The Confluence of Heavy Metal Biooxidation and Heavy Metal Resistance: Implications for Bioleaching by Extreme Thermoacidophiles
Abstract
:1. Introduction
2. Biodiversity of Extremely Thermoacidophilic Microorganisms
| Species | Isolated From | pH (Opt.) | T °C (Opt.) | ED | EA | Growth Modes | Seq. | Refs. |
|---|---|---|---|---|---|---|---|---|
| Sulfolobus acidocaldarius (98-3T, DSM639) | Locomotive Spring, Yellowstone National Park, USA | 1.0–5.9 (2.0–3.0) | 55–85 (70–75) | H2 ~, H2S ?, S0 ?, FeS ?, K2S4O6 ?, COR, Sugars, AA | O2 | Heterotrophic | Y | [16,17,18,19,20] |
| Sulfolobus solfataricus (P2, DSM1617) | Hot spring, Pisciarelli Solfatara, Italy | 2.0–4.0 (3.0) | 65–87 (80) | H2 ~, H2S ?, S0 ?, FeS ?, K2S4O6 ?, COR, Sugars, AA | O2 | Heterotrophic | Y | [17,18,19,21,22] |
| Sulfolobus shibatae (B12T, DSM 5389) | Geothermal pool, Beppu, Kiushu Island, Japan | ND (3.0) | ND–86 (81) | H2 ~, S0, Sugars, AA | O2 | Heterotrophic Mixotrophic | N | [17,18,23] |
| Sulfolobus metallicus (Kra23T, DSM 6482) | Continental solfataric fields, Iceland | 1.0–4.5 (ND) | 50–75 (65) | S0, Fe2+, FeS2, CuFeS2, ZnS, CdS | O2 | Chemolithoautotrophic | N | [18,24,25] |
| Sulfolobus tokodaii (7T, DSM 16993) | Beppu Hot Springs, Kyushu Island, Japan | 2.0–5.0 (2.5–3.0) | 70–85 (80) | S0, Fe2+, COR, AA | O2 | Heterotrophic Mixotrophic | Y | [25,26,27,28] |
| Sulfolobus yangmingensis (YM1T) | Acidic and muddy hot spring, Yang-Ming National Park, Taiwan | 2.0–6.0 (4.0) | 65–90 (80) | S0, FeS, K2S4O6, COR, Sugars, AA | O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | N | [29] |
| Sulfolobus tengchongensis (RT8-4T) | Sulfur-rich hot spring, Tengchong, China | 1.7–6.5 (3.5) | 65–95 (85) | S0, COR, Sugars, AA | O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | N | [30] |
| Sulfolobus islandicus (Ren1H1) | Solfataric fields, Iceland | ND | ND | ND | ND | Heterotrophic | N ^ | [31] |
| Metallosphaera sedula (TH2T, DSM 5348) | Thermal pond in Pisciarelli Solfatara, Italy | 1.0–4.5 (2.0) | 50–80 (75) | H2, S0, K2S4O6, K2SO4, Fe2+, FeS2, CuFeS2, CdS, SnS, ZnS, COR, Sugars, AA | O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | Y | [18,32,33,34] |
| Metallosphaera prunae (Ron 12/IIT, DSM 10039) | Smoldering slag heap, uranium mine, Thüringen, Germany | 1.0–4.5 (2.0) | 55–80 (75) | H2, So, FeS2, CuFeS2, ZnS, COR, Sugars , AA | O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | Y % | [35,36] |
| Metallosphaera hakonensis (HO1-1T, DSM 7519) | Geothermal field, Hakone National Park, Japan | 1.0–4.0 (3.0) | 50–80 (70) | H2S, S0, K2S4O6, Fe2+, FeS, FeS2, CuFeS2, COR, Sugars, AA | O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | N | [19,37,38,39,40] |
| Metallosphaera cuprina (Ar-4T) | Sulfuric hot spring in Tengchong, Yunnan, China | 2.5–5.5 (3.5) | 55–75 (65) | S0, K2S4O6, Fe2+, FeS2, CuFeS2, COR, Sugars, AA | O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | Y | [40,41] |
| Metallosphaera yellowstonensis (MK1T) | Acidic iron mat, Yellowstone National Park, USA | 1.0–4.5 (2.0–3.0) | 45–85 (65–75) | S0, Fe2+ sorbed, FeS, FeS2, CuFeS2, CuS, ZnS, COR | O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | Y | [42,43] |
| Acidianus hospitalis (W1) | Acidic hot spring, Yellowstone National Park, USA | 2.0 ? | 85 ? | ND | ND | ND | Y | [44,45,46] |
| Candidatus Acidianus copahuensis (ALE1) | Copahue geothermal area, Argentina | 1.0–5.0 (2.5–3.0) | 55–80 (75) | H2,S0, K2S4O6,Fe2+, FeS2, CuS, ZnS, COR, Sugars | Fe3+, S0, O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | Y | [47,48] |
| Acidianus infernus (So4aT, DSM 3191) | Solfatara Crater and Pisciarelli Solfatara, Naples | 1.0–5.5 (2.0) | 65–96 (90) | H2, H2S, S0 | S0, O2, MO42- | Mixotrophic Chemolithoautotrophic | N | [18,49,50] |
| Acidianus ambivalens (Lei 10T, DSM 3772) | Solfatara, Iceland | 1.0–3.5 (2.5) | 70–87 (80) | H2, H2S, S0 | S0, O2 | Mixotrophic Chemolithoautotrophic | N | [50,51,52] |
| Acidianus brierleyi (DSM 1651T) | Acid hot spring, Yellowstone National Park | 1.0–6.0 (1.5–2.0) | 45–75 (70) | H2 ?, H2S, S0, Fe2+,FeS2, CuFeS2, ZnS, MoS2, COR | Fe3+, S0, O2, MO42- | Heterotrophic Mixotrophic Chemolithoautotrophic | N | [14,18,21,49,50,53,54,55,56] |
| Acidianus sulfidivorans (JPTT, DSM 18786) | Solfatara on Lihir Island, Papua New Guinea | 0.35–3.0 (0.8–1.4) | 45–83 (74) | H2S, S0 ,Fe2+, FeS2,CuFeSs, FeAsS | Fe3+, S0, O2 | Mixotrophic Chemolithoautotrophic | N | [50] |
| Acidianus tengchongensis (S5T) | Acidothermal spring, Tengchong China | 1.0–5.5 (1.5–2.0) | 60–75 (70) | H2, S0, S2O32− | S0, O2 | Chemolithoautotrophic | N | [57] |
| Acidianus manzaensis (NA-1T) | Fumarole in Manza, Japan | 1.0–5.0 (1.2–1.5) | 60–90 (80) | H2, S0, COR, Sugars | Fe3+, O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | N | [58] |
| Acidianus manzaensis (YN25) | Acidothermal spring, Yunnan China | 1.0–6.0 (1.5–2.5) | 50–85 (65) | H2, S0, K2S4O6, Fe2+, CuFeS2 ,COR, Sugars, AA | S0, O2 | Heterotrophic Mixotrophic | N | [59] |
| Sulfurisphaera ohwakuensis (TA-1T, DSM 12421) | Acidic hot spring located in Ohwaku Valley, Hakone, Japan | 1.0–5.0 (2.0) | 63–92 (84) | H2, S0, COR | S0, O2 | Heterotrophic Mixotrophic | N | [60] |
| Stygiolobus azoricus (FC6T, DSM 6296) | Acidic geothermal spring (Furnas Caldeira), São Miguel Island, Azores | 1.0–5.5 (2.5–3.0) | 57–89 (80) | H2 | S0 | Mixotrophic Chemolithoautotrophic | N | [61] |
| Sulfurococcus yellowstonensis (Str6karT) | Thermal spring, Yellow Stone National Park, USA | 1.0–5.5 (2.0–2.6) | 40–80 (60) | S0, FeS2, ZnS, CuFeS2, Fe2+, COR, Sugars | O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | N | [13,62] |
| Sulfurococcus mirabilis (INMI AT-49T) | Crater, Uzon volcano in Kamchatka, Russia | 1.0–5.8 (2.0–2.6) | 50–86 (70–75) | S0, FeS2, ZnS, CuFeS2, COR, Sugars, AA | O2 | Heterotrophic Mixotrophic Chemolithoautotrophic | N | [13,62] |
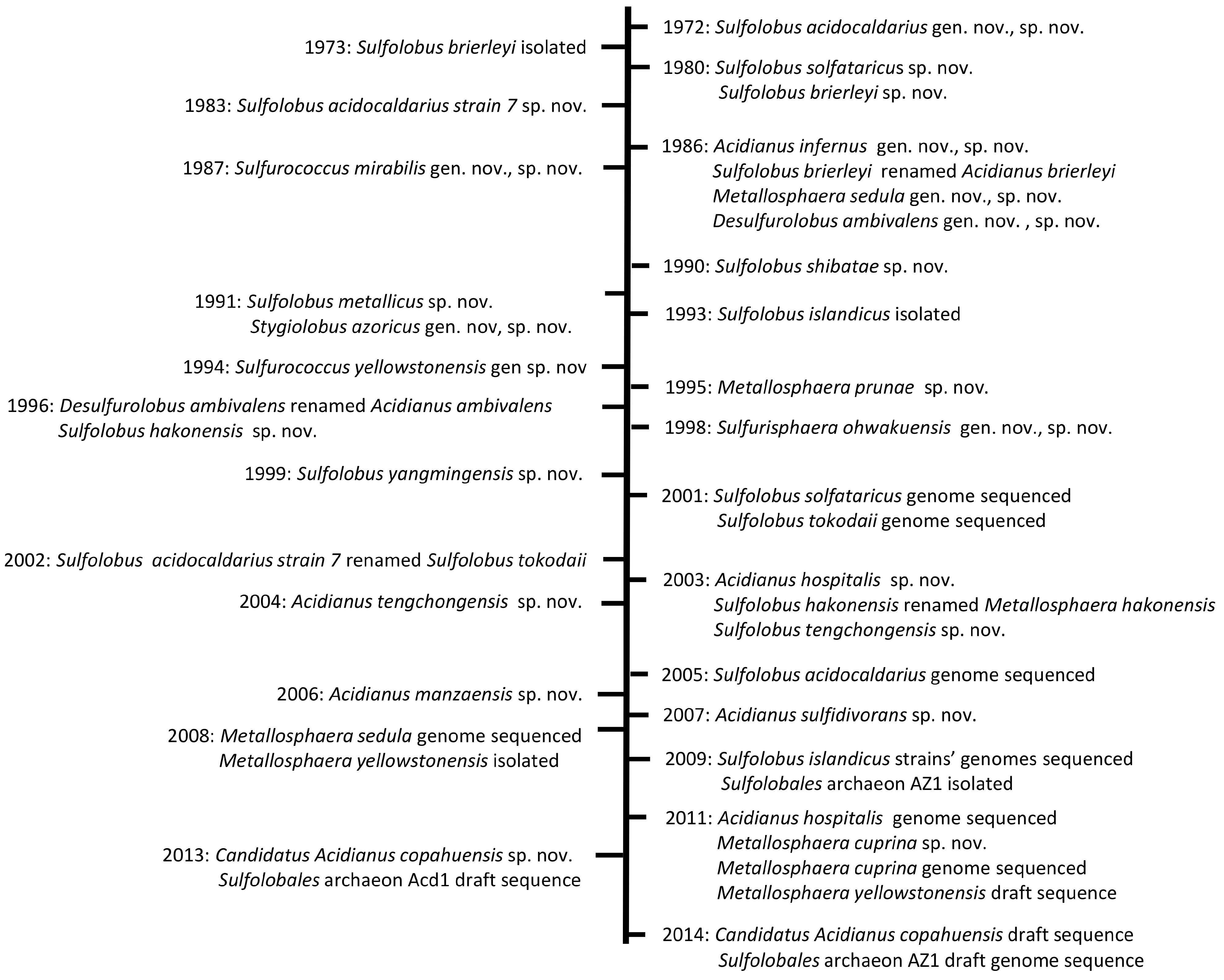
2.1. The Genus Sulfolobus
2.2. The Genus Metallosphaera
2.3. The Genus Acidianus
2.4. The Genera Sulfurisphaera, Stygiolobus and Sulfurococcus
2.5. Sequenced and Unclassified Sulfolobales
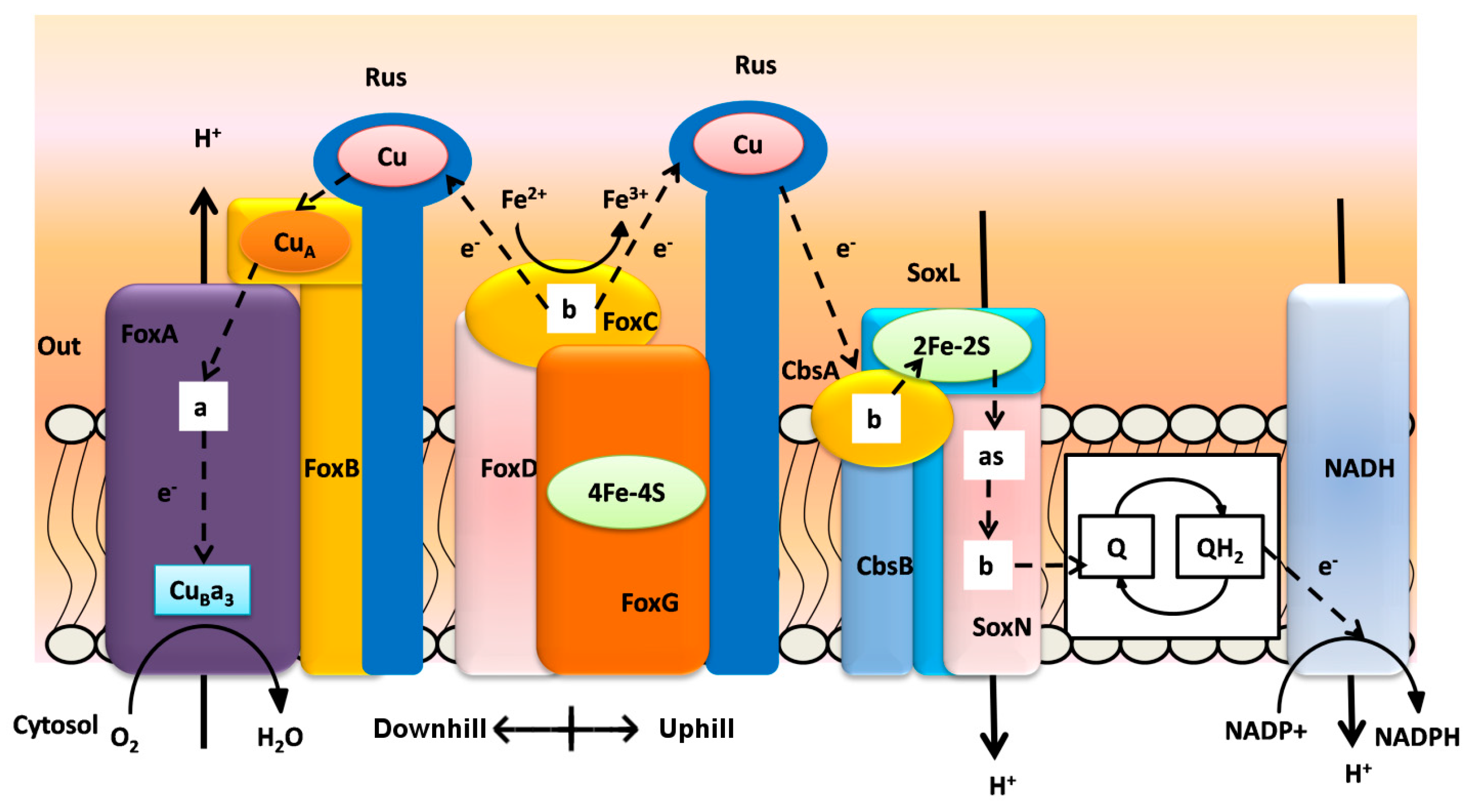
3. Biooxidation of Heavy Metals
| Terminal Oxidase | Fox ABCDEFG | Sox ABCDL | Sox EFGHIM | DoxBCE | CbsAB-SoxL2N | Rusticyanin | Sulfocyanin |
|---|---|---|---|---|---|---|---|
| Metallosphaera sedula | x | x | x | x | x | x, y | x, y |
| Metallosphaera prunae | x | x | x | x | x | x, y | x, y |
| Metallosphaera cuprina | - | x | x | x | x | - | x, y |
| Metallosphaera yellowstonensis | x | x | x | x | x | x, y | x, y |
| Sulfolobus solfataricus | - | x | x | x | x | x | x, y |
| Sulfolobus acidocaldarius | - | x | x | x | x | - | x, y |
| Sulfolobus tokodaii | x | x | x | x | x | - | x, y |
| Sulfolobus metallicus | x | ||||||
| Sulfolobus islandicus | - | x | x | x | x | x, y | x, y |
| Acidianus brierleyi | x | ||||||
| Acidianus ambivalens | x | x | |||||
| Acidianus hospitalis | - | - | - | x | x | - | x |
| Candidatus Acidianus copahuensis | x | x | - | x | x | x | x, y |
4. Heavy Metal Resistance Systems in Extreme Thermoacidophiles
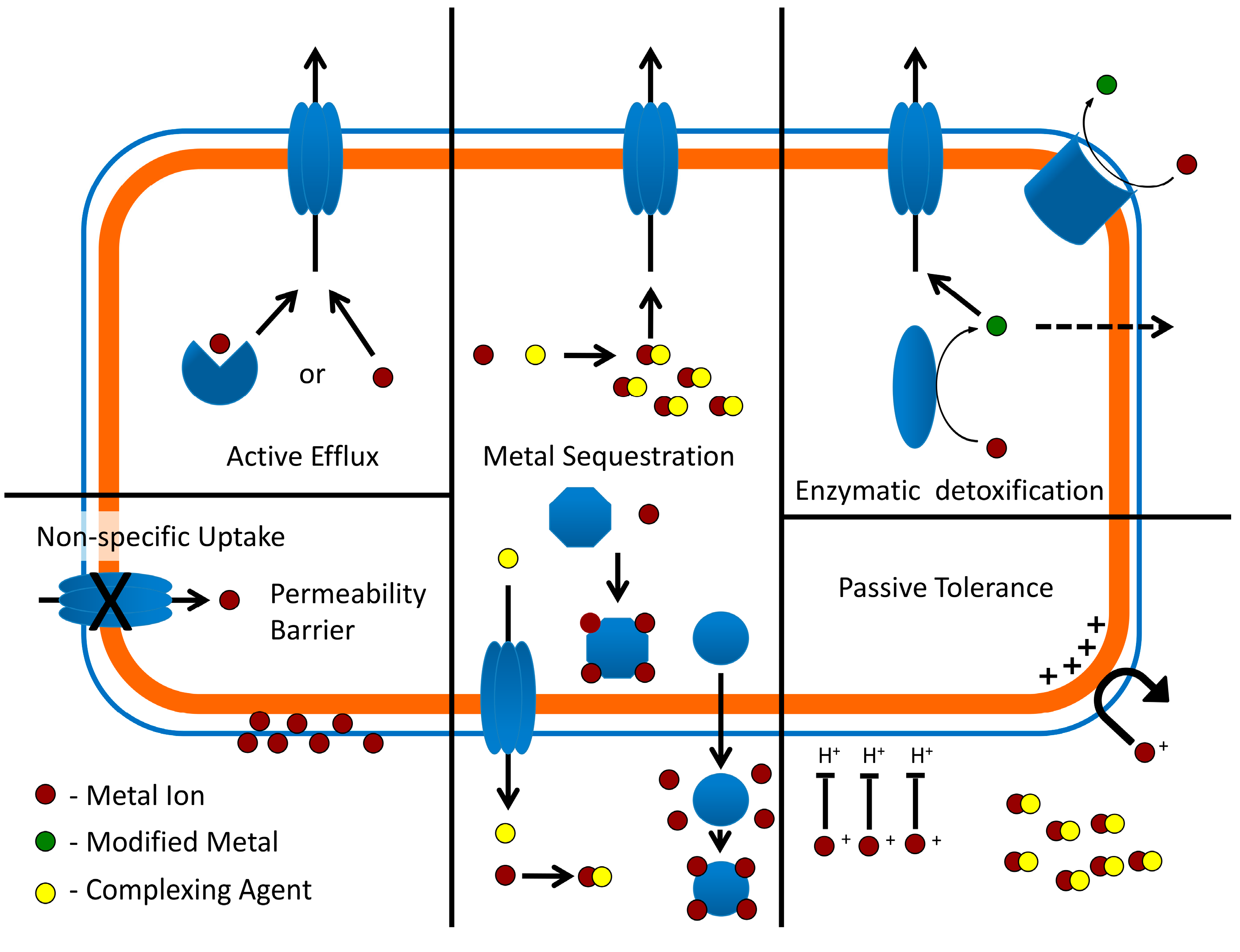
4.1. Passive Tolerance and Metal Exclusion
4.2. Copper
4.3. Mercury
4.4. Other Metals (Arsenic, Cadmium, Nickel, Uranium)
5. Attachment of Extreme Thermoacidophiles to Surfaces
6. Bioleaching
6.1. Current Biooxidation/Bioleaching Practices at Elevated Temperatures
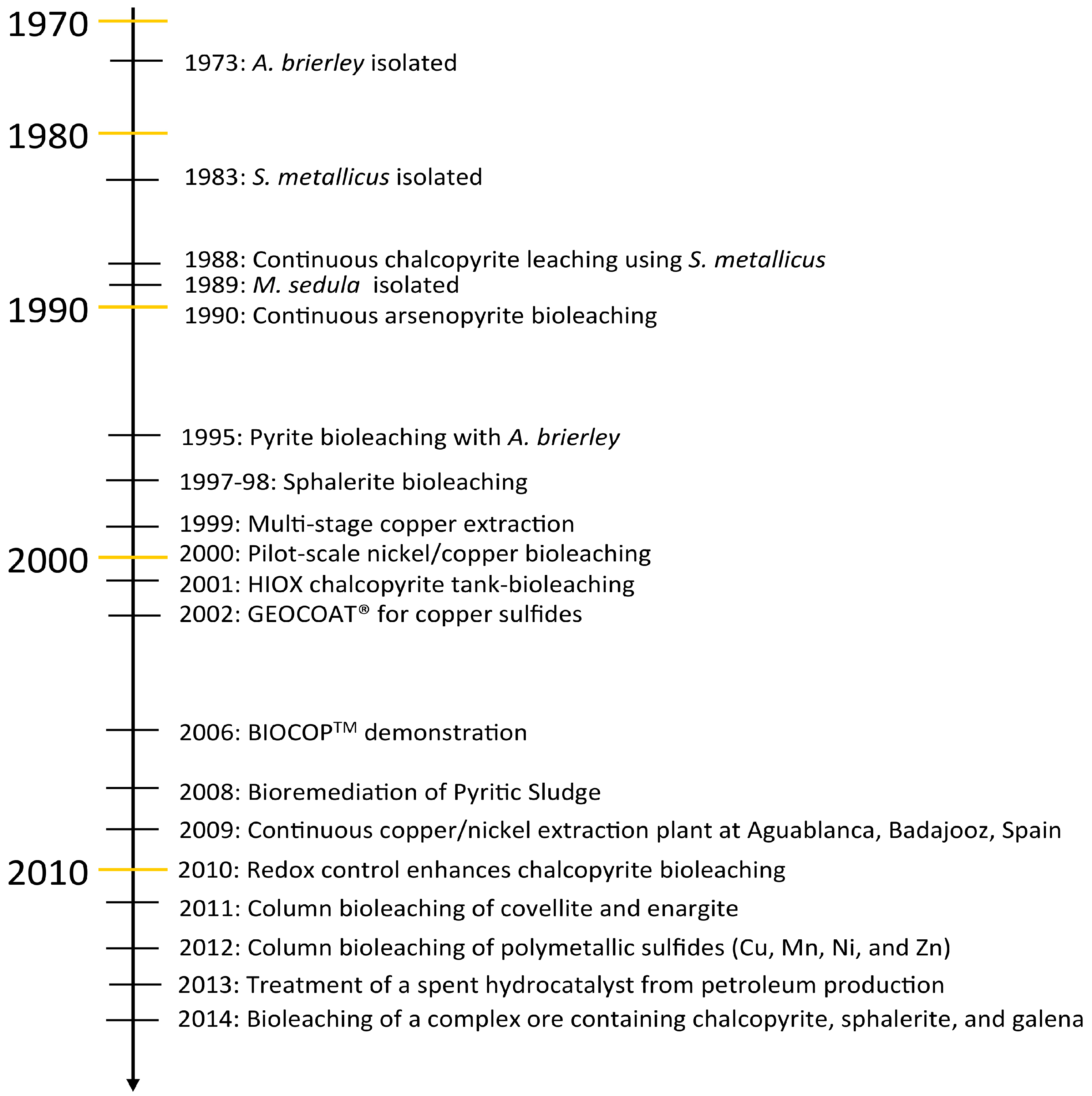
6.2. Extreme Thermoacidophile Process Challenges
6.3. Polymetallic Ores and Industrial Waste
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Brierley, J.A.; Brierley, C.L. Present and future commercial applications of biohydrometallurgy. Hydrometallurgy 2001, 59, 233–239. [Google Scholar] [CrossRef]
- Rawlings, D.E.; Dew, D.; du Plessis, C. Biomineralization of metal-containing ores and concentrates. Trends Biotechnol. 2003, 21, 38–44. [Google Scholar] [CrossRef]
