3.1. Identification of Plasmid pTC1
We previously reported the isolation and identification of
Sulfolobus tengchongensis strain RT8-4 from a hot spring in Tengchong in Southwestern China [
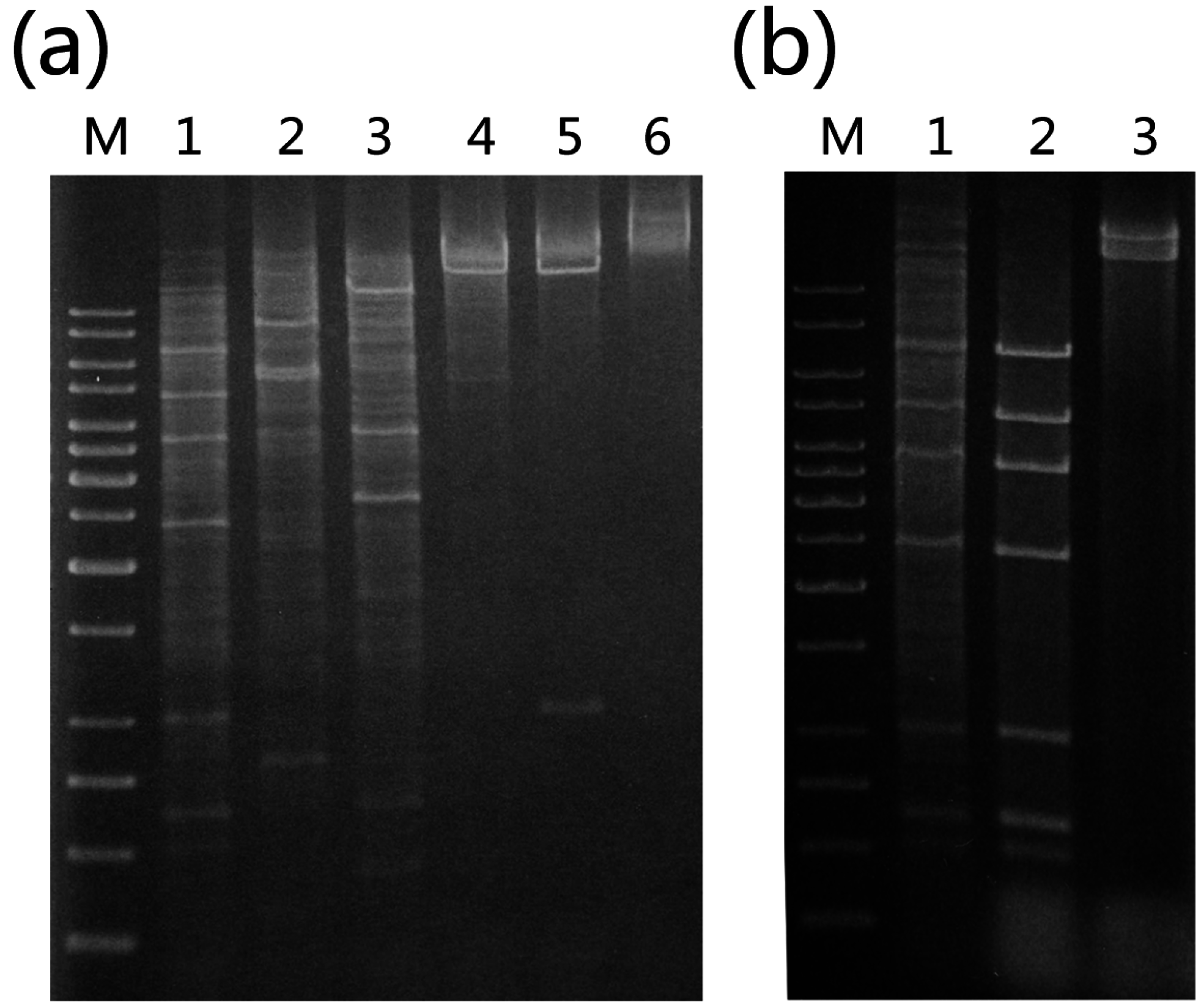
13]. In a subsequent study, we isolated total DNA from the strain and digested it with various restriction endonucleases. When the digests were subjected to agarose gel electrophoresis and ethidium bromide staining, a stoichiometric pattern of bands with intensity significantly higher than that of smearing bands derived from the genomic DNA of the host was observed, suggesting the presence of an extrachromasomal genetic element in the host cell (
Figure 1). This suggestion was supported by the finding that extraction of episomal DNA from
S. tengchongensis RT8-4 by using the alkaline lysis procedure yielded high molecular weight DNA species that migrated faster than the genomic DNA of the host. To determine whether the DNA was derived from a virus or a plasmid, we examined the culture supernatant of
S. tengchongensis RT8-4, and no virus-like particles were observed under an electron microscope. Based on these results, we conclude that a plasmid existed in
