 2.05 (2) | 90% (0.022) | 191 ± 9.37 (3) | 88% (0.237) | 239 ± 34.3 (3) | 110% (0.753) |
| Hydrophobic | |||||||
| l-Ala | 192 ± 1.09 (20) | 139 ± 13.4 (3) | 72% (0.145) | 203 ± 16.3 (4) | 105% (0.772) | 286 ± 6.03 (3) | 149% (0.004) |
| Gly | 200 ± 3.30 (15) | 107 ± 14.0 (3) | 54% (0.039) | 222 ± 8.84 (4) | 111% (0.337) | 252 ± 5.50 (3) | 126% (0.007) |
| l-Leu | 103 ± 0.99 (17) | 70.4 ± 5.34 (3) | 68% (0.051) | 117 ± 5.33 (4) | 113% (0.315) | 177 ± 12.0 (3) | 171% (0.067) |
| l-Pro | 135 ± 1.99 (16) | 82.8 ± 14.9 (3) | 61% (0.174) | 151 ± 10.0 (3) | 112% (0.449) | 198 ± 13.1 (4) | 147% (0.089) |
| l-Phe | 118 ± 1.87 (16) | 79.4 ± 1.78 (2) | 67% (<0.001) | 91.6 ± 7.79 (4) | 78% (0.197) | 109 ± 8.32 (3) | 92% (0.607) |
3.2. Predicted Amino Acid Transporters from Other Transporter Families
| Substrate | Transport activity (nmol [mg Chl]−1) | |||||
|---|---|---|---|---|---|---|
| BG11C | BG110C | |||||
| PCC 7120 | CSS4 (alr1519) | PCC 7120 | CSS4 (alr1519) | |||
| Mean ± SE (n) | Mean ± SE (n) | % (P) | Mean ± SE (n) | Mean ± SE (n) | % (P) | |
| Basic | ||||||
| l-Arg | 116 ± 7.64 (3) | 113 ± 3.16 (2) | 98% (0.864) | 187 ± 8.96 (3) | 167 ± 3.58 (2) | 89% (0.327) |
| Acidic | ||||||
| l-Asp | 30.3 ± 1.03 (4) | 42.0 ± 3.85 (2) | 139% (0.214) | 62.1 ± 0.98 (4) | 77.6 ± 3.57 (3) | 125% (0.119) |
| l-Glu | 10.4 ± 1.48 (4) | 10.3 ± 0.64 (3) | 99% (0.970) | 34.7 ± 1.63 (4) | 32.2 ± 1.90 (3) | 93% (0.619) |
| Neutral polar | ||||||
| l-Gln | 118 ± 4.46 (3) | 125 ± 2.15 (2) | 106% (0.487) | 216 ± 5.30 (3) | 214 ± 2.06 (2) | 99% (0.854) |
| Hydrophobic | ||||||
| l-Ala | 224 ± 12.6 (3) | 215 ± 17.8 (2) | 96% (0.826) | 309 ± 11.7 (3) | 299 ± 14.0 (2) | 108% (0.748) |
3.3. Release of Amino Acids from Vegetative Cells
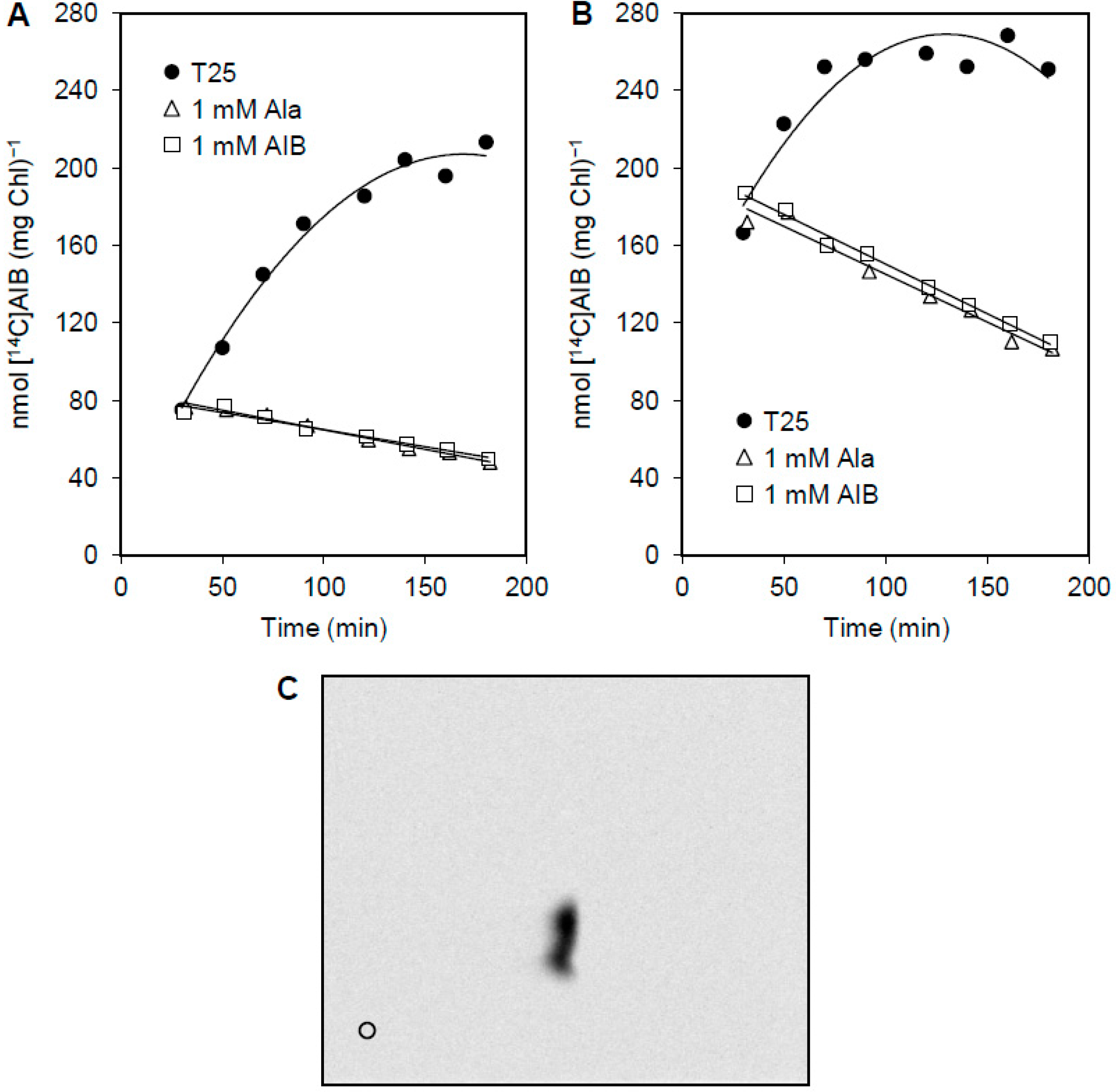
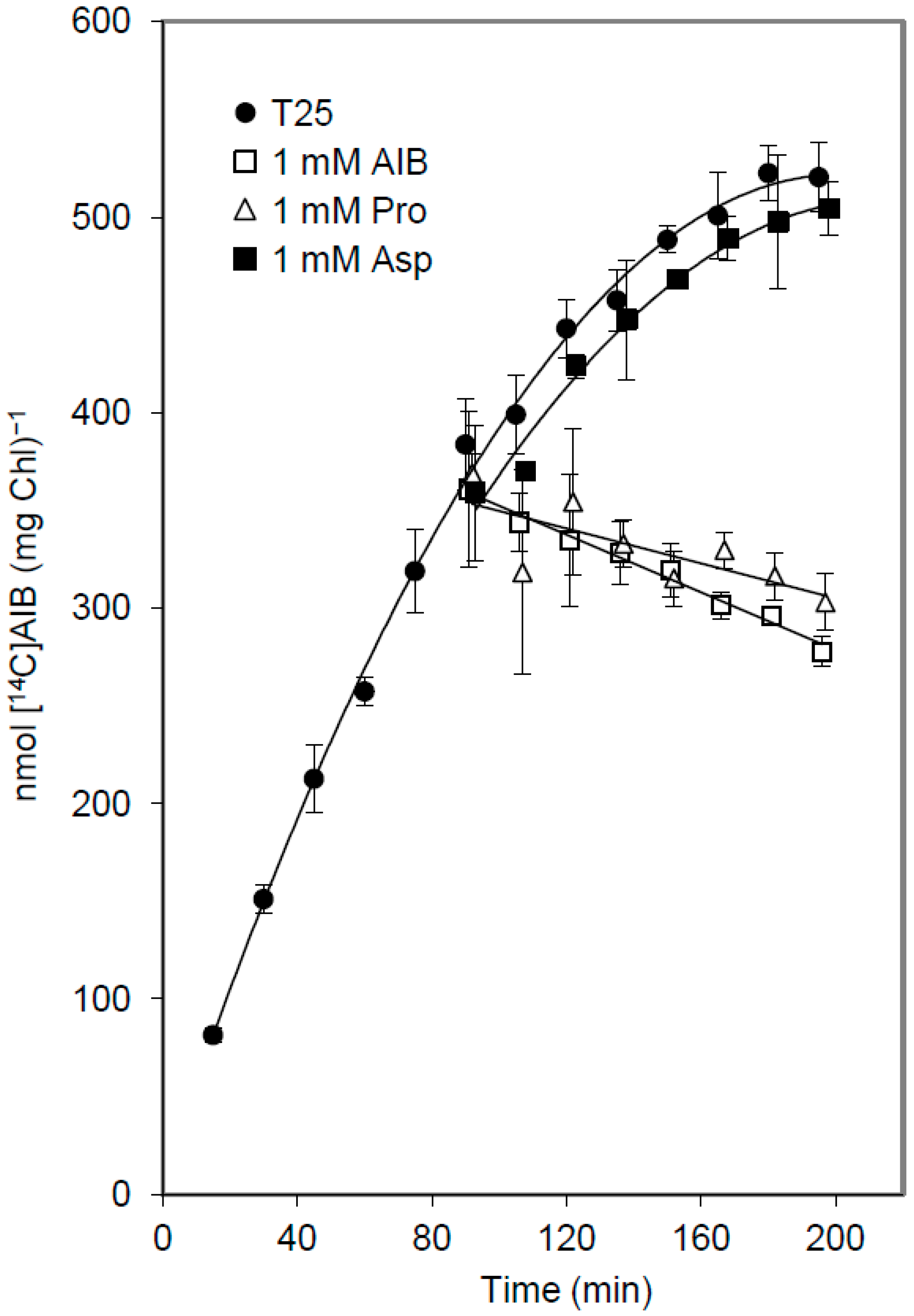

4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Flores, E.; Herrero, A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 2010, 8, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Meeks, J.C.; Wolk, C.P.; Shaffer, P.W.; Austin, S.M. Formation of glutamine from [13N]ammonia, [13N]dinitrogen, and [14C]glutamate by heterocysts isolated from Anabaena cylindrica. J. Bacteriol. 1977, 129, 1545–1555. [Google Scholar] [PubMed]
- Burnat, M.; Herrero, A.; Flores, E. Compartmentalized cyanophycin metabolism in the diazotrophic filaments of a heterocyst-forming cyanobacterium. Proc. Natl. Acad. Sci. USA 2014, 111, 3823–3828. [Google Scholar] [CrossRef] [PubMed]