- Bhakta, P.; Arthur, B. Heap bio-oxidation and gold recovery at newmont mining: First-year results. J. Miner. Met. Mater. Soc. 2002, 54, 31–34. [Google Scholar] [CrossRef]
- Rawlings, D.E. Heavy metal mining using microbes. Annu. Rev. Microbiol. 2002, 56, 65–91. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, D.E. Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Fact. 2005, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Acar, S.; Doerr, D.L.; Brierley, J.A. Comparative bioleaching and mineralogy of composited sulfide ores containing enargite, covellite and chalcocite by mesophilic and thermophilic microorganisms. Hydrometallurgy 2011, 105, 213–221. [Google Scholar] [CrossRef]
- Norris, P.R.; Burton, N.P.; Foulis, N.A. Acidophiles in bioreactor mineral processing. Extremophiles 2000, 4, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Acar, S.; Brierley, J.A.; Wan, R.Y. Conditions for bioleaching a covellite-bearing ore. Hydrometallurgy 2005, 77, 239–246. [Google Scholar] [CrossRef]
- Norris, P.R.; Calvo-Bado, L.A.; Brown, C.F.; Davis-Belmar, C.S. Ore column leaching with thermophiles: I, copper sulfide ore. Hydrometallurgy 2012, 127, 62–69. [Google Scholar] [CrossRef]
- Norris, P.R.; Brown, C.F.; Caldwell, P.E. Ore column leaching with thermophiles: II, polymetallic sulfide ore. Hydrometallurgy 2012, 127, 70–76. [Google Scholar] [CrossRef]
- Norris, P.R.; Burton, N.P.; Clark, D.A. Mineral sulfide concentrate leaching in high temperature bioreactors. Miner. Eng. 2013, 48, 10–19. [Google Scholar] [CrossRef]
- Pradhan, N.; Nathsarma, K.C.; Srinivasa Rao, K.; Sukla, L.B.; Mishra, B.K. Heap bioleaching of chalcopyrite: A review. Miner. Eng. 2008, 21, 355–365. [Google Scholar] [CrossRef]
- Schippers, A. Microorganisms involved in bioleaching and nucleic acid-based molecular methods for their identification and quantification. In Microbial Processing of Metal Sulfides; Donati, E.R., Sand, W., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 3–33. [Google Scholar]
- Norris, P.R. Acidophile Diversity in Mineral Sulfide Oxidation. In Biomining; Rawlings, D., Johnson, D.B., Eds.; Springer: Berlin, Germany, 2007; pp. 199–216. [Google Scholar]
- Ilbert, M.; Bonnefoy, V. Insight into the evolution of the iron oxidation pathways. Biochim. Biophys. Acta 2013, 1827, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Brock, T.D.; Brock, K.M.; Belly, R.T.; Weiss, R.L. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 1972, 84, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Grogan, D.W. Phenotypic characterization of the archaebacterial genus Sulfolobus: Comparison of five wild-type strains. J. Bacteriol. 1989, 171, 6710–6719. [Google Scholar] [PubMed]
- Huber, G.; Drobner, E.; Huber, H.; Stetter, K.O. Growth by Aerobic Oxidation of Molecular Hydrogen in Archaea—A Metabolic Property so far Unknown for this Domain. Syst. Appl. Microbiol. 1992, 15, 502–504. [Google Scholar] [CrossRef]
- Takayanagi, S.; Kawasaki, H.; Sugimori, K.; Yamada, T.; Sugai, A.; Ito, T.; Yamasato, K.; Shioda, M. Sulfolobus hakonensis sp. nov., a novel species of acidothermophilic archaeon. Int. J. Syst. Bacteriol. 1996, 46, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Brügger, K.; Skovgaard, M.; Redder, P.; She, Q.; Torarinsson, E.; Greve, B.; Awayez, M.; Zibat, A.; Klenk, H.; et al. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 2005, 187, 4992–4999. [Google Scholar] [CrossRef] [PubMed]
- Zillig, W.; Stetter, K.O.; Wunderl, S.; Schulz, W.; Priess, H.; Scholz, I. The Sulfolobus-“Caldariella” group: Taxonomy on the basis of the structure of DNA-dependent RNA polymerases. Arch. Microbiol. 1980, 125, 259–269. [Google Scholar] [CrossRef]
- She, Q.; Singh, R.K.; Confalonieri, F.; Zivanovic, Y.; Allard, G.; Awayez, M.J.; Chan-Weiher, C.C.; Clausen, I.G.; Curtis, B.A.; de Moors, A.; et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 2001, 98, 7835–7840. [Google Scholar] [CrossRef] [PubMed]
- Grogan, D.; Palm, P.; Zillig, W. Isolate B12, which harbours a virus-like element, represents a new species of the archaebacterial genus Sulfolobus, Sulfolobus shibatae, sp. nov. Arch. Microbiol. 1990, 154, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Huber, G.; Stetter, K.O. Sulfolobus metallicus, sp. nov., a novel strictly chemolithoautotrophic thermophilic archaeal species of metal-mobilizers. Syst. Appl. Microbiol. 1991, 14, 372–378. [Google Scholar] [CrossRef]
- Bathe, S.; Norris, P.R. Ferrous iron- and sulfur-induced genes in Sulfolobus metallicus. Appl. Environ. Microbiol. 2007, 73, 2491–2497. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Iwasaki, T.; Uzawa, T.; Hara, K.; Nemoto, N.; Kon, T.; Ueki, T.; Yamagishi, A.; Oshima, T. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 2002, 6, 39–44. [Google Scholar]
- Inatomi, K.; Ohba, M.; Oshima, T. Chemical properties of proteinaceus cell wall from an acido-thermophile, Sulfolobus acidocaldarius. Chem. Lett. 1983, 12, 1191–1194. [Google Scholar] [CrossRef]
- Kawarabayasi, Y.; Hino, Y.; Horikawa, H.; Jin-no, K.; Takahashi, M.; Sekine, M.; Baba, S.; Ankai, A.; Kosugi, H.; Hosoyama, A.; et al. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 2001, 8, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Jan, R.L.; Wu, J.; Chaw, S.M.; Tsai, C.W.; Tsen, S.D. A novel species of thermoacidophilic archaeon, Sulfolobus yangmingensis sp. nov. Int. J. Syst. Bacteriol. 1999, 49, 1809–1816. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Dong, X.; Huang, L. Sulfolobus tengchongensis sp. nov., a novel thermoacidophilic archaeon isolated from a hot spring in Tengchong, China. Extremophiles 2003, 7, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Zillig, W.; Kletzin, A.; Schleper, C.; Holz, I.; Janekovic, D.; Hain, J.; Lanzendörfer, M.; Kristjansson, J.K. Screening for Sulfolobales, their Plasmids and their Viruses in Icelandic Solfataras. Syst. Appl. Microbiol. 1994, 16, 609–628. [Google Scholar] [CrossRef]
- Huber, G.; Spinnler, C.; Gambacorta, A.; Stetter, K. Metallosphaera sedula gen, and sp. nov. represents a new genus of aerobic, metal-mobilizing, thermoacidophilic archaebacteria. Syst. Appl. Microbiol. 1989, 12, 38–47. [Google Scholar] [CrossRef]
- Auernik, K.S.; Kelly, R.M. Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl. Environ. Microbiol. 2008, 74, 7723–7732. [Google Scholar] [CrossRef] [PubMed]
- Auernik, K.S.; Maezato, Y.; Blum, P.H.; Kelly, R.M. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 2008, 74, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.; Huber, H.; Teiner, K.; Burggraf, S.; Stetter, K.O. Metallosphaera prunae, sp. nov., a novel metal-mobilizing, thermoacidophilic archaeum, isolated from a uranium mine in Germany. Syst. Appl. Microbiol. 1995, 18, 560–566. [Google Scholar] [CrossRef]
- Mukherjee, A.; Wheaton, G.H.; Blum, P.H.; Kelly, R.M. Uranium extremophily is an adaptive, rather than intrinsic, feature for extremely thermoacidophilic Metallosphaera species. Proc. Natl. Acad. Sci. USA 2012, 109, 16702–16707. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, N. Reclassification of Sulfolobus hakonensis Takayanagi et al. 1996 as Metallosphaera hakonensis comb. nov. based on phylogenetic evidence and DNA G+C content. Int. J. Syst. Evol. Microbiol. 2003, 53, 1607–1608. [Google Scholar] [CrossRef] [PubMed]
- Plumb, J.J.; Muddle, R.; Franzmann, P.D. Effect of pH on rates of iron and sulfur oxidation by bioleaching organisms. Miner. Eng. 2008, 21, 76–82. [Google Scholar] [CrossRef]
- Usher, K.M.; Shaw, J.A.; Kaksonen, A.H.; Saunders, M. Elemental analysis of extracellular polymeric substances and granules in chalcopyrite bioleaching microbes. Hydrometallurgy 2010, 104, 376–381. [Google Scholar] [CrossRef]
