The Adaptive Immune System of Haloferax volcanii
Abstract
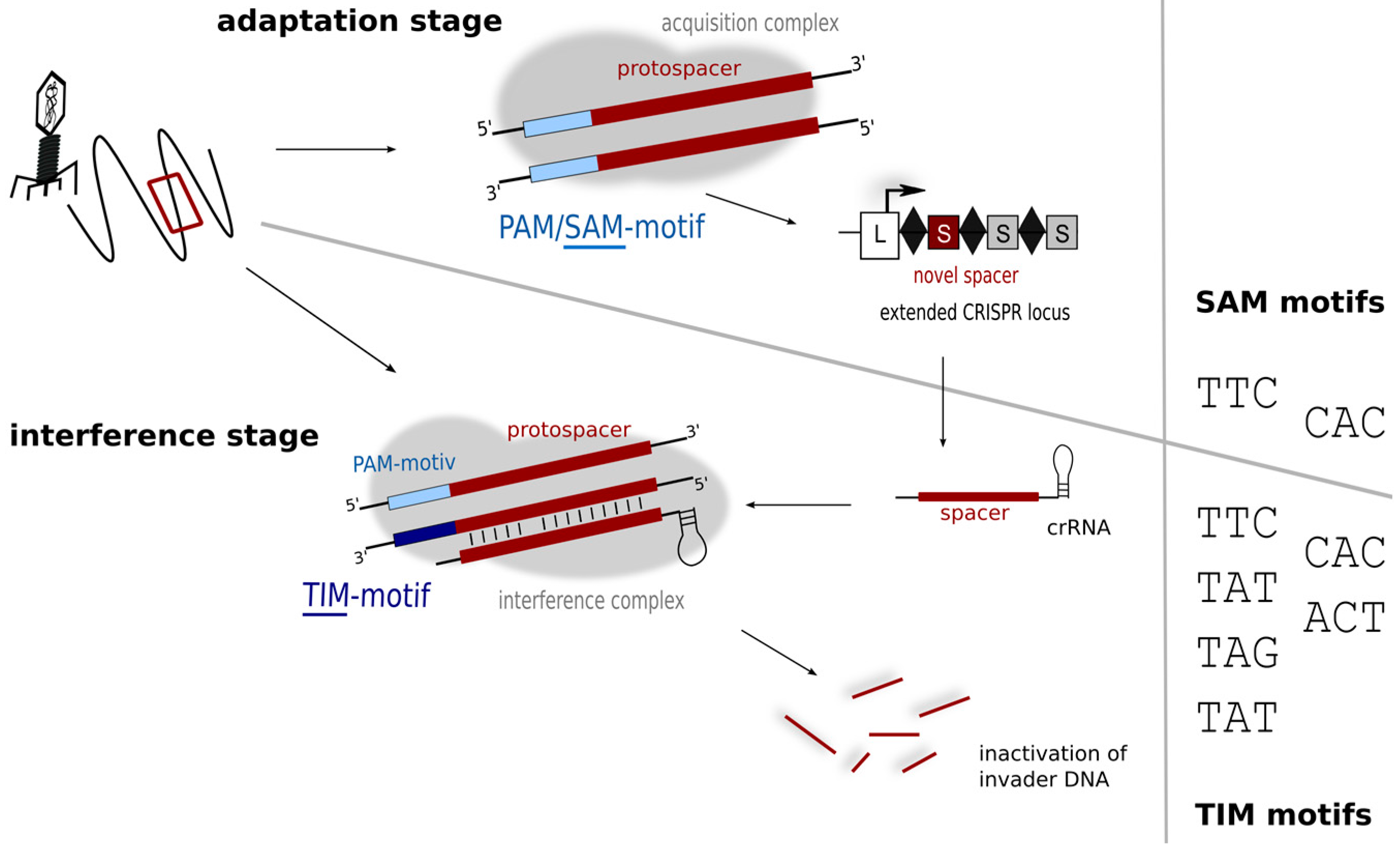
:1. The CRISPR-Cas Immune System
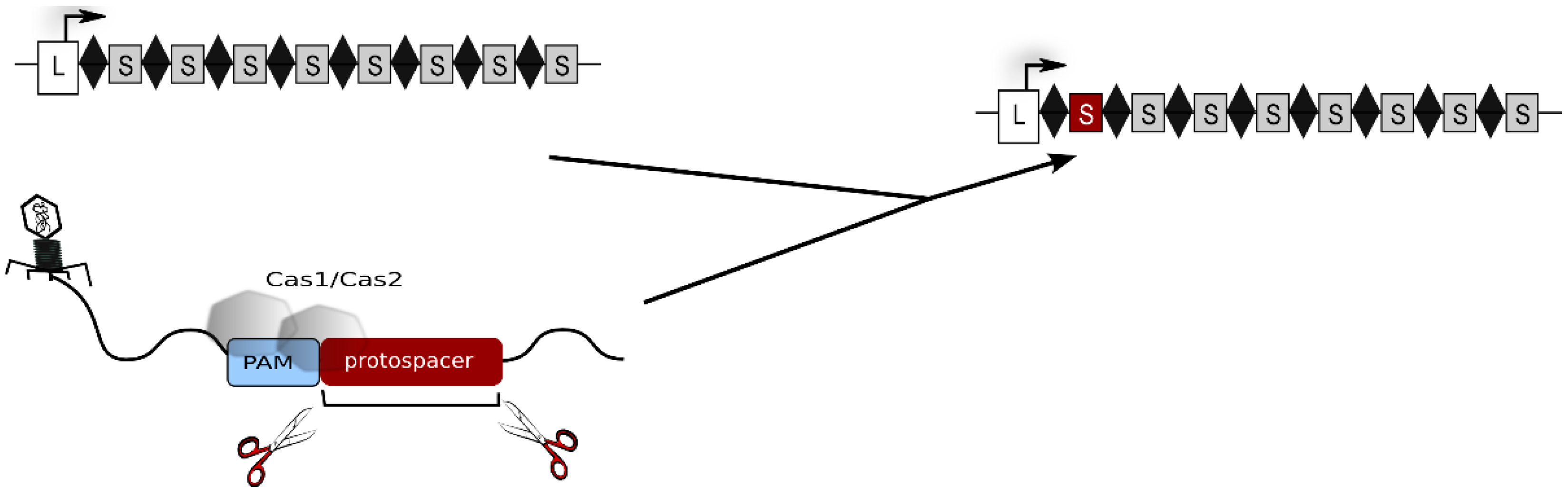
2. The Type I-B CRISPR-Cas System of Haloferax volcanii

| Spacer | Alignment of spacer/matching sequencea | Matching sequence |
|---|---|---|
| C-4 (nt 2386433:2386398) |  | Hrr. lacusprofundi chromosome 1 (nt 759433:759468), within ORF Hlac_0754. Predicted translation of spacer is identical to protein Hlac_0754 but for one conservative (L/F) change (i.e., MPDLVRDNIVDV/MPDFVRDNIVDV). Hlac_0754 is part of a 28.7 kb region (nt 750728:779675) containing genes related to halovirus BJ1, and previously denoted by Krupovic et al. [23] as provirus Hlac-Pro1. |
| C-4 (nt 2386433:2386398) |  | Hfx. elongans ATCC BAA-1513: AOLK01000020. Within ORF C453_12906 (nt 35718:35683), a homolog of Hlac_0754. Predicted translation of spacer exactly matches the protein sequence of C453_12906 (MPDLVRDNIVDV). Contig AOLK01000020 is likely to represent a halovirus genome, with many genes related to BJ1 or other haloviruses/plasmids. We denote this contig as HeloV2. A related virus appears to be represented by a contig (AOLN01000009) of Haloferax mucosum PA12, ATCC BAA-1512 (i.e., HmucV2, Supplementary Figure S2) |
| C-14 (nt 2385742:2385778) |  | b Hfx. volcanii (chromosome, nt 333928:333984), within ORF HVO_0372 (hypothetical protein). HVO_0372 occurs in a ~12kb region of foreign DNA flanked at one end by a tRNA-ala, and at the other end by an integrase and two partial repeats of the tRNA-ala gene. This region appears to be a provirus (we denote as Hvol-IV1). Related provirues are found in the genomes of at least four other Haloferax species (see Supplementary Figure S1). A nearby gene, HVO_0375 (CPxCG-related zinc finger protein), is the likely target of a CRISPR spacer of Hfx. sp. ATCC BAA-645 (contig_24). |
| C-14 (nt 2385742:2385778) |  | Line 2: Haloferax sp. ATB1 (JPES01000108.1) scaffold108 (nt 9103: 9067). Line 3: Haloferax sp. BAB2207: ANPG01000768 (nt 1559–1596). Both matches occur within a homolog of HVO_0372, and are likely to be part of proviruses (Hatb-IV1 and Hbab-IV1, see Figure S1). Elsewhere in this gene (and in HVO_0372) is a target sequence matching a CRISPR spacer carried by Hfx.denitrificans d. |
| P1-2 (nt 205072:205108) |  | Line 2: Lake Tyrrell metagenome (contig 1101968716470, library GS84-02-2-3kb, nt 851:887) b. Line 3: metavirome (assembly from SRR402046). Line 4: Haloterrigena jeotgali A29, HL44_contig00019.19 (nt 18864:18136), within locus tag HL44_04258. BLASTX predicts a COG1475 (ParBc domain) protein (plasmid partition protein). Closest relatives are bacterial (e.g., WP_021624091). The predicted aa sequences are identical i.e., HKSIKEDGYTQP. |
| P1-3 (nt 205139:205173) |  | Line 2: Lake Tyrrell metagenome (49037 1101497529448, library GS84-02-2-3kb, nt 190:224). Line 3: metavirome (assembly from SRR402046). BLASTX of matching contigs show matches to Hbor_29150 of Hgm. borinquense. The adjacent gene, Hbor_29160, on the genome most closely matches M201_gp84 of halovirus HCTV-2. The predicted aa sequences over the matching region (left) are identical but for one conservative change (F/L), i.e., VLDEAGVQFGNR / VLDEAGVQLGNR |
