Protocells: At the Interface of Life and Non-Life
Abstract
:1. Why Is Life Based on the Cellular Form?
2. Protocells: The Primitive Cellular Form
3. Pseudo-Protocells and True-Protocells: About the Membrane Synthesis
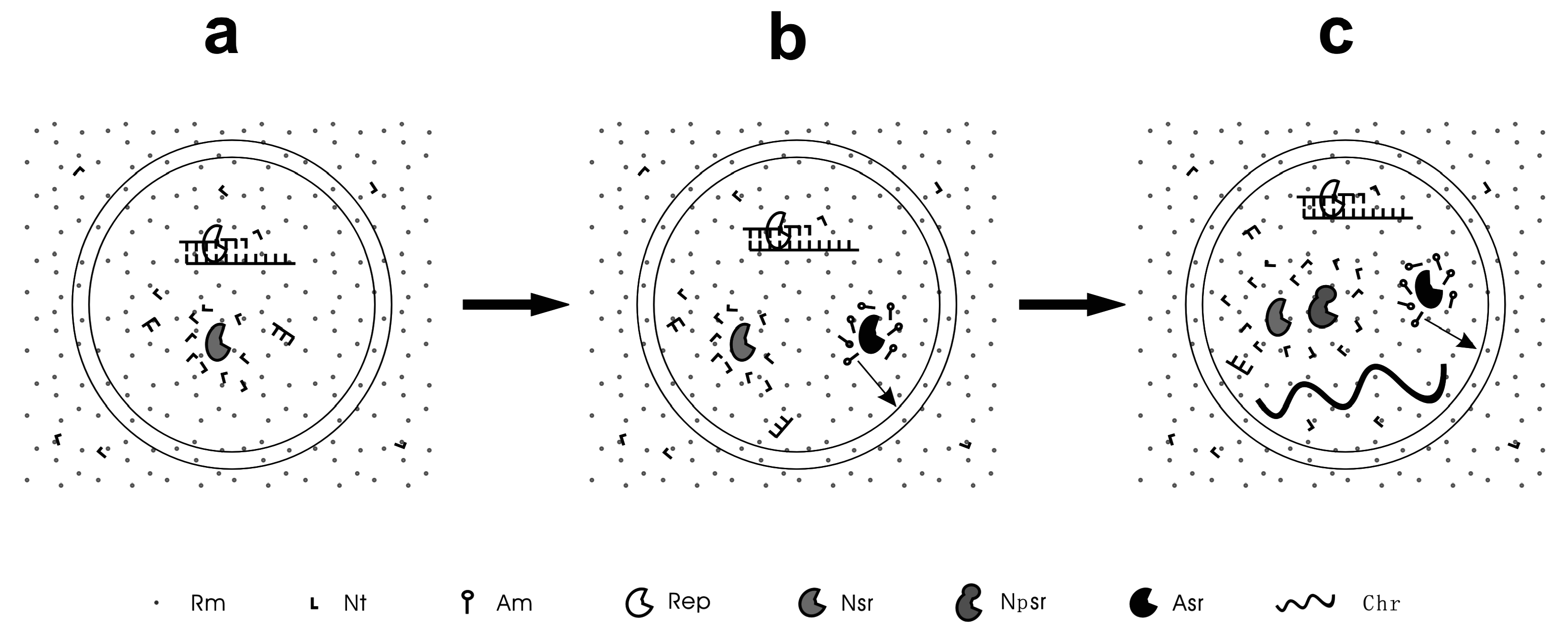
4. Unitary-Protocells: Linked Genes and the Starting Point of Real Life
5. Understanding Life by Synthesizing Protocells
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Luisi, P.L. About Various Definitions of Life. Orig. Life Evol. Biosph. 1998, 28, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Benner, S.A. Defining Life. Astrobiology 2010, 10, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Trifonov, E.N. Vocabulary of Definitions of Life Suggests a Definition. J. Biomol. Struct. Dyn. 2011, 29, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Kruger, K.; Grabowski, P.E.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-Splicing RNA: Autoexcision and Autocyclization of Ribosomal RNA Intervening Sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA Moiety of Ribonuclease P Is the Catalytic Subunit of the Enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W. The RNA World. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Joyce, G.F. The Antiquity of RNA-Based Evolution. Nature 2002, 418, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R. RNA Chemistry—Ribozyme Self-Replication. Nature 1989, 339, 507–508. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F.; Orgel, L.E. Prospects for Understanding the Origin of the RNA World. In The RNA World; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1999; Chapter 2; pp. 49–77. [Google Scholar]
- Robertson, M.P.; Joyce, G.F. The Origins of the RNA World. Cold Spring Harb. Perspect. Biol. 2012, 4, a003608. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.T.; Yu, C.W. Intramolecular RNA Replicase: Possibly the First Self-Replicating Molecule in the RNA World. Orig. Life Evol. Biosph. 2006, 36, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, O.L.; Orgel, L.E. Template-directed oligonucleotide ligation on hydroxylapatite. Nature 1986, 321, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Franchi, M.; Gallori, E. A Surface-Mediated Origin of the RNA World: Biogenic Activities of Clay-Adsorbed RNA Molecules. Gene 2005, 346, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.P. Montmorillonite Catalysis of 30–50 Mer Oligonucleotides: Laboratory Demonstration of Potential Steps in the Origin of the RNA World. Orig. Life Evol. Biosph. 2002, 32, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.P.; Hill, A.R.; Liu, R.; Orgel, L.E. Synthesis of Long Prebiotic Oligomers on Mineral Surfaces. Nature 1996, 381, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Ertem, G. Montmorillonite, Oligonucleotides, RNA and Origin of Life. Orig. Life Evol. Biosph. 2004, 34, 549–570. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Johnston, B.H.; Landweber, L.F.; Kazakov, S.A. Ligation Activity of Fragmented Ribozymes in Frozen Solution: Implications for the RNA World. Nucleic Acids Res. 2004, 32, 2966–2974. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Kazakov, S.A.; Johnston, B.H.; Landweber, L.F. The RNA World on Ice: A New Scenario for the Emergence of RNA Information. J. Mol. Evol. 2005, 61, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Trinks, H.; Schroder, W.; Biebricher, C.K. Ice and the Origin of Life. Orig. Life Evol. Biosph. 