- Liu, L.-J.; You, X.-Y.; Guo, X.; Liu, S.-J.; Jiang, C.-Y. Metallosphaera cuprina sp. nov., an acidothermophilic, metal-mobilizing archaeon. Int. J. Syst. Evol. Microbiol. 2011, 61, 2395–2400. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-J.; You, X.-Y.; Zheng, H.; Wang, S.; Jiang, C.-Y.; Liu, S.-J. Complete genome sequence of Metallosphaera cuprina, a metal sulfide-oxidizing archaeon from a hot spring. J. Bacteriol. 2011, 193, 3387–3388. [Google Scholar] [CrossRef] [PubMed]
- Kozubal, M.; Macur, R.E.; Korf, S.; Taylor, W.P.; Ackerman, G.G.; Nagy, A.; Inskeep, W.P. Isolation and distribution of a novel iron-oxidizing crenarchaeon from acidic geothermal springs in Yellowstone National Park. Appl. Environ. Microbiol. 2008, 74, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Kozubal, M.A.; Dlakic, M.; Macur, R.E.; Inskeep, W.P. Terminal oxidase diversity and function in “Metallosphaera yellowstonensis”: Gene expression and protein modeling suggest mechanisms of Fe(II) oxidation in the sulfolobales. Appl. Environ. Microbiol. 2011, 77, 1844–1853. [Google Scholar] [CrossRef] [PubMed]
- Bettstetter, M.; Peng, X.; Garrett, R.A.; Prangishvili, D. AFV1, a novel virus infecting hyperthermophilic archaea of the genus acidianus. Virology 2003, 315, 68–79. [Google Scholar] [CrossRef]
- Basta, T.; Smyth, J.; Forterre, P.; Prangishvili, D.; Peng, X. Novel archaeal plasmid pAH1 and its interactions with the lipothrixvirus AFV1. Mol. Microbiol. 2009, 71, 23–34. [Google Scholar] [CrossRef] [PubMed]
- You, X.Y.; Liu, C.; Wang, S.Y.; Jiang, C.Y.; Shah, S.A.; Prangishvili, D.; She, Q.; Liu, S.J.; Garrett, R.A. Genomic analysis of Acidianus hospitalis W1 a host for studying crenarchaeal virus and plasmid life cycles. Extremophiles 2011, 15, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Giaveno, M.A.; Urbieta, M.S.; Ulloa, J.R.; González Toril, E.; Donati, E.R. Physiologic Versatility and Growth Flexibility as the Main Characteristics of a Novel Thermoacidophilic Acidianus Strain Isolated from Copahue Geothermal Area in Argentina. Microb. Ecol. 2013, 65, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Urbieta, M.S.; Rascovan, N.; Castro, C.; Revale, S.; Giaveno, M.A.; Vazquez, M.; Donati, R. Draft Genome Sequence of the Novel Thermoacidophilic Archaeon Acidianus copahuensis Strain ALE1, Isolated from the Copahue Volcanic Area in Neuquén, Argentina. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Segerer, A.; Neuner, A.; Kristjansson, J.K.; Stetter, K.O. Acidianus infernus gen. nov., sp. nov., and Acidianus brierleyi Comb. nov.: Facultatively aerobic, extremely acidophilic thermophilic sulfur-metabolizing archaebacteria. Int. J. Syst. Bacteriol. 1986, 36, 559–564. [Google Scholar] [CrossRef]
- Plumb, J.J.; Haddad, C.M.; Gibson, J.A.E.; Franzmann, P.D. Acidianus sulfidivorans sp. nov., an extremely acidophilic, thermophilic archaeon isolated from a solfatara on Lihir Island, Papua New Guinea, and emendation of the genus description. Int. J. Syst. Evol. Microbiol. 2007, 57, 1418–1423. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.; Huber, H.; Burggraf, S.; Stetter, K.O. 16S rDNA-based Phylogeny of the Archaeal Order Sulfolobales and Reclassification of Desulfurolobus ambivalens as Acidianus ambivalens comb. nov. Syst. Appl. Microbiol. 1996, 19, 56–60. [Google Scholar] [CrossRef]
- Zillig, W.; Yeats, S.; Holz, I.; Böck, A.; Rettenberger, M.; Gropp, F.; Simon, G. Desulfurolobus ambivalens, gen. nov., sp. nov., an autotrophic archaebacterium facultatively oxidizing or reducing sulfur. Syst. Appl. Microbiol. 1986, 8, 197–203. [Google Scholar] [CrossRef]
- Brierley, C.L.; Brierley, J.A. A chemoautotrophic and thermophilic microorganism isolated from an acid hot spring. Can. J. Microbiol. 1973, 19, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Konishi, Y.; Nishimura, H.; Asai, S. Bioleaching of sphalerite by the acidophilic thermophile Acidianus brierleyi. Hydrometallurgy 1998, 47, 339–352. [Google Scholar] [CrossRef]
- Dinkla, I.J.T.; Gericke, M.; Geurkink, B.K.; Hallberg, K.B. Acidianus brierleyi is the dominant thermoacidophile in a bioleaching community processing chalcopyrite containing concentrates at 70 °C. Adv. Mater. Res. 2009, 71, 67–70. [Google Scholar] [CrossRef]
- Brierley, J.A. Acdidophilic thermophilic archaebacteria: Potential application for metals recovery. FEMS Microbiol. Rev. 1990, 75, 287–292. [Google Scholar] [CrossRef]
- He, Z.G.; Zhong, H.; Li, Y. Acidianus tengchongensis sp. nov., a new species of acidothermophilic archaeon isolated from an Acidothermal Spring. Curr. Microbiol. 2004, 48, 159–163. [Google Scholar] [PubMed]
- Yoshida, N.; Nakasato, M.; Ohmura, N.; Ando, A.; Saiki, H.; Ishii, M.; Igarashi, Y. Acidianus manzaensis sp. nov., a novel thermoacidophilic Archaeon growing autotrophically by the oxidation of H2 with the reduction of Fe3+. Curr. Microbiol. 2006, 53, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, R.; Yu, Y.; Jin, D.; Liang, C.; Yi, Y.; Zhu, W.; Xia, J. A novel acidophilic, thermophilic iron and sulfur-oxidizing archaeon isolated from a hot spring of tengchong, Yunnan, China. Brazilian J. Microbiol. 2011, 42, 514–525. [Google Scholar] [CrossRef]
- Kurosawa, N.; Itoh, Y.H.; Iwai, T.; Sugai, A.; Uda, I.; Kimura, N.; Horiuchi, T.; Itoh, T. Sulfurisphaera ohwakuensis gen. nov., sp. nov., a novel extremely thermophilic acidophile of the order Sulfolobales. Int. J. Syst. Bacteriol. 1998, 48, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Segerer, A.H.; Trincone, A.; Gahrtz, M.; Stetter, K.O. Stygiolobus azoricus gen. nov., sp. nov. represents a novel genus of anaerobic, extremely thermoacidophilic archaebacteria of the order sulfolobales. Int. J. Syst. Bacteriol. 1991, 41, 495–501. [Google Scholar] [CrossRef]
- Huber, H.; Prangishvili, D. Sulfolobales. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrant, E., Eds.; Springer Science: New York, NY, USA, 2006; pp. 1028–1049. [Google Scholar]
- Albers, S.-V.; Siebers, B. The family sulfolobaceae. In The Prokaryotes; Eugene, R., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin, Germany, 2014; pp. 323–346. [Google Scholar]
- Rawlings, D.E.; Johnson, D.B. The microbiology of biomining: Development and optimization of mineral-oxidizing microbial consortia. Microbiology 2007, 153, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Albers, S.-V.; Jarrell, K.F. The archaellum: How archaea swim. Front. Microbiol. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Grogan, D. Genomic evidence of rapid, global-scale gene flow in a Sulfolobus species. ISME J. 2012, 6, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Inskeep, W.P.; Jay, Z.J.; Tringe, S.G.; Herrgård, M.J.; Rusch, D.B. The YNP metagenome project: Environmental parameters responsible for microbial distribution in the yellowstone geothermal ecosystem. Front. Microbiol. 2013, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Reno, M.L.; Held, N.L.; Fields, C.J.; Burke, P.V.; Whitaker, R.J. Biogeography of the Sulfolobus islandicus pan-genome. Proc. Natl. Acad. Sci. USA 2009, 106, 8605–8610. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Brügger, K.; Liu, C.; Shah, S.A.; Zheng, H.; Zhu, Y.; Wang, S.; Lillestøl, R.K.; Chen, L.; Frank, J.; et al. Genome analyses of icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J. Bacteriol. 2011, 193, 1672–1680. [Google Scholar] [CrossRef] [PubMed]
- Cadillo-Quiroz, H.; Didelot, X.; Held, N.L.; Herrera, A.; Darling, A.; Reno, M.L.; Krause, D.J.; Whitaker, R.J. Patterns of gene flow define species of thermophilic Archaea. PLoS Biol. 2012, 10. [Google Scholar] [CrossRef] [PubMed]
- Jaubert, C.; Danioux, C.; Oberto, J.; Cortez, D.; Bize, A.; Krupovic, M.; She, Q.; Forterre, P.; Prangishvili, D.; Sezonov, G. Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol. 2013, 3, 130010. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; van Wolferen, M.; Wagner, A.; Lassak, K.; Meyer, B.H.; Reimann, J.; Albers, S.-V. Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Albers, S.V.; Driessen, A.J.M. Conditions for gene disruption by homologous recombination of exogenous DNA into the Sulfolobus solfataricus genome. Archaea 2008, 2, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Maezato, Y.; Dana, K.; Blum, P. Engineering thermoacidophilic archaea using linear DNA recombination. In Strain Engineering; Williams, J.A., Ed.; Humana Press: New York, NY, USA, 2011; Volume 765, pp. 435–445. [Google Scholar]