- Jüttner, F. 14C-labeled metabolites in heterocysts and vegetative cells of Anabaena cylindrica filaments and their presumptive function as transport vehicles of organic carbon and nitrogen. J. Bacteriol. 1983, 155, 628–633. [Google Scholar] [PubMed]
- López-Igual, R.; Flores, E.; Herrero, A. Inactivation of a heterocyst-specific invertase indicates a principal role of sucrose catabolism in heterocysts of Anabaena sp. J. Bacteriol. 2010, 192, 5526–5533. [Google Scholar] [CrossRef] [PubMed]
- Pernil, R.; Herrero, A.; Flores, E. Catabolic function of compartmentalized alanine dehydrogenase in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 2010, 192, 5165–5172. [Google Scholar] [CrossRef] [PubMed]
- Vargas, W.A.; Nishi, C.N.; Giarrocco, L.E.; Salerno, G.L. Differential roles of alkaline/neutral invertases in Nostoc sp. PCC 7120: Inv-B isoform is essential for diazotrophic growth. Planta 2011, 233, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Ayre, B.G. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 2011, 4, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Kolman, M.A.; Nishi, C.N.; Perez-Cenci, M.; Salerno, G.L. Sucrose in cyanobacteria: From a salt-response molecule to play a key role in nitrogen fixation. Life 2015, 5, 102–126. [Google Scholar] [CrossRef] [PubMed]
- Martín-Figueroa, E.; Navarro, F.; Florencio, F.J. The GS-GOGAT pathway is not operative in the heterocysts. Cloning and expression of glsF gene from the cyanobacterium Anabaena sp. PCC 7120. FEBS Lett. 2000, 476, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Sherman, D.M.; Tucker, D.; Sherman, L.A. Heterocyst development and localization of cyanophycin in N2-fixing cultures of Anabaena sp. PCC 7120 (Cyanobacteria). J. Phycol. 2000, 36, 932–941. [Google Scholar] [CrossRef]
- Wolk, C.P.; Thomas, J.; Shaffer, P.W.; Austin, S.M.; Galonsky, A. Pathway of nitrogen metabolism after fixation of 13N-labeled nitrogen gas by the cyanobacterium, Anabaena cylindrica. J. Biol. Chem. 1976, 251, 5027–5034. [Google Scholar] [PubMed]
- Mariscal, V.; Herrero, A.; Flores, E. Continuous periplasm in a filamentous, heterocyst-forming cyanobacterium. Mol. Microbiol. 2007, 65, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Haselkorn, R. Cell-cell communication in filamentous cyanobacteria. Mol. Microbiol. 2008, 70, 783–785. [Google Scholar] [PubMed]