S. tengchongensis RT8-4, and designated this plasmid as pTC1.
pTC1 plasmid appeared to exist in high copy number in the original enrichment cultures, but was unstable to plating or sub-culturing. Upon repeated culture transfers, the copy number of pTC1 gradually decreased to levels undetectable even by Southern hybridization. A similar result was obtained for pTC2, the only other pTC plasmid obtained in a colony-purified strain, from a close relative of
S. tengchongenesis RT8-4 based on 16S rRNA sequence similarity. The conjugational transferability of pTC1 and pTC2 was tested from the appropriate donor strains using the assays described by Schleper
et al. [
1,
14] with a plasmid-free
S. tengchongenesis or
S. solfataricus strain as a recipient. Despite repeated efforts, however, no conjugation was detected. It is possible that the tested recipient strains were not appropriate for the assays. In addition, pTC plasmids were not integrated into the host genomes, as revealed by Southern hybridization.
Figure 1.
Identification of plasmid pTC1 from Sulfolobus tengchongensis RT8-4. (a) Restriction digestion of the total DNA from S. tengchongensis RT8-4 containing pTC1. The total DNA from the cells was digested with indicated restriction enzymes. Restriction fragments were subjected to electrophoresis in agarose gel. Lane M, 1-kb DNA ladder with sizes of 10, 8, 6, 5, 4, 3.5, 3, 2.5, 2, 1.5, 1, 0.75, 0.5, and 0.25 kb (from top to bottom); lane 1, EcoRI; lane 2, EcoRV; lane 3, PstI; lane 4, XhoI; lane 5 SalI; lane 6, total DNA; (b) Restriction digestion of pTC1 DNA. pTC1 DNA was extracted from the cells by alkaline lysis and purified using a plasmid purification kit. The DNA was digested with EcoRI. The restriction digest was subjected to electrophoresis in agarose gel. Lane M, 1-kb DNA ladder; lane 1, EcoRI digest of the total DNA from S. tengchongensis RT8-4 containing pTC1; lane 2, EcoRI digest of purified pTC1 DNA; lane 3, purified pTC1 DNA.
Figure 1.
Identification of plasmid pTC1 from Sulfolobus tengchongensis RT8-4. (a) Restriction digestion of the total DNA from S. tengchongensis RT8-4 containing pTC1. The total DNA from the cells was digested with indicated restriction enzymes. Restriction fragments were subjected to electrophoresis in agarose gel. Lane M, 1-kb DNA ladder with sizes of 10, 8, 6, 5, 4, 3.5, 3, 2.5, 2, 1.5, 1, 0.75, 0.5, and 0.25 kb (from top to bottom); lane 1, EcoRI; lane 2, EcoRV; lane 3, PstI; lane 4, XhoI; lane 5 SalI; lane 6, total DNA; (b) Restriction digestion of pTC1 DNA. pTC1 DNA was extracted from the cells by alkaline lysis and purified using a plasmid purification kit. The DNA was digested with EcoRI. The restriction digest was subjected to electrophoresis in agarose gel. Lane M, 1-kb DNA ladder; lane 1, EcoRI digest of the total DNA from S. tengchongensis RT8-4 containing pTC1; lane 2, EcoRI digest of purified pTC1 DNA; lane 3, purified pTC1 DNA.