- Deng, L.; Zhu, H.; Chen, Z.; Liang, Y.X.; She, Q. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 2009, 13, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Whitaker, R.J. A broadly applicable gene knockout system for the thermoacidophilic archaeon Sulfolobus islandicus based on simvastatin selection. Microbiology 2012, 158, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Berkner, S.; Wlodkowski, A.; Albers, S.V.; Lipps, G. Inducible and constitutive promoters for genetic systems in Sulfolobus acidocaldarius. Extremophiles 2010, 14, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Berkner, S.; Grogan, D.; Albers, S.V.; Lipps, G. Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea. Nucl. Acids Res. 2007, 35, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Krause, D.J.; Whitaker, R.J. Sulfolobus islandicus: A model system for evolutionary genomics. Biochem. Soc. Trans. 2013, 41, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Huang, Q.; Zhang, C.; Ni, J.; She, Q.; Shen, Y. Development of a simvastatin selection marker for a hyperthermophilic acidophile, Sulfolobus islandicus. Appl. Environ. Microbiol. 2012, 78, 568–574. [Google Scholar] [CrossRef] [PubMed]
- López-García, P.; Forterre, P. Control of DNA topology during thermal stress in hyperthermophilic archaea: DNA topoisomerase levels, activities and induced thermotolerance during heat and cold shock in Sulfolobus. Mol. Microbiol. 1999, 33, 766–777. [Google Scholar] [CrossRef] [PubMed]
- Zillig, W.; Arnold, H.P.; Holz, I.; Prangishvili, D.; Schweier, A.; Stedman, K.; She, Q.; Phan, H.; Garrett, R.; Kristjansson, J.K. Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles 1998, 2, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, R.J.; Grogan, D.W.; Taylor, J.W. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 2003, 301, 976–978. [Google Scholar] [CrossRef] [PubMed]
- Grogan, D.W.; Ozarzak, M.A.; Bernander, R. Variation in gene content among geographically diverse Sulfolobus isolates. Environ. Microbiol. 2008, 10, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Held, N.L.; Herrera, A.; Whitaker, R.J. Reassortment of CRISPR repeat-spacer loci in Sulfolobus islandicus. Environ. Microbiol. 2013, 15, 3065–3076. [Google Scholar] [PubMed]
- Bautista, M.A.; Zhang, C.; Whitaker, R.J. Virus-Induced Dormancy in the Archaeon Sulfolobus islandicus. MBio 2015, 6, e02565-14. [Google Scholar] [CrossRef] [PubMed]
- Peeples, T.L.; Kelly, R.M. Bioenergetics of the metal/sulfur-oxidizing extreme thermoacidophile, Metallosphaera sedula. Fuel 1993, 72, 1619–1624. [Google Scholar] [CrossRef]
- Berg, I.A.; Kockelkorn, D.; Buckel, W.; Fuchs, G. A 3-Hydroxypropionate/4-Hydroxybutyrate Autotrophic Carbon Dioxide Assimilation Pathway in Archaea. Science 2007, 318, 1782–1786. [Google Scholar] [CrossRef] [PubMed]
- Laska, S.; Lottspeich, F.; Kletzin, A. Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology 2003, 149, 2357–2371. [Google Scholar] [CrossRef] [PubMed]
- Podar, M.; Makarova, K.S.; Graham, D.E.; Wolf, Y.I.; Koonin, E.V.; Reysenbach, A.-L. Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park. Biol. Direct 2013, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Servín-Garcidueñas, L.E.; Martínez-Romero, E. Draft Genome Sequence of the Sulfolobales Archaeon AZ1, Obtained through Metagenomic Analysis of a Mexican Hot Spring. Genome Announc. 2014, 2, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Ingledew, W.J. Thiobacillus Ferrooxidans the bioenergetics of an acidophilic chemolithotroph. Biochim. Biophys. Acta 1982, 683, 89–117. [Google Scholar] [CrossRef]
- Bird, L.J.; Bonnefoy, V.; Newman, D.K. Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol. 2011, 19, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Bonnefoy, V.; Holmes, D.S. Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments. Environ. Microbiol. 2012, 14, 1597–1611. [Google Scholar] [CrossRef] [PubMed]
- Bonnefoy, V. Bioinformatics and Genomics of Iron- and Sulfur-Oxidizing Acidophiles. In Geomicrobiology: Molecular and Environmental Perspective; Barton, L.L., Mandl, M., Loy, A., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2010; pp. 169–192. [Google Scholar]
- Quatrini, R.; Appia-Ayme, C.; Denis, Y.; Jedlicki, E.; Holmes, D.S.; Bonnefoy, V. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 2009, 10, 394. [Google Scholar] [CrossRef] [PubMed]
- Blake, R.C.; Shute, E.A.; Greenwood, M.M.; Spencer, G.H.; Ingledew, W.J. Enzymes of aerobic respiration on iron. FEMS Microbiol. Rev. 1993, 11, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Barr, D.W.; Ingledew, W.J.; Norris, P.R. Respiratory chain components of iron-oxidizing acidophilic bacteria. FEMS Microbiol. Lett. 1990, 70, 85–89. [Google Scholar] [CrossRef]
- Schäfer, G.; Engelhard, M.; Müller, V. Bioenergetics of the Archaea. Microbiol. Mol. Biol. Rev. 1999, 63, 570–620. [Google Scholar] [PubMed]
- Pereira, M.M.; Bandeiras, T.M.; Fernandes, A.S.; Lemos, R.S.; Melo, A.M.; Teixeira, M. Respiratory chains from aerobic thermophilic prokaryotes. J. Bioenerg. Biomembr. 2004, 36, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.L. Rieske Iron-Sulfur Proteins from Extremophilic Organisms. J. Bioenerg. Biomembr. 2004, 36, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Lübben, M.; Kolmerer, B.; Saraste, M. An archaebacterial terminal oxidase combines core structures of two mitochondrial respiratory complexes. EMBO J. 1992, 11, 805–812. [Google Scholar] [PubMed]
- Lübben, M.; Warne, A.; Albracht, S.P.; Saraste, M. The purified SoxABCD quinol oxidase complex of Sulfolobus acidocaldarius contains a novel haem. Mol. Microbiol. 1994, 13, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Gleißner, M.; Kaiser, U.; Antonopoulos, E.; Schäfer, G. The archaeal SoxABCD complex is a proton pump in Sulfolobus acidocaldarius. J. Biol. Chem. 1997, 272, 8417–8426. [Google Scholar] [CrossRef] [PubMed]
- Lübben, M.; Arnaud, S.; Castresana, J.; Warne, A.; Albracht, S.P.; Saraste, M. A second terminal oxidase in Sulfolobus acidocaldarius. Eur. J. Biochem. 1994, 224, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J.; Lübben, M.; Saraste, M. New archaebacterial genes coding for redox proteins: Implications for the evolution of aerobic metabolism. J. Mol. Biol. 1995, 250, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Komorowski, L.; Schäfer, G. Sulfocyanin and subunit II, two copper proteins with novel features, provide new insight into the archaeal SoxM oxidase supercomplex. FEBS Lett. 2001, 487, 351–355. [Google Scholar] [CrossRef]
- Komorowski, L.; Verheyen, W.; Schäfer, G. The archaeal respiratory supercomplex SoxM from S. acidocaldarius combines features of quinole and cytochrome c oxidases. Biol. Chem. 2002, 383, 1791–1799. [Google Scholar] [CrossRef] [PubMed]
- Hiller, A.; Henninger, T.; Schäfer, G.; Schmidt, C.L. New genes encoding subunits of a cytochrome bc1-analogous complex in the respiratory chain of the hyperthermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J. Bioenerg. Biomembr. 2003, 35, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Bandeiras, T.M.; Refojo, P.N.; Todorovic, S.; Murgida, D.H.; Hildebrandt, P.; Bauer, C.; Pereira, M.M.; Kletzin, A.; Teixeira, M. The cytochrome ba complex from the thermoacidophilic crenarchaeote Acidianus ambivalens is an analog of bc(1) complexes. Biochim. Biophys. Acta 2009, 1787, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Purschke, W.G.; Schmidt, C.L.; Petersen, A.; Schäfer, G. The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J. Bacteriol. 1997, 179, 1344–1353. [Google Scholar] [PubMed]