| P1-38 (nt 207450:207485) |  | Lake Tyrrell metavirome (assembly from SRR402046). BLASTX of matching contig shows a match (E = 10−13) to the integrase of halovirus HCTV-5 (M200_gp113). The predicted aa sequences of spacer and matching contig sequence differ by one conservative (D/E) change i.e., RLDDDYFALEAR/RLDDEYFALEAR. |
| P2-1 (nt 217843:217879) |  | Great Salt Lake metagenome sequence 162854 GSLNARP_GFPJP1N02GIUFX (nt 107:73). BLASTX shows strong similarity (E = 10−24) to MCM/cdc46 family proteins (e.g., Natrialba taiwanensis (ELY91445), and the spacer sequence translates to an identical aa sequence, i.e., YIAYARQNVHP to that of the matching GSL sequence. |
| P2-2 (nt 217911:217945) |  | Haloferax sp. ATB1: ATB1DRAFT_JPES01000050_1.50(nt 15622:15655). Match occurs within a hypothetical protein gene (ATB1DRAFT_01921), with homologs in other species, e.g., Hfx. mucosum (ELZ94997.1), Hfx. mediterranei (AFK19012.1) and Hgm. borinquense Hbor_29150 (see P1-3, above). The predicted aa sequence of spacer and matching genome are similar, i.e., AESMEAETEQL/AESKEAEAEQL. |
| P2-10 (nt 218436:218471) |  | Haloferax sp. ATB1: ATB1DRAFT_JPES01000088_1.88, nt 33483:33448. Matches a surface glycoprotein-like ORF (ATB1DRAFT_03295), with many haloarchaeal homologs (e.g., D320_03738, C456_06592 and HVO_2072). |
| P2-11 (nt 218503:218535) |  | CRISPR spacer of Haloferax lucentense DSM 14919: AOLH01000022 (nt 8242–8273). |
| P2-11 (nt 218503:218535) |  | CRISPR spacer of Haloferax sp. BAB2207: ANPG01000305 (nt 8,276–8,244). |
| P2-11 (nt 218503:218535) |  | CRISPR spacer of Haloferax alexandrinus JCM 10717: AOLL01000024 (nt 43672–43639). |


3. Generation of crRNAs and Composition of the Interference Complex

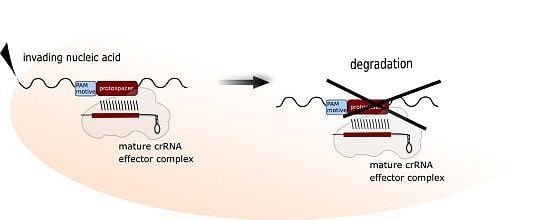
4. Characteristics of a Functional crRNA and Its Interaction with the Invader


5. Motifs for Detecting Invaders
6. Selection Pressure for Retention of the cas Genes
7. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Conflict of Interests
References
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Deveau, H.; Garneau, J.E.; Moineau, S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Terns, M.P.; Terns, R.M. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011, 14, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Van der Oost, J.; Westra, E.R.; Jackson, R.N.; Wiedenheft, B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014, 12, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Westra, E.R.; Buckling, A.; Fineran, P.C. CRISPR-Cas systems: Beyond adaptive immunity. Nat. Rev. Microbiol. 2014, 12, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 2014, 54, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef] [PubMed]