- Mariscal, V.; Flores, E. Multicellularity in a heterocyst-forming cyanobacterium: Pathways for intercellular communication. Adv. Exp. Med. Biol. 2010, 675, 123–135. [Google Scholar] [PubMed]
- Mariscal, V. Cell-cell joining proteins in heterocyst-forming cyanobacteria. In The Cell Biology of Cyanobacteria; Flores, E., Herrero, A., Eds.; Caister Academic Press: Norfolk, UK, 2014; pp. 293–304. [Google Scholar]
- Mullineaux, C.W.; Mariscal, V.; Nenninger, A.; Khanum, H.; Herrero, A.; Flores, E.; Adams, D.G. Mechanism of intercellular molecular exchange in heterocyst-forming cyanobacteria. EMBO J. 2008, 27, 1299–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merino-Puerto, V.; Schwarz, H.; Maldener, I.; Mariscal, V.; Mullineaux, C.W.; Herrero, A.; Flores, E. FraC/FraD-dependent intercellular molecular exchange in the filaments of a heterocyst-forming cyanobacterium, Anabaena sp. Mol. Microbiol. 2011, 82, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.; Pernil, R.; Muro-Pastor, A.M.; Mariscal, V.; Maldener, I.; Lechno-Yossef, S.; Fan, Q.; Wolk, C.P.; Herrero, A. Septum-localized protein required for filament integrity and diazotrophy in the heterocyst forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 2007, 189, 3884–3890. [Google Scholar] [CrossRef] [PubMed]
- Merino-Puerto, V.; Mariscal, V.; Mullineaux, C.W.; Herrero, A.; Flores, E. Fra proteins influencing filament integrity, diazotrophy and localization of septal protein SepJ in the heterocyst-forming cyanobacterium Anabaena sp. Mol. Microbiol. 2010, 75, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.; Herrero, A.; Wolk, C.P.; Maldener, I. Is the periplasm continuous in filamentous multicellular cyanobacteria? Trends Microbiol. 2006, 14, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Nicolaisen, K.; Mariscal, V.; Bredemeier, R.; Pernil, R.; Moslavac, S.; López-Igual, R.; Maldener, I.; Herrero, A.; Schleiff, E.; Flores, E. The outer membrane of a heterocyst-forming cyanobacterium is a permeability barrier for uptake of metabolites that are exchanged between cells. Mol. Microbiol. 2009, 74, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, M.L.; Herrero, A.; Flores, E. Amino acid transport systems required for diazotrophic growth in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1995, 177, 3150–3157. [Google Scholar] [PubMed]
- Flores, E.; Muro-Pastor, M.I. Uptake of glutamine and glutamate by the dinitrogen-fixing cyanobacterium Anabaena sp. PCC 7120. FEMS Microbiol. Lett. 1988, 56, 127–130. [Google Scholar] [CrossRef]