- Giuffrè, A.; Gomes, C.M.; Antonini, G.; D’Itri, E.; Teixeira, M.; Brunori, M. Functional properties of the quinol oxidase from Acidianus ambivalens and the possible catalytic role of its electron donor—Studies on the membrane-integrated and purified enzyme. Eur. J. Biochem. 1997, 250, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Das, T.K.; Gomes, C.M.; Bandeiras, T.M.; Pereira, M.M.; Teixeira, M.; Rousseau, D.L. Active site structure of the aa3 quinol oxidase of Acidianus ambivalens. Biochim. Biophys. Acta 2004, 1655, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Kappler, U.; Sly, L.I.; McEwan, A.G. Respiratory gene clusters of Metallosphaera sedula—Differential expression and transcriptional organization. Microbiology 2005, 151, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Hettmann, T.; Schmidt, C.L.; Anemüller, S.; Zähringer, U.; Moll, H.; Petersen, A.; Schäfer, G. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius. A novel highly glycosylated, membrane-bound b-type hemoprotein. J. Biol. Chem. 1998, 273, 12032–12040. [Google Scholar] [CrossRef] [PubMed]
- Schoepp-Cothenet, B.; Schütz, M.; Baymann, F.; Brugna, M.; Nitschke, W.; Myllykallio, H.; Schmidt, C. The membrane-extrinsic domain of cytochrome b558/566 from the Archaeon Sulfolobus acidocaldarius performs pivoting movements with respect to the membrane surface. FEBS Lett. 2001, 487, 372–376. [Google Scholar] [CrossRef]
- Gomes, C.M.; Bandeiras, T.M.; Teixeira, M. A new type-II NADH dehydrogenase from the archaeon acidianus ambivalens: Characterization and in vitro reconstitution of the respiratory chain. J. Bioenerg. Biomembr. 2001, 33, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lemos, R.S.; Gomes, C.M.; Teixeira, M. Acidianus ambivalens Complex II typifies a novel family of succinate dehydrogenases. Biochem. Biophys. Res. Commun. 2001, 281, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, T.; Kounosu, A.; Aoshima, M.; Ohmori, D.; Imai, T.; Urushiyama, A.; Cosper, N.J.; Scott, R.A. Novel [2Fe-2S]-type redox center C in sdhC of archaeal respiratory complex II from Sulfolobus tokodaii strain 7. J. Biol. Chem. 2002, 277, 39642–39648. [Google Scholar] [CrossRef] [PubMed]
- Kanao, T.; Kamimura, K.; Sugio, T. Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans. J. Biotechnol. 2007, 132, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Bugaytsova, Z.; Lindström, E.B. Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus. Eur. J. Biochem. 2004, 271, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.H.; Bandeiras, T.M.; Urich, T.; Teixeira, M.; Gomes, C.M.; Kletzin, A. Coupling of the pathway of sulphur oxidation to dioxygen reduction: Characterization of a novel membrane-bound thiosulphate: Quinone oxidoreductase. Mol. Microbiol. 2004, 53, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Castelle, C.; Guiral, M.; Malarte, G.; Ledgham, F.; Leroy, G.; Brugna, M.; Giudici-Orticoni, M.-T. A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J. Biol. Chem. 2008, 283, 25803–25811. [Google Scholar] [CrossRef] [PubMed]
- Yarzábal, A.; Brasseur, G.; Ratouchniak, J.; Lund, K.; Lemesle-Meunier, D.; DeMoss, J.A.; Bonnefoy, V. The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J. Bacteriol. 2002, 184, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Bengrine, A.; Guiliani, N.; Appia-Ayme, C.; Jedlicki, E.; Holmes, D.S.; Chippaux, M.; Bonnefoy, V. Sequence and expression of the rusticyanin structural gene from Thiobacillus ferrooxidans ATCC33020 strain. Biochim. Biophys. Acta 1998, 1443, 99–112. [Google Scholar] [CrossRef]
- Giudici-Orticoni, M.T.; Guerlesquin, F.; Bruschi, M.; Nitschke, W. Interaction-induced redox switch in the electron transfer complex rusticyanin-cytochrome c(4). J. Biol. Chem. 1999, 274, 30365–30369. [Google Scholar] [CrossRef] [PubMed]
- Malarte, G.; Leroy, G.; Lojou, E.; Abergel, C.; Bruschi, M.; Giudici-Orticoni, M.T. Insight into molecular stability and physiological properties of the diheme cytochrome CYC41 from the acidophilic bacterium Acidithiobacillus ferrooxidans. Biochemistry 2005, 44, 6471–6481. [Google Scholar] [CrossRef] [PubMed]
- Kai, M.; Yano, T.; Tamegai, H.; Fukumori, Y.; Yamanaka, T. Thiobacillus ferrooxidans cytochrome c oxidase: Purification, and molecular and enzymatic features. J. Biochem. 1992, 112, 816–821. [Google Scholar] [PubMed]
- Appia-Ayme, C.; Guiliani, N.; Ratouchniak, J.; Bonnefoy, V. Characterization of an operon encoding two c-type cytochromes, an aa(3)-type cytochrome oxidase, and rusticyanin in Thiobacillus ferrooxidans ATCC 33020. Appl. Environ. Microbiol. 1999, 65, 4781–4787. [Google Scholar] [PubMed]
- Elbehti, A.; Brasseur, G.; Lemesle-Meunier, D. First evidence for existence of an uphill electron transfer through the bc(1) and NADH-Q oxidoreductase complexes of the acidophilic obligate chemolithotrophic ferrous ion-oxidizing bacterium Thiobacillus ferrooxidans. J. Bacteriol. 2000, 182, 3602–3606. [Google Scholar] [CrossRef] [PubMed]
- Brasseur, G.; Bruscella, P.; Bonnefoy, V.; Lemesle-Meunier, D. The bc1 complex of the iron-grown acidophilic chemolithotrophic bacterium Acidithiobacillus ferrooxidans functions in the reverse but not in the forward direction. Is there a second bc1 complex? Biochim. Biophys. Acta 2002, 1555, 37–43. [Google Scholar] [CrossRef]
- Brasseur, G.; Levican, G.; Bonnefoy, V.; Holmes, D.; Jedlicki, E.; Lemesle-Meunier, D. Apparent redundancy of electron transfer pathways via bc1 complexes and terminal oxidases in the extremophilic chemolithoautotrophic Acidithiobacillus ferrooxidans. Biochim. Biophys. Acta 2004, 1656, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Dispirito, A.A.; Tuovinen, O.H. Uranous ion oxidation and carbon dioxide fixation by Thiobacillus ferrooxidans. Arch. Microbiol. 1982, 133, 28–32. [Google Scholar] [CrossRef]
- Finneran, K.T.; Housewright, M.E.; Lovley, D.R. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 2002, 4, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Beller, H.R. Anaerobic, nitrate-dependent oxidation of U(IV) oxide minerals by the chemolithoautotrophic bacterium Thiobacillus denitrificans. Appl. Environ. Microbiol. 2005, 71, 2170–2174. [Google Scholar] [CrossRef] [PubMed]
- Beller, H.R.; Legler, T.C.; Bourguet, F.; Letain, T.E.; Kane, S.R.; Coleman, M. A Identification of c-type cytochromes involved in anaerobic, bacterial U(IV) oxidation. Biodegradation 2009, 20, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Dispirito, A.A.; Tuovinen, O.H. Kinetics of uranous ion and ferrous iron oxidation by Thiobacillus ferrooxidans. Arch. Microbiol. 1982, 133, 33–37. [Google Scholar] [CrossRef]
- Nies, D.; Silver, S. Molecular Microbiology of Heavy Metals; Springer-Verlag: Berlin, Germany, 2007. [Google Scholar]
- Bruins, M.R.; Kapil, S.; Oehme, F.W. Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 2000, 45, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H.; Silver, S. Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. 1995, 14, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Holmes, D.S. Metal resistance in acidophilic microorganisms and its significance for biotechnologies. Appl. Microbiol. Biotechnol. 2014, 98, 8133–8144. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.A.; von Bernath, D.; Jerez, C.A. Heavy metal resistance strategies of acidophilic bacteria and their acquisition: Importance for biomining and bioremediation. Biol. Res. 2013, 46, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Silver, S.; Phung, L.T. A bacterial view of the periodic table: Genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 2005, 32, 587–605. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Orell, A.; Navarro, C.A.; Arancibia, R.; Mobarec, J.C.; Jerez, C.A. Life in blue: Copper resistance mechanisms of bacteria and archaea used in industrial biomining of minerals. Biotechnol. Adv. 2010, 28, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Ossandon, F.J.; Lövgren, L.; Holmes, D.S. Metal resistance or tolerance? Acidophiles confront high metal loads via both abiotic and biotic mechanisms. Front. Microbiol. 2014, 5, 157. [Google Scholar] [CrossRef] [PubMed]