- Marchfelder, A.; Fischer, S.; Brendel, J.; Stoll, B.; Maier, L.K.; Jager, D.; Prasse, D.; Plagens, A.; Schmitz, R.A.; Randau, L. Small RNAs for defence and regulation in archaea. Extremophiles 2012, 16, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Ferrer, C.; Juez, G.; Rodriguez-Valera, F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 1995, 17, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Diez-Villasenor, C.; Soria, E.; Juez, G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000, 36, 244–246. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Diez-Villasenor, C.; Garcia-Martinez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.; Erdmann, S.; Mojica, F.J.; Garrett, R.A. Protospacer recognition motifs: Mixed identities and functional diversity. RNA Biol. 2013, 10, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Chylinski, K.; Makarova, K.S.; Charpentier, E.; Koonin, E.V. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014, 42, 6091–6105. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. The basic building blocks and evolution of CRISPR-Cas systems. Biochem. Soc. Trans. 2013, 41, 1392–1400. [Google Scholar] [PubMed]
- Vestergaard, G.; Garrett, R.A.; Shah, S.A. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014, 11, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Westra, E.R.; Swarts, D.C.; Staals, R.H.; Jore, M.M.; Brouns, S.J.; van der Oost, J. The CRISPRs, they are a-changin’: How prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012, 46, 311–339. [Google Scholar] [CrossRef] [PubMed]
- Mullakhanbhai, M.F.; Larsen, H. Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch. Microbiol. 1975, 104, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Mevarech, M.; Werczberger, R. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 1985, 162, 461–462. [Google Scholar] [PubMed]
- Fischer, S.; Maier, L.K.; Stoll, B.; Brendel, J.; Fischer, E.; Pfeiffer, F.; Dyall-Smith, M.; Marchfelder, A. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J. Biol. Chem. 2012, 287, 33351–33365. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.K.; Fischer, S.; Stoll, B.; Brendel, J.; Pfeiffer, F.; Dyall-Smith, M.; Marchfelder, A. The immune system of halophilic archaea. Mob. Genet. Elem. 2012, 2, 228–232. [Google Scholar] [CrossRef]
- Hartman, A.L.; Norais, C.; Badger, J.H.; Delmas, S.; Haldenby, S.; Madupu, R.; Robinson, J.; Khouri, H.; Ren, Q.; Lowe, T.M.; et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PloS One 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Forterre, P.; Bamford, D.H. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J. Mol. Biol. 2010, 397, 144–160. [Google Scholar] [CrossRef] [PubMed]
- Reeks, J.; Naismith, J.H.; White, M.F. CRISPR interference: A structural perspective. Biochem. J. 2013, 453, 155–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charpentier, E.; van der Oost, J.; White, M.F. crRNA biogenesis. In CRISPR-Cas Systems; Barrangou, R., van der Oost, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 115–144. [Google Scholar]
- Jackson, R.N.; Golden, S.M.; van Erp, P.B.; Carter, J.; Westra, E.R.; Brouns, S.J.; van der Oost, J.; Terwilliger, T.C.; Read, R.J.; Wiedenheft, B. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 2014, 345, 1473–1479. [Google Scholar] [CrossRef] [PubMed]