3.2. Genomic Analysis of pTC1
Both strands of plasmid pTC1 were sequenced. The restriction patterns of the plasmid, predicted from the genomic sequence, agreed well with the results of restriction digestion. As revealed by sequencing, pTC1 is a circular double-stranded DNA molecule of 20,417 bp in length. The G+C content of pTC1 was 41.4%, which was higher than that of its host (34.4%) [
13] as well as those of other known
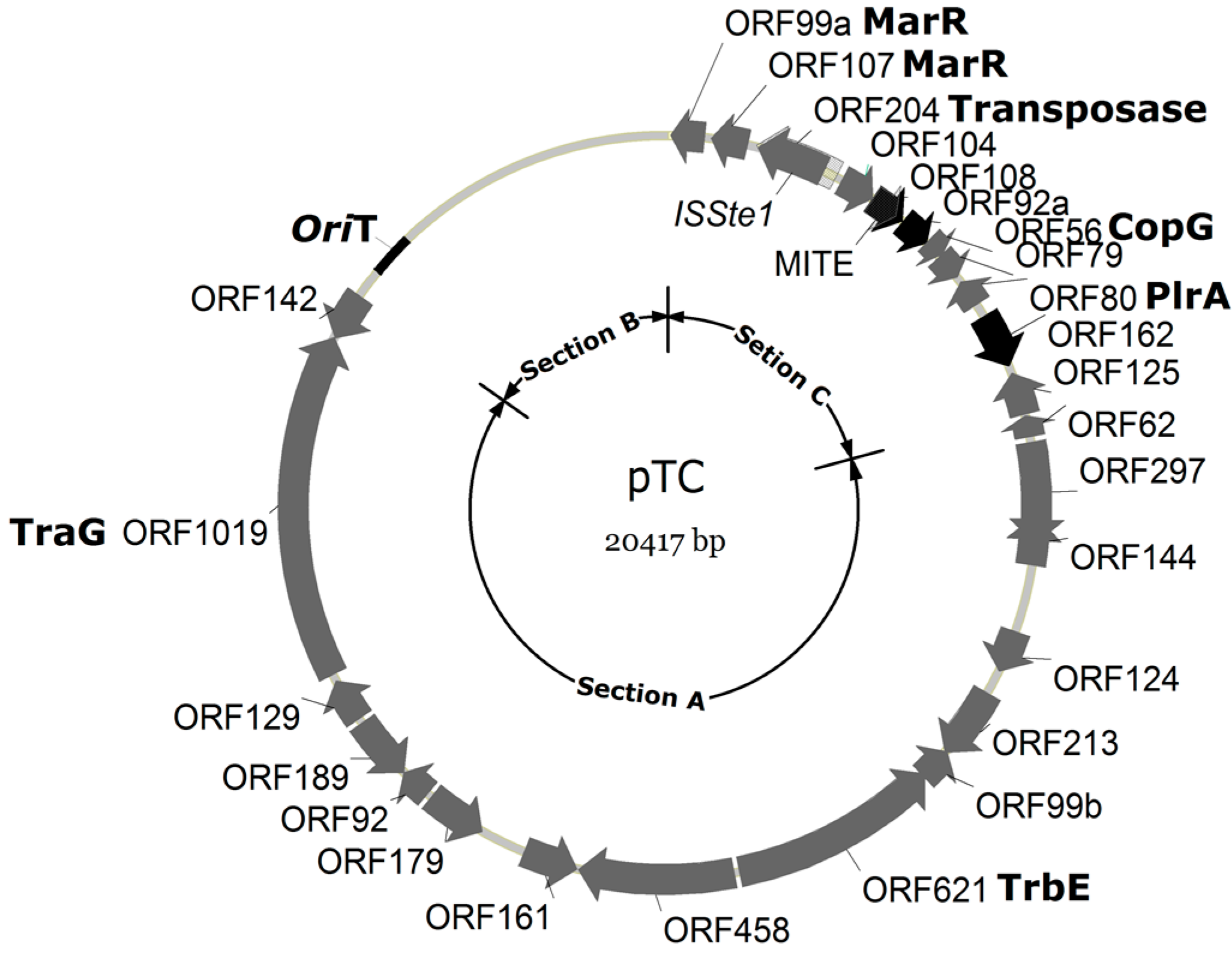
Sulfolobus CPs. The plasmid contains 26 open reading frames (ORFs) (
Figure 2;
Table 1). In most of the plasmid, genes are tightly packed or even overlapping. Among all known
Sulfolobus CPs, pTC1 is the smallest and carries fewest ORFs.
Figure 2.
A map of pTC1. ORFs are shown by arrows. ORFs with no homologues in the other known Sulfolobus CPs are indicated with blank arrows, whereas those with homologues in other CPs are shown with black arrows. Functional sections A, B and C are indicated.
Figure 2.
A map of pTC1. ORFs are shown by arrows. ORFs with no homologues in the other known Sulfolobus CPs are indicated with blank arrows, whereas those with homologues in other CPs are shown with black arrows. Functional sections A, B and C are indicated.
Table 1.
Genome annotation of pTC1.
Table 1.
Genome annotation of pTC1.