- Di Toro, D.M.; Allen, H.E.; Bergman, H.L.; Meyer, J.S.; Paquin, P.R.; Santore, R.C. Biotic ligand model of the acute toxicity of metals. 1. Technical basis. Environ. Toxicol. Chem. 2001, 20, 2383–2396. [Google Scholar] [CrossRef] [PubMed]
- Moberly, J.G.; Staven, A.R.I.; Sani, R.K. Influence of pH and inorganic phosphate on toxicity of zinc to Arthrobacter sp. isolated from sediments. Environ. Sci. Technol. 2010, 44, 7302–7308. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Dopson, M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007, 15, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Slonczewski, J.L.; Fujisawa, M.; Dopson, M.; Krulwich, T.A. Cytoplasmic pH Measurement and Homeostasis in Bacteria and Archaea. Adv. Microb Physiol. 2009, 55, 1–79. [Google Scholar] [PubMed]
- Heijerick, D.G.; de Schamphelaere, K.A.C.; Janssen, C.R. Biotic ligand model development predicting Zn toxicity to the alga Pseudokirchneriella subcapitata: Possibilities and limitations. Comp. Biochem. Physiol. 2002, 133, 207–218. [Google Scholar] [CrossRef]
- Mertens, J.; Degryse, F.; Springael, D.; Smolders, E. Zinc toxicity to nitrification in soil and soilless culture can be predicted with the same biotic ligand model. Environ. Sci. Technol. 2007, 41, 2992–2997. [Google Scholar] [CrossRef] [PubMed]
- Mangold, S.; Potrykus, J.; Björn, E.; Lövgren, L.; Dopson, M. Extreme zinc tolerance in acidophilic microorganisms from the bacterial and archaeal domains. Extremophiles 2013, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Maezato, Y.; Johnson, T.; McCarthy, S.; Dana, K.; Blum, P. Metal Resistance and Lithoautotrophy in the Extreme Thermoacidophile Metallosphaera Sedula. J. Bacteriol. 2012, 194, 6856–6863. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, C.; Ji, G.; Ramírez, J.L.; Silver, S. Resistance to arsenic compounds in microorganisms. FEMS Microbiol. Rev. 1994, 15, 355–367. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.; Ai, C.; Wheaton, G.; Tevatia, R.; Eckrich, V.; Kelly, R.; Blum, P. Role of an archaeal PitA transporter in the copper and arsenic resistance of Metallosphaera sedula, an extreme thermoacidophile. J. Bacteriol. 2014, 196, 3562–35670. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.L.; Malamy, M.H. Arsenate resistant mutants of Escherichia coli and phosphate transport. Biochem. Biophys. Res. Commun. 1970, 40, 496–503. [Google Scholar] [CrossRef]
- Willsky, G.R.; Malamy, M.H. Effect of arsenate on inorganic phosphate transport in Escherichia coli. J. Bacteriol. 1980, 144, 356–365. [Google Scholar] [PubMed]
- Nelson, D.L.; Kennedy, E.P. Magnesium transport in Escherichia coli. Inhibition by cobaltous ion. J. Biol. Chem. 1971, 246, 3042–3049. [Google Scholar] [PubMed]
- Park, M.H.; Wong, B.B.; Lusk, J.E. Mutants in three genes affecting transport of magnesium in Escherichia coli: Genetics and physiology. J. Bacteriol. 1976, 126, 1096–1103. [Google Scholar] [PubMed]
- Bini, E. Archaeal transformation of metals in the environment. FEMS Microbiol. Ecol. 2010, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Chivers, P.T.; Sauer, R.T. Regulation of high affinity nickel uptake in bacteria. Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J. Biol. Chem. 2000, 275, 19735–19741. [Google Scholar] [CrossRef] [PubMed]
- LaPaglia, C.; Hartzell, P.L. Stress-induced production of biofilm in thehyperthermophile Archaeoglobus fulgidus. Appl. Environ. Microbiol. 1997, 63, 3158–3163. [Google Scholar] [PubMed]
- Teitzel, G.M.; Parsek, M.R. Heavy Metal Resistance of Biofilm and Planktonic Pseudomonas aeruginosa Heavy Metal Resistance of Biofilm and Planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2003, 69, 2313–2320. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.J.; Ceri, H.; Turner, R.J. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 2007, 5, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Macaskie, L.E.; Bonthrone, K.M.; Yong, P.; Goddard, D.T. Enzymically mediated bioprecipitation of uranium by a Citrobacter sp.: A concerted role for exocellular lipopolysaccharide and associated phosphatase in biomineral formation. Microbiology 2000, 146, 1855–1867. [Google Scholar] [PubMed]
- Merroun, M.; Hennig, C.; Rossberg, A.; Geipel, G.; Reich, T. Molecular and atomic analysis of uranium complexes formed by three eco-types of Acidithiobacillus ferrooxidans. Biochem. Soc. Trans. 2002, 30, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Merroun, M.L.; Raff, J.; Rossberg, A.; Hennig, C.; Reich, T.; Selenska-pobell, S. Complexation of Uranium by Cells and S-Layer Sheets of Bacillus sphaericus JG-A12. Appl. Environ. Microbiol. 2005, 71, 5532–5543. [Google Scholar] [CrossRef] [PubMed]
- Sleytr, U.B.; Schuster, B.; Egelseer, E.M.; Pum, D. S-layers: Principles and applications. FEMS Microbiol. Rev. 2014, 38, 823–864. [Google Scholar] [CrossRef] [PubMed]
- Palmgren, M.G.; Nissen, P. P-type ATPases. Annu. Rev. Biophys. 2011, 40, 243–266. [Google Scholar] [CrossRef] [PubMed]
- Argüello, J.M. Identification of Ion-Selectivity Determinants in Heavy-Metal Transport P1B-type ATPases. J. Membr. Biol. 2003, 195, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Argüello, J.M.; Eren, E.; González-Guerrero, M. The structure and function of heavy metal Transport P1B-ATPases. Biometals 2007, 20, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Argüello, J.M.; González-Guerrero, M.; Raimunda, D. Bacterial transition metal P(1B)-ATPases: Transport mechanism and roles in virulence. Biochemistry 2011, 50, 9940–9949. [Google Scholar] [CrossRef] [PubMed]
- Moraleda-Muñoz, A.; Pérez, J.; Extremera, A.L.; Muñoz-Dorado, J. Expression and physiological role of three Myxococcus xanthus copper-dependent P1B-type ATPases during bacterial growth and development. Appl. Environ. Microbiol. 2010, 76, 6077–6084. [Google Scholar] [CrossRef] [PubMed]
- Rensing, C.; Ghosh, M.; Rosen, B.P. Families of soft-metal-ion-transporting ATPases. J. Bacteriol. 1999, 181, 5891–5897. [Google Scholar] [PubMed]
- Solioz, M.; Vulpe, C. CPx-type ATPases: A class of P-type ATPases that pump heavy metals. Trends Biochem. Sci. 1996, 21, 237–241. [Google Scholar] [CrossRef]
- Völlmecke, C.; Drees, S.L.; Reimann, J.; Albers, S.-V.; Lübben, M. The ATPases CopA and CopB both contribute to copper resistance of the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 2012, 158, 1622–1633. [Google Scholar] [CrossRef] [PubMed]
- Chávez, F.P.; Lünsdorf, H.; Jerez, C.A. Growth of polychlorinated-biphenyl-degrading bacteria in the presence of biphenyl and chlorobiphenyls generates oxidative stress and massive accumulation of inorganic polyphosphate. Appl. Environ. Microbiol. 2004, 70, 3064–3072. [Google Scholar] [CrossRef] [PubMed]
- Seufferheld, M.J.; Alvarez, H.M.; Farias, M.E. Role of polyphosphates in microbial adaptation to extreme environments. Appl. Environ. Microbiol. 2008, 74, 5867–5874. [Google Scholar] [CrossRef] [PubMed]
- Rao, N.N.; Gómez-García, M.R.; Kornberg, A. Inorganic polyphosphate: Essential for growth and survival. Annu. Rev. Biochem. 2009, 78, 605–647. [Google Scholar] [CrossRef] [PubMed]
- Keasling, J.D. Regulation of intracellular toxic metals and other cations by hydrolysis of polyphosphate. Ann. N. Y. Acad. Sci. 1997, 829, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.; Jerez, C.A. Copper ions stimulate polyphosphate degradation and phosphate efflux in Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2004, 70, 5177–5182. [Google Scholar] [CrossRef] [PubMed]