- Herrero, A.; Flores, E. Transport of basic amino acids by the dinitrogen-fixing cyanobacterium Anabaena PCC 7120. J. Biol. Chem. 1990, 265, 3931–3935. [Google Scholar] [PubMed]
- Xu, P.; McAuley, P.J. Uptake of amino acids by the cyanobacterium Anabaena ATCC 27893. New Phytol. 1990, 115, 581–585. [Google Scholar] [CrossRef]
- Picossi, S.; Montesinos, M.L.; Pernil, R.; Lichtlé, C.; Herrero, A.; Flores, E. ABC-type neutral amino acid permease N-I is required for optimal diazotrophic growth and is repressed in the heterocysts of Anabaena sp. strain PCC 7120. Mol. Microbiol. 2005, 57, 1582–1592. [Google Scholar] [CrossRef] [PubMed]
- Pernil, R.; Picossi, S.; Mariscal, V.; Herrero, A.; Flores, E. ABC-type amino acid uptake transporters Bgt and N-II of Anabaena sp. strain PCC 7120 share an ATPase subunit and are expressed in vegetative cells and heterocysts. Mol. Microbiol. 2008, 67, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Busch, W.; Saier, M.H., Jr. The transporter classification (TC) system. Crit. Rev. Biochem. Mol. Biol. 2002, 37, 287–337. [Google Scholar] [CrossRef] [PubMed]
- Transporter Classification Database. Available online: http://www.tcdb.org (accessed on 15 April 2015).
- Labarre, J.; Thuriaux, P.; Chauvat, F. Genetic analysis of amino acid transport in the facultatively heterotrophic cyanobacterium Synechocystis sp. strain 6803. J. Bacteriol. 1987, 169, 4668–4673. [Google Scholar] [PubMed]
- Montesinos, M.L.; Herrero, A.; Flores, E. Amino acid transport in taxonomically diverse cyanobacteria and identification of two genes encoding elements of a neutral amino acid permease putatively involved in recapture of leaked hydrophobic amino acids. J. Bacteriol. 1997, 179, 853–862. [Google Scholar] [PubMed]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Cai, Y.P.; Wolk, C.P. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 1990, 172, 3138–3145. [Google Scholar] [PubMed]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current Protocols in Molecular Biology; Greene Publishing and Wiley-Interscience: New York, NY, USA, 2015. [Google Scholar]
- Elhai, J.; Wolk, C.P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 1988, 68, 119–138. [Google Scholar] [CrossRef] [PubMed]