| Name | Predicted function | Conserved motifs | Homologues in other Sulfolobus CPs | Section |
|---|
| ORF62 | Transmembrane protein involved in conjugation | Signal peptide and transmembrane regions (1)1 | pAH1_p35, pARN3_p40, pARN4_p39, pHVE14_p53, pSOG1_p44, pSOG2_p41, pYN01_p2 | A |
| ORF297 | Signal peptide involved in conjugation | Signal peptide | pAH1_p1, pARN3_p1, pARN4_p1, pLD8501_p25, pYN01_p50, pHVE14_p3, pING1_p34, pKEF9_p3, pNOB8_p33, pSOG1_p5, pSOG2_p4, pMGB1_p1 | A |
| ORF144 | Unknown | - | pAH1_p1, pARN3_p1, pARN4_p1, pLD8501_p25, pYN01_p50, pMGB1_p1 | A |
| ORF124 | Unknown | - | pAH1_p1, pARN3_p1, pARN4_p1, pLD8501_p25, pYN01_p50, pMGB1_p1 | A |
| ORF213 | The transcription factor TFIIB | TF_Zn_Ribbon superfamily | pAH1_p2, pARN3_p2, pARN4_p2, pLD8501_p24, pYN01_p49, pSOG1_p45 | A |
| ORF99c | Unknown | - | pAH1_p3, pARN3_p3, pARN4_p3, pLD8501_p23, pYN01_p48, pMGB1_p3 | A |
| ORF621 | TrbE-like protein involved in conjugation transfer | AAA_10 | pAH1_p4, pARN3_p4, pARN4_p4, pHVE14_p1, pING1_p32, pKEF9_p1, pLD8501_p22, pNOB8_p31, pSOG1_p2, pSOG2_p2, pYN01_p47, pMGB1_p4 | A |
| ORF458 | Transmembrane protein involved in conjugation | Signal peptide and transmembrane regions (9) | pAH1_p5, pARN3_p5, pARN4_p5, pLD8501_p6, pSOG1_p7, pYN01_p36, pMGB1_p5 | A |
| ORF161 | Transmembrane protein involved in conjugation | Transmembrane regions (2) | pAH1_p6, pARN3_p6, pARN4_p6, pYN01_p35 | A |
| ORF179 | Transmembrane protein involved in conjugation | Transmembrane regions (3) | pAH1_p9, pARN3_p7, pARN4_p8, pLD8501_p1, pYN01_p33 | A |
| ORF92 | Transmembrane protein involved in conjugation | Transmembrane regions (2) | pAH1_p10, pARN3_p8, pARN4_p9, pYN01, pMGB1_p8 | A |
| ORF189 | Transmembrane protein involved in conjugation | Signal peptide and transmembrane regions (1) | pAH1_p11, pARN3_p9, pARN4_p10, pSOG1_p4, pYN01_p31, pMGB1_p9 | A |
| ORF129 | Transmembrane protein involved in conjugation | Signal peptide and transmembrane regions (2) | pAH1_p12, pARN3_p10, pARN4_p11, pYN01_p30, pMGB1_p10 | A |
| ORF1019 | VirD/TraG-like protein | TraG_VirD4/AAA_10 | pAH1_p13, pARN3_p11, pARN4_p12, pHVE14_p14, pING1_p5, pKEF9_p12, pNOB8_p10, pSOG1_p12, pSOG1_p2, pSOG2_p12, pYN01_p29, pMGB1_p11 | A |
| ORF142 | Unknown | - | pAH1_p14, pARN3_p12, pARN4_p13, pING1_p6, pKEF9_p13, pSOG1_p13, pSOG2_p13 | A |
| ORF99a | Predicted transcriptional regulators | HTH_ARSR superfamily | pING_p18, pNOB8_p18, pNOB8_p44, pYN01_p41, pMGB1_p20 | C |
| ORF107 | Predicted transcriptional regulators | HTH_MARR | pAH1_p22, pKEF9_p24, pNOB8_p18, pNOB8_p2, pNOB8_p19, pYN01_p42, pLD8501_p16, pMGB1_p20, pMGB1_21 | C |
| ORF204 | Transposase | COG3316 | pARN4_p16 | C |
| ORF104 | Unknown | - | pARN3_p27, pARN4_p25, pING1_p19, pNOB8_p20, pYN01_p17, pHVE14_p30, pKEF9_p25, pSOG1_p23, pSOG2_p26 | C |
| ORF108 | Unknown | - | - | C |
| ORF92a | Unknown | - | - | C |
| ORF79 | Unknown | - | pSOG1_p24, pSOG2_p27, pMGB1_p7 | C |
| ORF80 | PlrA-like protein | Sulfolobus_pRN | pING1_p26, pARN4_p33, pNOB8_p28, pKEF9_p33, pSOG1_p34, pLD8501_p29, pHVE14_p41, pAH1_p33, pYN01_p6 | C |
| ORF56 | CopG family transcriptional regulator | PHA01748/RHH_1 superfamily | pAH1_p28, pKEF9_p28, pYN01_p11, pNOB8_24, pMGB1_p26 | C |
| ORF162 | Unknown | - | | C |
| ORF125 | Unknown | - | pARN3_p38, pARN4_p37, pKEF9_p38, pHVE14_p50, pSOG1_p42, pSOG2_p39, pYN01_p4 | C |
To determine the similarity of pTC1 ORFs to homologous ORFs from other CPs, we assembled a database consisting of ORFs from 12 other known Sulfolobus CPs (i.e., pNOB8, pING1, pARN3, pARN4, pAH1, pSOG1, pSOG2, pLD8501, pYN01, pKEF9, pHVE14 and pMGB1). As revealed by BLASTP with an E-value cutoff of <10−3, only three ORFs from pTC1 (i.e., ORF108, ORF92a and ORF162) have no homologues in the other twelve Sulfolobus CPs. All the three ORFs are located in section C of the plasmid genome (see below). The rest of the pTC1 ORFs share 22.50%~69.99% similarity at the amino acid sequence level with their respective best matches in the other 12 CPs.