- Remonsellez, F.; Orell, A.; Jerez, C.A. Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: Possible role of polyphosphate metabolism. Microbiology 2006, 152, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Schurig-Briccio, L.A.; Gennis, R.B. Characterization of the PIB-Type ATPases Present in Thermus thermophilus. J. Bacteriol. 2012, 194, 4107–4113. [Google Scholar] [CrossRef] [PubMed]
- González-Guerrero, M.; Raimunda, D.; Cheng, X.; Argüello, J.M. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 2010, 78, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.G.; Winterstein, C.; Saribas, A.S.; Alben, J.O.; Daldal, F. Roles of the ccoGHIS gene products in the biogenesis of the cbb(3)-type cytochrome c oxidase. J. Mol. Biol. 2000, 297, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Hassani, B.K.; Astier, C.; Nitschke, W.; Ouchane, S. CtpA, a copper-translocating P-type ATPase involved in the biogenesis of multiple copper-requiring enzymes. J. Biol. Chem. 2010, 285, 19330–19337. [Google Scholar] [CrossRef] [PubMed]
- Villafane, A.; Voskoboynik, Y.; Ruhl, I.; Sannino, D.; Maezato, Y.; Blum, P.; Bini, E. CopR of Sulfolobus solfataricus represents a novel class of archaeal-specific copper-responsive activators of transcription. Microbiology 2011, 157, 2808–2817. [Google Scholar] [CrossRef] [PubMed]
- Ettema, T.J.G.; Huynen, M.A.; de Vos, W.M.; van der Oost, J. TRASH: A novel metal-binding domain predicted to be involved in heavy-metal sensing, trafficking and resistance. Trends Biochem. Sci. 2003, 28, 170–173. [Google Scholar] [CrossRef]
- Villafane, A.A.; Voskoboynik, Y.; Cuebas, M.; Ruhl, I.; Bini, E. Response to excess copper in the hyperthermophile Sulfolobus solfataricus strain 98/2. Biochem. Biophys. Res. Commun. 2009, 385, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Ettema, T.J.G.; Brinkman, A.B.; Lamers, P.P.; Kornet, N.G.; de Vos, W.M.; van der Oost, J. Molecular characterization of a conserved archaeal copper resistance (cop) gene cluster and its copper-responsive regulator in Sulfolobus solfataricus P2. Microbiology 2006, 152, 1969–1979. [Google Scholar] [CrossRef] [PubMed]
- Deigweiher, K.; Drell, T.L.; Prutsch, A.; Scheidig, A.J.; Lübben, M. Expression, isolation, and crystallization of the catalytic domain of CopB, a putative copper transporting ATPase from the thermoacidophilic archaeon Sulfolobus solfataricus. J. Bioenerg. Biomembr. 2004, 36, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Mana-Capelli, S.; Mandal, A.K.; Argüello, J.M. Archaeoglobus fulgidus CopB is a thermophilic Cu2+-ATPase: Functional role of its histidine-rich-N-terminal metal binding domain. J. Biol. Chem. 2003, 278, 40534–40541. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.K.; Cheung, W.D.; Argüello, J.M. Characterization of a thermophilic P-type Ag+/Cu+-ATPase from the extremophile Archaeoglobus fulgidus. J. Biol. Chem. 2002, 277, 7201–7208. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.K.; Argüello, J.M. Functional roles of metal binding domains of the Archaeoglobus fulgidus Cu(+)-ATPase CopA. Biochemistry 2003, 42, 11040–11047. [Google Scholar] [CrossRef] [PubMed]
- Sazinsky, M.H.; Agarwal, S.; Argüello, J.M.; Rosenzweig, A.C. Structure of the actuator domain from the Archaeoglobus fulgidus Cu(+)-ATPase. Biochemistry 2006, 45, 9949–9955. [Google Scholar] [CrossRef] [PubMed]
- Sazinsky, M.H.; Mandal, A.K.; Argüello, J.M.; Rosenzweig, A.C. Structure of the ATP binding domain from the Archaeoglobus fulgidus Cu+-ATPase. J. Biol. Chem. 2006, 281, 11161–11166. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Hong, D.; Desai, N.K.; Sazinsky, M.H.; Argüello, J.M.; Rosenzweig, A.C. Structure and interactions of the C-terminal metal binding domain of Archaeoglobus fulgidus CopA. Proteins 2010, 78, 2450–2458. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Benavides, T.; McCann, C.J.; Arguello, J.M. The mechanism of Cu+ transport ATPases: Interaction with Cu+ chaperones and the role of transient metal binding sites. J. Biol. Chem. 2012, 288, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.S.; Wu, C.-C.; Cardozo, T.; Stokes, D.L. The architecture of CopA from Archeaoglobus fulgidus studied by cryo-electron microscopy and computational docking. Structure 2011, 19, 1219–1232. [Google Scholar] [CrossRef] [PubMed]
- González-Guerrero, M.; Argüello, J.M. Mechanism of Cu+-transporting ATPases: Soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc. Natl. Acad. Sci. USA 2008, 105, 5992–5997. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Dopson, M.; Wexler, M.; Sawers, R.G.; Bond, P.L. Molecular insight into extreme copper resistance in the extremophilic archaeon “Ferroplasma acidarmanus” Fer1. Microbiology 2005, 151, 2637–2646. [Google Scholar] [CrossRef] [PubMed]
- Orell, A.; Remonsellez, F.; Arancibia, R.; Jerez, C. A Molecular characterization of copper and cadmium resistance determinants in the biomining thermoacidophilic archaeon Sulfolobus metallicus. Archaea 2013, 2013, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, A.; Rao, N.N.; Ault-Riché, D. Inorganic polyphosphate: A molecule of many functions. Annu. Rev. Biochem. 1999, 68, 89–125. [Google Scholar] [CrossRef] [PubMed]
- Orell, A.; Navarro, C.A.; Rivero, M.; Aguilar, J.S.; Jerez, C.A. Inorganic polyphosphates in extremophiles and their possible functions. Extremophiles 2012, 16, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.A.; Orellana, L.H.; Mauriaca, C.; Jerez, C.A. Transcriptional and functional studies of Acidithiobacillus ferrooxidans genes related to survival in the presence of copper. Appl. Environ. Microbiol. 2009, 75, 6102–6109. [Google Scholar] [CrossRef] [PubMed]
- Rensing, C.; Grass, G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 2003, 27, 197–213. [Google Scholar] [CrossRef]
- Cardona, S.T.; Chávez, F.P.; Jerez, C.A. The exopolyphosphatase gene from sulfolobus solfataricus: Characterization of the first gene found to be involved in polyphosphate metabolism in archaea. Appl. Environ. Microbiol. 2002, 68, 4812–4819. [Google Scholar] [CrossRef] [PubMed]
- Mathema, V.B.; Thakuri, B.C.; Sillanpää, M. Bacterial mer operon-mediated detoxification of mercurial compounds: A short review. Arch. Microbiol. 2011, 193, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.S.; Barkay, T. The mercury resistance operon: From an origin in a geothermal environment to an efficient detoxification machine. Front. Microbiol. 2012, 3, 349. [Google Scholar] [CrossRef] [PubMed]
- Barkay, T.; Kritee, K.; Boyd, E.; Geesey, G. A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ. Microbiol. 2010, 12, 2904–2917. [Google Scholar] [CrossRef] [PubMed]
- Barkay, T.; Miller, S.M.; Summers, A.O. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 2003, 27, 355–384. [Google Scholar] [CrossRef]
- Simbahan, J.; Kurth, E.; Schelert, J.; Dillman, A.; Moriyama, E.; Jovanovich, S.; Blum, P. Community analysis of a mercury hot spring supports occurrence of domain-specific forms of mercuric reductase. Appl. Environ. Microbiol. 2005, 71, 8836–8845. [Google Scholar] [CrossRef] [PubMed]
- King, J.K.; Kostka, J.E.; Frischer, M.E.; Saunders, F.M. Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments. Appl. Environ. Microbiol. 2000, 66, 2430–2437. [Google Scholar] [CrossRef] [PubMed]
- Hintelmann, H. Organomercurials. Their formation and pathways in the environment. Met. Ions Life Sci. 2010, 7, 365–401. [Google Scholar] [PubMed]
- Lin, C.; Yee, N.; Barkay, T. Microbial Transformations in the Mercury Cycle; Liu, G., Cai, Y., O’Driscoll, N., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Pak, K.; Bartha, R. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Appl. Environ. Microbiol. 1998, 64, 1987–1990. [Google Scholar] [PubMed]
- Silver, S.; Phung, L.T. Bacterial heavy metal resistance: New surprises. Annu. Rev. Microbiol. 1996, 50, 753–789. [Google Scholar] [CrossRef] [PubMed]
- Osborn, A.M.; Bruce, K.D.; Strike, P.; Ritchie, D.A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 2006, 19, 239–262. [Google Scholar] [CrossRef]