2005, 35, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Monnard, P.A.; Szostak, J.W. Metal-Ion Catalyzed Polymerization in the Eutectic Phase in Water-Ice: A Possible Approach to Template-Directed RNA Polymerization. J. Inorg. Biochem. 2008, 102, 1104–1111. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, S.A.; Balatskaya, S.V.; Johnston, B.H. Ligation of the Hairpin Ribozyme in Cis Induced by Freezing and Dehydration. RNA 2006, 12, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Monnard, P.A.; Ziock, H. Eutectic Phase in Water-Ice: A Self-Assembled Environment Conducive to Metal-Catalyzed Non-Enzymatic RNA Polymerization. Chem. Biodiver. 2008, 5, 1521–1539. [Google Scholar] [CrossRef]
- Attwater, J.; Wochner, A.; Pinheiro, V.B.; Coulson, A.; Holliger, P. Ice As a Protocellular Medium for RNA Replication. Nature Comm. 2010, 1. [Google Scholar] [CrossRef]
- Russell, M.J.; Hall, A.J.; Cairns-Smith, A.G.; Braterman, P.S. Submarine Hot Spring and Origin of Life. Nature 1988, 336, 117. [Google Scholar] [CrossRef]
- Koonin, E.V.; Martin, W. On the Origin of Genomes and Cells within Inorganic Compartments. Trends Genet. 2005, 21, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.; Baross, J.; Kelley, D.; Russell, M.J. Hydrothermal Vents and the Origin of Life. Nat. Rev. Microbiol. 2008, 6, 805–814. [Google Scholar] [PubMed]
- Morowitz, H. Beginnings of Cellular Life; Yale University Press: New Haven, CT, USA; London, UK, 1992. [Google Scholar]
- Bachmann, P.A.; Luisi, P.L.; Lang, J. Autocatalytic Self-Replicating Micelles as Models for Prebiotic Structures. Nature 1992, 357, 57–59. [Google Scholar] [CrossRef]
- Chen, I.A.; Hanczyc, M.M.; Sazani, P.L.; Szostak, J.W. Protocells: Genetic Polymers inside Membrane Vesicles. In The RNA World; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2006; pp. 57–88. [Google Scholar]
- Chen, I.A.; Walde, P. From Self-Assembled Vesicles to Protocells. Cold Spring Harb. Perspect. Biol. 2010, 2, a002170. [Google Scholar] [CrossRef] [PubMed]
- Hanczyc, M.M.; Fujikawa, S.M.; Szostak, J.W. Experimental Models of Primitive Cellular Compartments: Encapsulation, Growth, and Division. Science 2003, 302, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Hanczyc, M.M.; Mansy, S.S.; Szostak, J.W. Mineral Surface Directed Membrane Assembly. Orig. Life Evol. Biosph. 2007, 37, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Shenhav, B.; Bar-Even, A.; Kafri, R.; Lancet, D. Polymer GARD: Computer Simulation of Covalent Bond Formation in Reproducing Molecular Assemblies. Orig. Life Evol. Biosph. 2005, 35, 111–133. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.M.; Gocayne, J.D.; White, O.; Adams, M.D.; Clayton, R.A.; Fleischmann, R.D.; Bult, C.J.; Kerlavage, A.R.; Sutton, G.; Kelley, J.M.; et al. The Minimal Gene Complement of Mycoplasma Genitalium. Science 1995, 270, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Mushegian, A.R.; Koonin, E.V. A Minimal Gene Set for Cellular Life Derived by Comparison of Complete Bacterial Genomes. Proc. Natl. Acad. Sci. USA 1996, 93, 10268–10273. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. How Many Genes can Make a Cell: The Minimal-gene-set Concept. Annu. Rev. Genomics Hum. Genet. 2000, 1, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Gil, R.; Silva, F.J.; Pereto, J.; Moya, A. Determination of the Core of a Minimal Bacterial Gene Set. Microbiol. Mol. Biol. Rev. 2004, 68, 518–537. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Madrigal, S.; Latorre, A.; Porcar, M.; Moya, A.; Gil, R. Complete Genome Sequence of “Candidatus Tremblaya Princeps” Strain PCVAL, an Intriguing Translational Machine below the Living-Cell Status. J. Bacteriol. 2011, 193, 5587–5588. [Google Scholar] [CrossRef] [PubMed]
- Bennett, G.M.; Moran, N.A. Small, Smaller, Smallest: The Origins and Evolution of Ancient Dual Symbioses in a Phloem-Feeding Insect. Genome Biol. Evol. 2013, 5, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Szathmary, E.; Santos, M.; Fernando, C. Evolutionary Potential and Requirements for Minimal Protocells. Top. Curr. Chem. 2005, 259, 167–211. [Google Scholar]
- Mansy, S.S. Membrane Transport in Primitive Cells. Cold Spring Harb. Perspect. Biol. 2010, 2, a002188. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, H.S. The RNA World Hypothesis: The Worst Theory of the Early Evolution of Life (Except for All the Others). Biol. Direct 2012, 7. [Google Scholar] [CrossRef]
- Szathmary, E. Life—In Search of the Simplest Cell. Nature 2005, 433, 469–470. [Google Scholar] [CrossRef] [PubMed]
- Szostak, J.W.; Bartel, D.P.; Luisi, P.L. Synthesizing Life. Nature 2001, 409, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Schrum, J.P.; Zhu, T.F.; Szostak, J.W. The Origins of Cellular Life. Cold Spring Harb. Perspect. Biol. 2010, 2, a002212. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Re-Creating an RNA Replicase. In The RNA World; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1999; Chapter 5; pp. 143–162. [Google Scholar]
- McGinness, K.E.; Joyce, G.F. In Search of an RNA Replicase Ribozyme. Chem. Biol. 2003, 10, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.K.L.; Unrau, P.J. Closing the Circle: Replicating RNA with RNA. Cold Spring Harb. Perspect. Biol. 2010, 2, a002204. [Google Scholar] [CrossRef] [PubMed]
- Wochner, A.; Attwater, J.; Coulson, A.; Holliger, P. Ribozyme-Catalyzed Transcription of an Active Ribozyme. Science 2011, 332, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Attwater, J.; Wochner, A.; Holliger, P. In-Ice Evolution of RNA Polymerase Ribozyme Activity. Nat. Chem. 2013, 5, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Murtas, G. Early Self-Reproduction, the Emergence of Division Mechanisms in Protocells. Mol. Biosyst. 2013, 9, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.A.; Roberts, R.W.; Szostak, J.W. The Emergence of Competition between Model Protocells. Science 2004, 305, 1474–1476. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.T.; Hu, J.M. Computer Simulation on the Cooperation of Functional Molecules during the Early Stages of Evolution. PLoS One 2012, 7, e35454. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.T.; Yu, C.W.; Zhang, W.T.; Zhou, P.; Hu, J.M. The Emergence of Ribozymes Synthesizing Membrane Components in RNA-Based Protocells. Biosystems 2010, 99, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Szathmary, E. The Origin of Replicators and Reproducers. Philos. Trans. R. Soc. B Biol. Sci. 2006, 361, 1761–1776. [Google Scholar] [CrossRef]