- Frías, J.E.; Flores, E.; Herrero, A. Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and NtcB (LysR family) transcription factors. Mol. Microbiol. 2000, 38, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Black, T.A.; Cai, Y.; Wolk, C.P. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 1993, 9, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Elhai, J.; Vepritskiy, A.; Muro-Pastor, A.M.; Flores, E.; Wolk, C.P. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 1997, 179, 1998–2005. [Google Scholar] [PubMed]
- Markwell, M.A.K.; Hass, S.M.; Bieber, L.L.; Tolbert, N.E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 1978, 87, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Mackinney, G. Absorption of light by chlorophyll solutions. J. Biol. Chem. 1941, 140, 109–112. [Google Scholar]
- Hoshino, T.; Kose, K. Cloning and nucleotide sequence of braC, the structural gene for the leucine-, isoleucine-, and valine-binding protein of Pseudomonas aeruginosa PAO. J. Bacteriol. 1989, 171, 6300–6306. [Google Scholar] [PubMed]
- Okamoto, S.; Yamanishi, Y.; Ehira, S.; Kawashima, S.; Tonomura, K.; Kanehisa, M. Prediction of nitrogen metabolism-related genes in Anabaena by kernel-based network analysis. Proteomics 2007, 7, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Quintero, M.J.; Montesinos, M.L.; Herrero, A.; Flores, E. Identification of genes encoding amino acid permeases by inactivation of selected ORFs from the Synechocystis genomic sequence. Genome Res. 2001, 11, 2034–2040. [Google Scholar] [CrossRef] [PubMed]
- Slotboom, D.J.; Konings, W.N.; Lolkema, J.S. Structural features of the glutamate transporter family. Microbiol. Mol. Biol. Rev. 1999, 63, 293–307. [Google Scholar] [PubMed]
- Wong, F.H.; Chen, J.S.; Reddy, V.; Day, J.L.; Shlykov, M.A.; Wakabayashi, S.T.; Saier, M.H., Jr. The amino acid-polyamine-organocation superfamily. J. Mol. Microbiol. Biotechnol. 2012, 22, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Rudnick, G.; Krämer, R.; Blakely, R.D.; Murphy, D.L.; Verrey, F. The SLC6 transporters: Perspectives on structure, functions, regulation, and models for transporter dysfunction. Pflugers Arch. 2014, 466, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Ihlenfeldt, M.J.A.; Gibson, J. CO2 fixation and its regulation in Anacystis nidulans (Synechococcus). Arch. Microbiol. 1975, 102, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Raboy, B.; Padan, E. Active transport of glucose and α-methylglucoside in the cyanobacterium Plectonema boryanum. J. Biol. Chem. 1978, 253, 3287–3291. [Google Scholar] [PubMed]