Genomic comparison shows that pTC1 resembles known
Sulfolobus CPs in architecture [
6,
9]. Like all other
Sulfolobus CPs, pTC1 comprises three conserved and functionally distinct genomic sections (
Figure 2 and
Table 1). Section A of pTC1 accounts for about half of the plasmid (~12.5 kb) and contains 15 ORFs that are conserved among
Sulfolobus CPs. All section A proteins, except for those encoded by ORF144, 124, 213, 142 and 99b, are predicted to possess transmembrane helix motifs, and are believed to be involved in plasmid conjugation. These include two highly conserved
Sulfolobus CP proteins that are likely homologues of the bacterial conjugative ATPase proteins VirD/TraG (ORF621) and TrbE (ORF1019). Bacterial TraG is the coupling protein that connects the membrane-spanning mating pair formation (Mpf) complex with the cytoplasmic nucleoprotein relaxosome complex at the cytoplasmic membrane during conjugative transfer of DNA [
15]. However, it is worth noting that no genes encoding relaxase, a bacterial homologue which is involved in the mobilization of the conjugational plasmid transfer [
9], were found in the pTC1 genome. Also unusual was the observation that ORFs probably involved in transcriptional regulation (
i.e., ORF144, 124, 213 and 99b) interrupted the otherwise contiguous cluster of ORFs encoding transmembrane proteins. The contiguous array of these proteins is highly conserved among other
Sulfolobus CPs.
Section B contains multiple sequence repeats and no ORFs. Based on Z-curve and GC-skew analyses, the origin of replication for pTC1 is located in a 209-bp stretch, downstream of ORF142, in a large intergenic region (
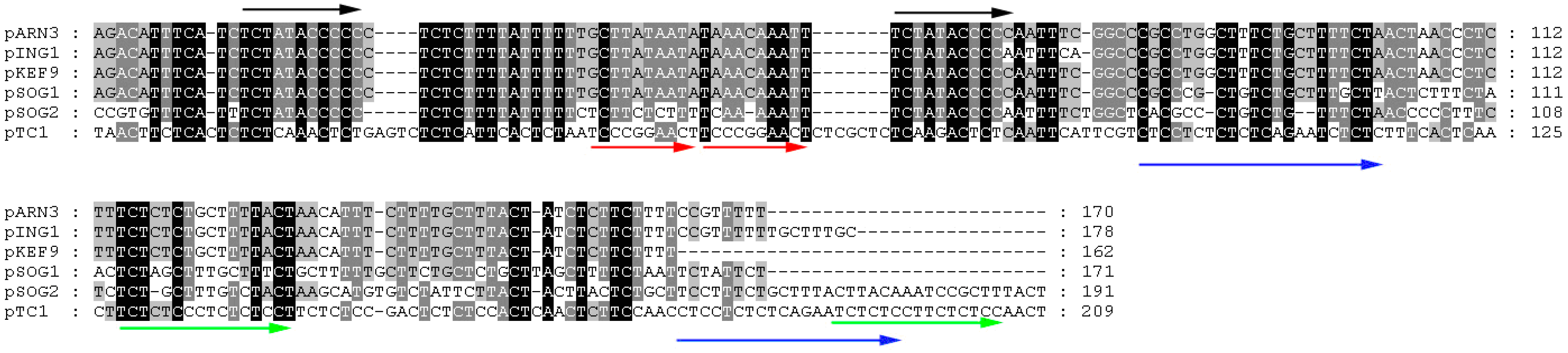
Figure 2). The 209-bp region shares limited sequence similarity to the replication origin of pSOG2, pKEF9, pING1, pARN3, and pSOG1 (
Figure 3). However, the conserved sequence TCTATACCCCC, found in the putative origin of the other CPs [
9], is not present in pTC1 (
Figure 3). On the other hand, there are two perfect direct repeats (TCCCCGGAACT) and three imperfect direct repeats (TCTCTCCNNCT) in this region as well as four imperfect repeats (AGCAGCGCTTGYCCT) 529–959 bp downstream of this region. The presence of these direct and inverted repeating sequences supports the possibility that pTC1 initiates replication in this region [
16].
Figure 3.
Alignment of the putative origins of replication of pARN3, pING1, pKEF9, pSOG1, pSOG2 and pTC1. Repeating sequences are indicated by arrows. Identical or similar sequences are shown in dark or grey background, respectively.
Figure 3.
Alignment of the putative origins of replication of pARN3, pING1, pKEF9, pSOG1, pSOG2 and pTC1. Repeating sequences are indicated by arrows. Identical or similar sequences are shown in dark or grey background, respectively.