- Maynard-Smith, J.; Szathmary, E. The Origin of Chromosomes I. Selection for Linkage. J. Theor. Biol. 1993, 164, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.T.; Yu, C.W.; Zhang, W.T. Circularity and Self-cleavage as a Strategy for the Emergence of a Chromosome in the RNA-based Protocell. Biol. Direct 2013, 8. [Google Scholar] [CrossRef]
- Diener, T.O. Circular RNAs: Relics of Precellular Evolution? Proc. Natl. Acad. Sci. USA. 1989, 86, 9370–9374. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, C.J.; Rathjen, P.D.; Forster, A.C.; Symons, R.H. Self-Cleavage of Plus and Minus RNA Transcripts of Avocado Sunblotch Viroid. Nucleic Acids Res. 1986, 14, 3627–3640. [Google Scholar] [CrossRef] [PubMed]
- Duran-Vila, N.; Elena, S.F.; Daros, J.A.; Flores, R. Structure and Evolution of Viroids. In Origin and Evolution of Viruses, 2nd ed.; Domingo, E., Parish, C.R., Holland, J.J., Eds.; Elsevier: Oxford, UK, 2008; Chapter 2; pp. 43–64. [Google Scholar]
- Ma, W.T. The Origin of Life: A Problem of History, Chemistry and Evolution. Chem. Biodiver. 2014, 11, 1998–2010. [Google Scholar] [CrossRef]
- Ruiz-Mirazo, K.; Pereto, J.; Moreno, A. A Universal Definition of Life: Autonomy and Open-Ended Evolution. Orig. Life Evol. Biosph. 2004, 34, 323–346. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H.C. A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Berry, A. DNA: The Secret of Life; The Random House Group Limited: London, UK, 2003. [Google Scholar]
- Schrodinger, E. What Is Life; Cambridge University Press: Cambridge, UK, 1944. [Google Scholar]
- Deamer, D. Special Collection of Essays: What Is Life?—Introduction. Astrobiology 2010, 10, 1001–1002. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P.; Unrau, P.J. Constructing an RNA World. Trends Biochem. Sci. 1999, 24, M9–M13. [Google Scholar] [CrossRef]
- Luisi, P.L. Chemical Aspects of Synthetic Biology. Chem. Biodiver. 2007, 4, 603–621. [Google Scholar] [CrossRef]
- Sole, R.V.; Rasmussen, S.; Bedau, M. Introduction. Artificial Protocells. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362. [Google Scholar] [CrossRef] [Green Version]
- Monnard, P.A.; Ziock, H.J. Prospects for the Construction of Artificial Cells or Protocells. Orig. Life Evol. Biosph. 2007, 37, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Mansy, S.S.; Szostak, J.W. Reconstructing the Emergence of Cellular Life through the Synthesis of Model Protocells. Cold Spring Harbor Symp. Quant. Biol. 2009, 74, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Blain, J.C.; Szostak, J.W. Progress toward synthetic cells. Annu. Rev. Biochem. 2014, 83, 615–640. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, W.; Feng, Y. Protocells: At the Interface of Life and Non-Life. Life 2015, 5, 447-458. https://doi.org/10.3390/life5010447
Ma W, Feng Y. Protocells: At the Interface of Life and Non-Life. Life. 2015; 5(1):447-458. https://doi.org/10.3390/life5010447
Chicago/Turabian StyleMa, Wentao, and Yu Feng. 2015. "Protocells: At the Interface of Life and Non-Life" Life 5, no. 1: 447-458. https://doi.org/10.3390/life5010447