- Klein, R.A.; Moore, M.J.; Smith, M.W. Selective diffusion of neutral amino acids across lipid bilayers. Biochim. Biophys. Acta 1971, 233, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.C. Permeability of membranes to amino acids and modified amino acids: Mechanisms involved in translocation. Amino Acids 1994, 6, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Krämer, R. Secretion of amino acids by bacteria: Physiology and mechanism. FEMS Microbiol. Rev. 1994, 13, 75–93. [Google Scholar] [CrossRef]
- CyanoBase. Available online: http://genome.microbedb.jp/cyanobase/Anabaena (accessed on 15 April 2015).
- TransportDB. Available online: http://www.membranetransport.org/index_v2_rc1.html (accessed on 15 April 2015).
- Paulsen, I.T.; Nguyen, L.; Sliwinski, M.K.; Rabus, R.; Saier, M.H., Jr. Microbial genome analyses: Comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 2000, 301, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Schleiff, E. The cell envelope. In The Cell Biology of Cyanobacteria; Flores, E., Herrero, A., Eds.; Caister Academic Press: Norfolk, UK, 2014; pp. 29–87. [Google Scholar]
- Stebegg, R.; Wurzinger, B.; Mikulic, M.; Schmetterer, G. Chemoheterotrophic growth of the cyanobacterium Anabaena sp. strain PCC 7120 dependent on a functional cytochrome c oxidase. J. Bacteriol. 2012, 194, 4601–607. [Google Scholar] [CrossRef] [PubMed]
- Pernil, R.; Herrero, A.; Flores, E. A TRAP transporter for pyruvate and other monocarboxylate 2-oxoacids in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 2010, 192, 6089–6092. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pernil, R.; Picossi, S.; Herrero, A.; Flores, E.; Mariscal, V. Amino Acid Transporters and Release of Hydrophobic Amino Acids in the Heterocyst-Forming Cyanobacterium Anabaena sp. Strain PCC 7120. Life 2015, 5, 1282-1300. https://doi.org/10.3390/life5021282
Pernil R, Picossi S, Herrero A, Flores E, Mariscal V. Amino Acid Transporters and Release of Hydrophobic Amino Acids in the Heterocyst-Forming Cyanobacterium Anabaena sp. Strain PCC 7120. Life. 2015; 5(2):1282-1300. https://doi.org/10.3390/life5021282
Chicago/Turabian StylePernil, Rafael, Silvia Picossi, Antonia Herrero, Enrique Flores, and Vicente Mariscal. 2015. "Amino Acid Transporters and Release of Hydrophobic Amino Acids in the Heterocyst-Forming Cyanobacterium Anabaena sp. Strain PCC 7120" Life 5, no. 2: 1282-1300. https://doi.org/10.3390/life5021282