- Mulepati, S.; Heroux, A.; Bailey, S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 2014, 345, 1479–1484. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Sheng, G.; Wang, J.; Wang, M.; Bunkoczi, G.; Gong, W.; Wei, Z.; Wang, Y. Crystal structure of the RNA-guided immune surveillance cascade complex in Escherichia coli. Nature 2014, 515, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Garside, E.L.; Schellenberg, M.J.; Gesner, E.M.; Bonanno, J.B.; Sauder, J.M.; Burley, S.K.; Almo, S.C.; Mehta, G.; MacMillan, A.M. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA 2012, 18, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H.; Haitjema, C.; Liu, X.; Ding, F.; Wang, H.; DeLisa, M.P.; Ke, A. Cas5d protein processes pre-CRRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure 2012, 20, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Kunin, V.; Sorek, R.; Hugenholtz, P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zheng, H.; Preamplume, G.; Shao, Y.; Li, H. The impact of CRISPR repeat sequence on structures of a Cas6 protein-RNA complex. Protein Sci. 2012, 21, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Structural principles of CRISPR RNA processing. Structure 2015, 23, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Brendel, J.; Stoll, B.; Lange, S.J.; Sharma, K.; Lenz, C.; Stachler, A.E.; Maier, L.K.; Richter, H.; Nickel, L.; Schmitz, R.A.; et al. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (CRISPR)-derived RNAs (crRNAs) in Haloferax volcanii. J. Biol. Chem. 2014, 289, 7164–7177. [Google Scholar] [CrossRef] [PubMed]
- Richter, H.; Zoephel, J.; Schermuly, J.; Maticzka, D.; Backofen, R.; Randau, L. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012, 40, 9887–9896. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.K.; Lange, S.J.; Stoll, B.; Haas, K.A.; Fischer, S.; Fischer, E.; Duchardt-Ferner, E.; Wohnert, J.; Backofen, R.; Marchfelder, A. Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B. RNA Biol. 2013, 10, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.K.; Stachler, A.E.; Saunders, S.J.; Backofen, R.; Marchfelder, A. An active immune defence with a minimal CRISPR (clustered regularly interspaced short palindromic repeats) RNA and without the Cas6 protein. J. Biol. Chem. 2014, 15. [Google Scholar] [CrossRef]
- Nickel, L.; Weidenbach, K.; Jäger, D.; Backofen, R.; Lange, S.J.; Heidrich, N.; Schmitz, R.A. Two CRISPR-Cas systems in Methanosarcina mazei strain Gö1 display common processing features despite belonging to different types I and III. RNA Biol. 2013, 10, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Scholz, I.; Lange, S.J.; Hein, S.; Hess, W.R.; Backofen, R. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PloS One 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Rouillon, C.; Kerou, M.; Reeks, J.; Brugger, K.; Graham, S.; Reimann, J.; Cannone, G.; Liu, H.; Albers, S.V.; et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell 2012, 45, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Kenchappa, C.S.; Peng, X.; She, Q.; Garrett, R.A. Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012, 40, 2470–2480. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.R.; Majumdar, S.; Elmore, J.; Pfister, N.; Compton, M.; Olson, S.; Resch, A.M.; Glover, C.V., III; Graveley, B.R.; Terns, R.M.; et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 2012, 45, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Delmas, S.; Shunburne, L.; Ngo, H.P.; Allers, T. Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination. PloS Genet. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Norais, C.; Hawkins, M.; Hartman, A.L.; Eisen, J.A.; Myllykallio, H.; Allers, T. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PloS Genet. 2007, 3. [Google Scholar] [CrossRef] [PubMed]
- Woods, W.G.; Dyall-Smith, M.L. Construction and analysis of a recombination-deficient (radA) mutant of Haloferax volcanii. Mol. Microbiol. 1997, 23, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Charlebois, R.L.; Lam, W.L.; Cline, S.W.; Doolittle, W.F. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc. Natl. Acad. Sci. USA 1987, 84, 8530–8534. [Google Scholar] [CrossRef] [PubMed]
- Künne, T.; Swarts, D.C.; Brouns, S.J. Planting the seed: Target recognition of short guide RNas. Trends Microbiol. 2014, 22, 74–83. [Google Scholar] [PubMed]
- Semenova, E.; Jore, M.M.; Datsenko, K.A.; Semenova, A.; Westra, E.R.; Wanner, B.; van der Oost, J.; Brouns, S.J.; Severinov, K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. USA 2011, 108, 10098–10103. [Google Scholar] [CrossRef] [PubMed]
- Wiedenheft, B.; van Duijn, E.; Bultema, J.B.; Waghmare, S.P.; Zhou, K.; Barendregt, A.; Westphal, W.; Heck, A.J.; Boekema, E.J.; Dickman, M.J.; et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 10092–10097. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.K.; Stoll, B.; Brendel, J.; Fischer, S.; Pfeiffer, F.; Dyall-Smith, M.; Marchfelder, A. The ring of confidence: A haloarchaeal CRISPR/Cas system. Biochem. Soc. Trans. 2013, 41, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Stoll, B.; Maier, L.K.; Lange, S.J.; Brendel, J.; Fischer, S.; Backofen, R.; Marchfelder, A. Requirements for a successful defence reaction by the CRISPR-Cas subtype I-B system. Biochem. Soc. Trans. 2013, 41, 1444–1448. [Google Scholar] [PubMed]
- Garcia-Heredia, I.; Martin-Cuadrado, A.B.; Mojica, F.J.; Santos, F.; Mira, A.; Anton, J.; Rodriguez-Valera, F. Reconstructing viral genomes from the environment using fosmid clones: The case of haloviruses. PloS One 2012, 7. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maier, L.-K.; Dyall-Smith, M.; Marchfelder, A. The Adaptive Immune System of Haloferax volcanii. Life 2015, 5, 521-537. https://doi.org/10.3390/life5010521
Maier L-K, Dyall-Smith M, Marchfelder A. The Adaptive Immune System of Haloferax volcanii. Life. 2015; 5(1):521-537. https://doi.org/10.3390/life5010521
Chicago/Turabian StyleMaier, Lisa-Katharina, Mike Dyall-Smith, and Anita Marchfelder. 2015. "The Adaptive Immune System of Haloferax volcanii" Life 5, no. 1: 521-537. https://doi.org/10.3390/life5010521