Section C, which includes a tightly packed cluster of six to nine genes, is conserved among previously characterized
Sulfolobus CPs and has been implicated in the initiation and regulation of plasmid DNA replication [
7,
9]. These genes encode hypothetical relaxase (e.g., some pING ORFs), putative RepA (pING1_ORF99, pKEF9_ORF106), …, various types of transcriptional regulators, such as CopG (pAH1_p28, pKEF9_p28, pYN01_p11, pNOB8_24, pMGB1_p26), MarR-like protein(pAH1_p22, pKEF9_p24,
etc.), Zinc finger protein (pAH1_p36),
etc., as well as 1–4 hypothetical proteins. Surprisingly, section C in pTC1 is not well conserved as compared to the corresponding region in other
Sulfolobus CPs. Most of the CPs encode a putative RepA protein. However, no homologue of a gene encoding putative RepA was found in pTC1. A
plrA gene was identified in pTC1. PlrA, a highly conserved transcriptional regulator in
Sulfolobus plasmids, is speculated to be involved in plasmid segregation [
9]. All previously described CPs contain an ORF for a pNOB8-type integrase of the tyrosine recombinase superfamily. However, no homologue of the integrase gene was found in pTC1. Instead, an insertion element and a miniature inverted-repeat transposable element (MITE) were detected in section C (see below).
The insertion sequence (IS), denoted
ISSte1 [
17], is 762 bp in length and located in the intergenic region between ORF107 and ORF104 (
Figure 4a). It contains a transposase (Tpase) gene (ORF204) flanked by 16-bp perfect inverted repeats (IR: GTGTGTGTCCAACAAT). Notably, there is a pair of perfect inverted repeats (IR': GGGTCAGGACGGG) between the left IR and ORF204. The insertion of
ISSte1 has led to the duplication of the target site of 8 bp (DR: ATCACAAA). Sequence analysis shows that ORF204-encoded Tpase contains the conserved DDE domain (
Figure S1) [
17]. This, together with the size and sequence of the IRs, places
ISSte1 in the
IS6 family [
17].
Figure 4.
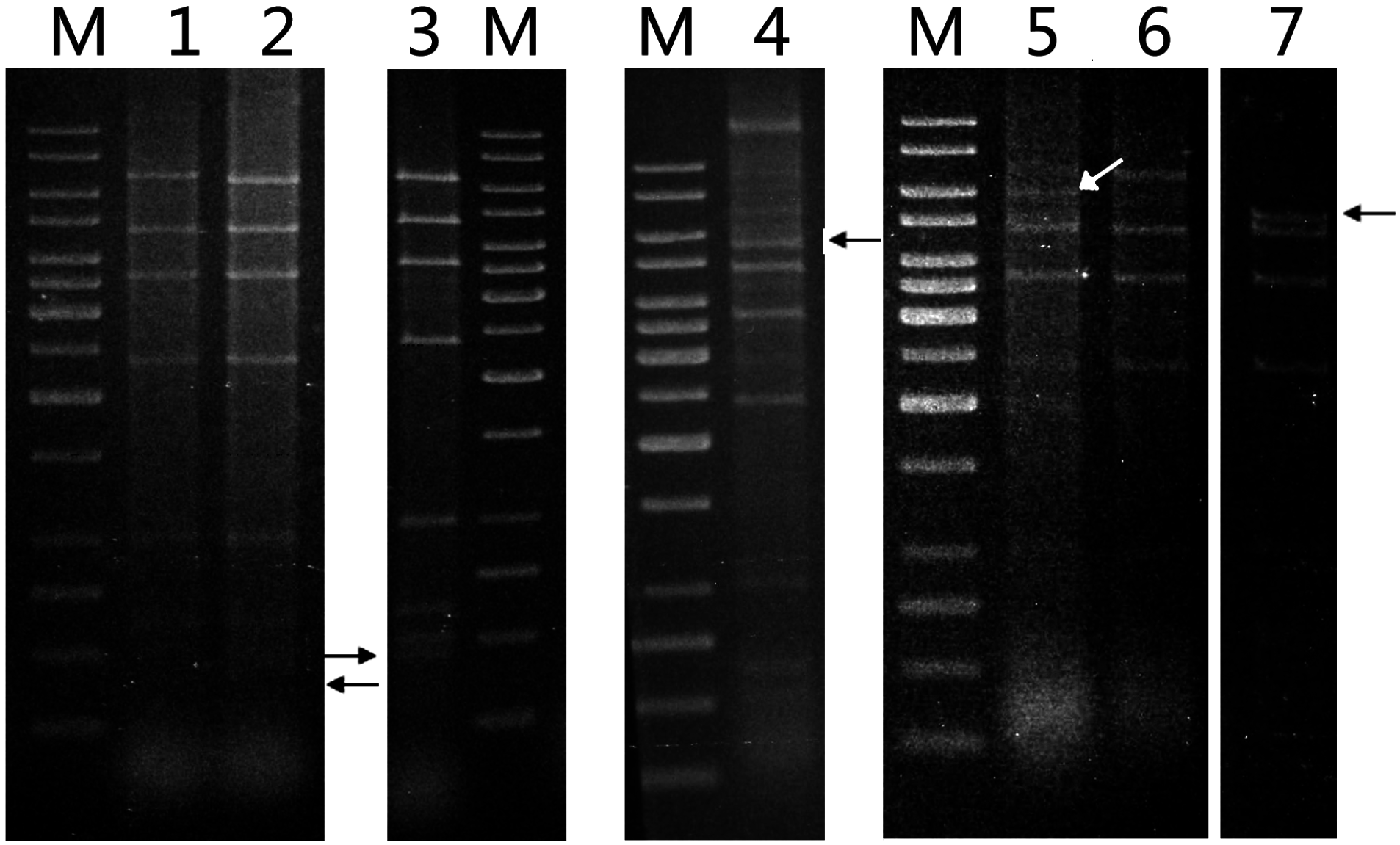
Patterns of deletion at the sites of transposition by ISSte1 (a) and MITE (b). Total DNAs or plasmid DNAs were isolated from enrichment cultures established with various hot spring and mud hole samples. PCR targeting the ISSte1 site or the MITE site was carried out. Fragments shorter than expected for the insertion of full-length ISStel or MITE into the pTC1 DNA were sequenced. Repeating sequences are labeled in color and specified. ORFs are shown with blank arrows. An additional deletion (AGGGGCTCT) occurred at 88 bp upstream of LDR in type 2 deletion variants, whereas an additional deletion (AGAGACCGAGAATGATA) was 86 bp downstream of RDR in types 3 and 8 deletion variants. The percentage of each deletion type for ISStel in all samples is indicated.
Figure 4.
Patterns of deletion at the sites of transposition by ISSte1 (a) and MITE (b). Total DNAs or plasmid DNAs were isolated from enrichment cultures established with various hot spring and mud hole samples. PCR targeting the ISSte1 site or the MITE site was carried out. Fragments shorter than expected for the insertion of full-length ISStel or MITE into the pTC1 DNA were sequenced. Repeating sequences are labeled in color and specified. ORFs are shown with blank arrows. An additional deletion (AGGGGCTCT) occurred at 88 bp upstream of LDR in type 2 deletion variants, whereas an additional deletion (AGAGACCGAGAATGATA) was 86 bp downstream of RDR in types 3 and 8 deletion variants. The percentage of each deletion type for ISStel in all samples is indicated.
MITEs are mobile genetic elements derived from ISs with DDE Tpases, and contain flanking terminal IRs but no internal Tpase gene [
17,
18,
19]. The MITE sequence identified in pTC1 is only 142 bp long (
Figure 4b). Intriguingly, the entire element resides in ORF108. It would be of interest to determine if ORF108 is functional. If the ORF encodes a functional protein, the presence or absence of the MITE at the site of transposition might be of significance to the biology of the plasmid. It is clear, however, that ORF108 is not essential for the plasmid since it is absent from all other known
Sulfolobus CPs. The MITE of pTC1 carries long imperfect IRs (LIR: AATCGAGAACGGTGTCAT, RIR: ATGAGACATTCCTCGTT) at the two ends and is bounded by long imperfect DRs (LDR: AAGTGAGGTGATGTAAATGGTCAC, RDR: CAGTATATCATGTACTGGTAC. The intervening sequence between the two flanking IRs shares no significant similarity with known sequences in the public database, indicating that the element is a type II MITE. Since both IRs and DRs of the MITE are not well conserved, it appears that the element has lost the mobility due to mutation.
3.3. pTC Plasmids in the Geothermal Area of Tengchong
To determine the distribution of pTC1 and its variants in acidic hot springs and mud holes in Tengchong, we collected 35 samples at various spots in a general area of ~0.1 km
2. Enrichment cultures from 27 of the samples were successfully established in Zillig’s medium supplemented with 0.2% tryptone [
3]. Plasmid DNAs were isolated from 11 of the enrichment cultures by using the alkaline lysis method. Restriction analysis showed that the
EcoRI cleavage patterns of seven of the plasmids resembled that of pTC1, whereas those of the other four were also indistinguishable from that of pTC1 except for one fragment (
Figure 5). An additional fragment of ~400 bp appeared in one of the pTC variants, whereas the 6.8-kb
EcoRI fragment, which harbored
ISStel, was shortened to 6.0, 5.5 and 5.2 kb, respectively, in the other three variants. The ~400-bp
EcoRI fragment of the former pTC variant, denoted pTC2, was sequenced. As shown in
Figure 4 and
Figure S2b in supplementary, pTC2 was 163 bp shorter than pTC1 in that fragment, and the MITE found in pTC1 was absent from pTC2 except for a single DR of the element. Therefore, pTC1 appears to have resulted from the insertion of the MITE in pTC2. These results suggest that pTC plasmids were widely distributed in the geothermal area of Tengchong, and pTC1 was likely a predominant species of these plasmids.
The presence of the insertion sequences in pTC1 prompted us to look for naturally-occurring variants of this plasmid. PCR reactions were carried out on total DNA or plasmid DNA samples isolated from each enrichment culture using a pair of primers targeting sequences upstream and downstream of
ISStel. The primers were designed based on the sequences inside the upstream ORF104 and downstream ORF107 to ensure specific amplification of the sequence from pTC plasmids. Because of the absence of the integration of pTC into the host genome, it would be most likely that the specific PCR products were derived from the sequences of free pTC plasmids. A total of 90 PCR fragments shorter than that expected for the transposition by a full-length
ISStel were obtained and subsequently sequenced. These sequences fall into nine categories (
Figure 4 and
Figure S2a in supplementary). Only a small fraction (14.4%) of the fragments contained the unaltered plasmid sequence, suggesting that either no transposition by
ISStel had occurred at the site or an inserted
ISStel had undergone precise excision, leaving behind a single copy of the target sequence (
i.e., DR). In any case, the target site appears to be efficiently used for
ISStel transposition. No perfect reversion of the transposition by
ISStel to the target site was observed. In the remainder of the fragments, transposition of
ISStel to the target site and subsequent excision of the inserted IS in an imprecise manner were detected. A large deletion spanning the entire
ISStel as well as both DRs and a 14-bp 3'-flanking sequence was found in 70.1% of these fragments. Some of them carried a small additional deletion either upstream (17.4%) or downstream (12.7%) of the target site for
ISStel transposition. The rest of these fragments (29.8%) had smaller deletions, which started 8–65 bp downstream of LIR and mostly ended 5–19 bp downstream of RDR. So all of them retained LDR and LIR, but lost the Tpase gene. Taken together, these observations suggest that deletion of
ISStel proceeded more frequently in an imprecise manner than in a precise manner in natural environments.
Figure 5.
Restriction patterns of pTC plasmids. Plasmid DNAs were extracted from various isolates by alkaline lysis and purified using a plasmid purification kit. The DNAs were digested with EcoRI. The restriction digests were subjected to electrophoresis in agarose gel. Lane M, 1-kb DNA ladder with sizes of 10, 8, 6, 5, 4, 3.5, 3, 2.5, 2, 1.5, 1, 0.75, 0.5, and 0.25 kb (from top to bottom); lane 1, pTC1 DNA from S. tengchongensis RT8-4; lane 2, plasmid DNA from isolate H7; lane 3, plasmid DNA from isolate H7; lane 4, plasmid DNA from isolate H3; lane 5, plasmid DNA from isolate D2; lane 6, plasmid DNA from isolate D4; lane 7, plasmid DNA from isolate H5. Restriction fragments containing a deletion are indicated by arrows.
Figure 5.
Restriction patterns of pTC plasmids. Plasmid DNAs were extracted from various isolates by alkaline lysis and purified using a plasmid purification kit. The DNAs were digested with EcoRI. The restriction digests were subjected to electrophoresis in agarose gel. Lane M, 1-kb DNA ladder with sizes of 10, 8, 6, 5, 4, 3.5, 3, 2.5, 2, 1.5, 1, 0.75, 0.5, and 0.25 kb (from top to bottom); lane 1, pTC1 DNA from S. tengchongensis RT8-4; lane 2, plasmid DNA from isolate H7; lane 3, plasmid DNA from isolate H7; lane 4, plasmid DNA from isolate H3; lane 5, plasmid DNA from isolate D2; lane 6, plasmid DNA from isolate D4; lane 7, plasmid DNA from isolate H5. Restriction fragments containing a deletion are indicated by arrows.
A diverse array of pTC variants associated with the transposition of MITE and subsequent deletion of the inserted element was also observed. As shown in
Figure 4 and
Figure S2b in supplementary, the MITE sequence was either entirely absent or deleted in different fashions. The sequence lacking the entire MITE may have yet to undergo transposition by the MITE. It is also possible that the sequence resulted from the precise deletion of the inserted MITE form a pTC1-like plasmid. Since the LIR of the pTC1 MITE is identical to the target sequence, the divergence between the two inverted repeats suggests that the MITE in pTC1, acquired through transposition, has probably become immobilized through mutation of the RIR. Imprecise deletion of the MITE apparently occurred as observed with
ISStel.