- Schelert, J.; Dixit, V.; Hoang, V.; Simbahan, J.; Drozda, M.; Blum, P. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J. Bacteriol. 2004, 186, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Schelert, J.; Drozda, M.; Dixit, V.; Dillman, A.; Blum, P. Regulation of mercury resistance in the crenarchaeote Sulfolobus solfataricus. J. Bacteriol. 2006, 188, 7141–7150. [Google Scholar] [CrossRef] [PubMed]
- Freedman, Z.; Zhu, C.; Barkay, T. Mercury Resistance and Mercuric Reductase Activities and Expression among Chemotrophic Thermophilic Aquificae. Appl. Environ. Microbiol. 2012, 78, 6568–6575. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Freedman, Z.; Lu-Irving, P.; Kaletsky, R.; Barkay, T. An initial characterization of the mercury resistance (mer) system of the thermophilic bacterium Thermus thermophilus HB27. FEMS Microbiol. Ecol. 2009, 67, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Sugio, T.; Komoda, T.; Okazaki, Y.; Takeda, Y.; Nakamura, S.; Takeuchi, F. Volatilization of Metal Mercury from Organomercurials by Highly Mercury-Resistant Acidithiobacillus ferrooxidans MON-1. Biosci. Biotechnol. Biochem. 2010, 74, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Sugio, T.; Iwahori, K.; Takeuchi, F.; Negishi, A.; Maeda, T.; Kamimura, K. Cytochrome c oxidase purified from a mercury-resistant strain of Acidithiobacillus ferrooxidans volatilizes mercury. J. Biosci. Bioeng. 2001, 92, 44–49. [Google Scholar] [CrossRef]
- Páez-Espino, D.; Tamames, J.; de Lorenzo, V.; Cánovas, D. Microbial responses to environmental arsenic. Biometals 2009, 22, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.P.; Liu, Z. Transport pathways for arsenic and selenium: A minireview. Environ. Int. 2009, 35, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, R.; Rosen, B.P.; Phung, L.T.; Silver, S. Microbial arsenic: From geocycles to genes and enzymes. FEMS Microbiol. Rev. 2002, 26, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhou, T.; Kuroda, M.; Rosen, B.P. Metalloid resistance mechanisms in prokaryotes. J. Biochem. 1998, 123, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, R.; Rosen, B.P. Arsenate reductases in prokaryotes and eukaryotes. Environ. Health Perspect. 2002, 110, 745–748. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.-L.; Liu, Z.; Rosen, B.P. As(III) and Sb(III) uptake by GlpF and efflux by ArsB in Escherichia coli. J. Biol. Chem. 2004, 279, 18334–18341. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Rosen, B.P. Dual mode of energy coupling by the oxyanion-translocating ArsB protein. J. Bacteriol. 1995, 177, 385–389. [Google Scholar] [PubMed]
- Lin, Y.-F.; Walmsley, A.R.; Rosen, B.P. An arsenic metallochaperone for an arsenic detoxification pump. Proc. Natl. Acad. Sci. USA 2006, 103, 15617–15622. [Google Scholar] [CrossRef] [PubMed]
- Kotze, A.A.; Tuffin, I.M.; Deane, S.M.; Rawlings, D.E. Cloning and characterization of the chromosomal arsenic resistance genes from Acidithiobacillus caldus and enhanced arsenic resistance on conjugal transfer of ars genes located on transposon TnAtcArs. Microbiology 2006, 152, 3551–3560. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, S.; Xiao, X.; Huang, X.; You, D.; Zhou, X.; Deng, Z. arsRBOCT arsenic resistance system encoded by linear plasmid pHZ227 in Streptomyces sp. strain FR-008. Appl. Environ. Microbiol. 2006, 72, 3738–3742. [Google Scholar] [CrossRef] [PubMed]
- Sekine, M.; Tanikawa, S.; Omata, S.; Saito, M.; Fujisawa, T.; Tsukatani, N.; Tajima, T.; Sekigawa, T.; Kosugi, H.; Matsuo, Y.; et al. Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environ. Microbiol. 2006, 8, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Achour-Rokbani, A.; Cordi, A.; Poupin, P.; Bauda, P.; Billard, P. Characterization of the ars gene cluster from extremely arsenic-resistant Microbacterium sp. strain A33. Appl. Environ. Microbiol. 2010, 76, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Butcher, B.G.; Deane, S.M.; Rawlings, D.E. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl. Environ. Microbiol. 2000, 66, 1826–1833. [Google Scholar] [CrossRef] [PubMed]
- López-Maury, L.; Florencio, F.J.; Reyes, J.C. Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2003, 185, 5363–5371. [Google Scholar] [CrossRef] [PubMed]
- Vorontsov, I.I.; Minasov, G.; Brunzelle, J.S.; Shuvalova, L.; Kiryukhina, O.; Collart, F.R.; Anderson, W.F. Crystal structure of an apo form of Shigella flexneri ArsH protein with an NADPH-dependent FMN reductase activity. Protein Sci. 2007, 16, 2483–2490. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Dopson, M.; Wexler, M.; Sawers, R.G.; Stemmler, A.; Rosen, B.P.; Bond, P.L. Extreme arsenic resistance by the acidophilic archaeon “Ferroplasma acidarmanus” Fer1. Extremophiles 2007, 11, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Gihring, T.M.; Bond, P.L.; Peters, S.C.; Banfield, J.F. Arsenic resistance in the archaeon “Ferroplasma acidarmanus”: New insights into the structure and evolution of the ars genes. Extremophiles 2003, 7, 123–130. [Google Scholar] [PubMed]
- Ruepp, A.; Graml, W.; Santos-Martinez, M.L.; Koretke, K.K.; Volker, C.; Mewes, H.W.; Frishman, D.; Stocker, S.; Lupas, A.N.; Baumeister, W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 2000, 407, 508–513. [Google Scholar] [PubMed]
- Fütterer, O.; Angelov, A.; Liesegang, H.; Gottschalk, G.; Schleper, C.; Schepers, B.; Dock, C.; Antranikian, G.; Liebl, W. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc. Natl. Acad. Sci. USA 2004, 101, 9091–9096. [Google Scholar] [CrossRef] [PubMed]
- Cozen, A.E.; Weirauch, M.T.; Pollard, K.S.; Bernick, D.L.; Stuart, J.M.; Lowe, T.M. Transcriptional map of respiratory versatility in the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. J. Bacteriol. 2009, 191, 782–794. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Lindström, E.B.; Hallberg, K.B. Chromosomally encoded arsenical resistance of the moderately thermophilic acidophile Acidithiobacillus caldus. Extremophiles 2001, 5, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Wakao, N.; Kimura, T.; Sakka, K.; Ohmiya, K. Expression and regulation of the arsenic resistance operon of Acidiphilium multivorum AIU 301 plasmid pKW301 in Escherichia coli. Appl. Environ. Microbiol. 1998, 64, 411–418. [Google Scholar] [PubMed]
- Tuffin, I.M.; Hector, S.B.; Deane, S.M.; Rawlings, D.E. Resistance determinants of a highly arsenic-resistant strain of Leptospirillum ferriphilum isolated from a commercial biooxidation tank. Appl. Environ. Microbiol. 2006, 72, 2247–2253. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kennedy, S.P.; Fasiludeen, S.; Rensing, C.; DasSarma, S. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J. Bacteriol. 2004, 186, 3187–3194. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Rosen, B.P.; Zhang, Y.; Wang, G.; Franke, S.; Rensing, C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 2075–2080. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Liu, G.; Rensing, C.; Wang, G. Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 2009, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Sehlin, H.M.; Lindstrom, E.B. Oxidation and reduction of arsenic by Sulfolobus acidocaldarius strain BC. FEMS Microbiol. Lett. 1992, 93, 87–92. [Google Scholar] [CrossRef]
- Okibe, N.; Koga, M.; Sasaki, K.; Hirajima, T.; Heguri, S.; Asano, S. Simultaneous oxidation and immobilization of arsenite from refinery waste water by thermoacidophilic iron-oxidizing archaeon, Acidianus brierleyi. Miner. Eng. 2013, 48, 126–134. [Google Scholar] [CrossRef]
- Battaglia-Brunet, F.; Crouzet, C.; Breeze, D.; Tris, H.; Morin, D. Decreased leachability of arsenic linked to biological oxidation of As(III) in solid wastes from bioleaching liquors. Hydrometallurgy 2011, 107, 34–39. [Google Scholar] [CrossRef]
- Van Lis, R.; Nitschke, W.; Duval, S.; Schoepp-Cothenet, B. Arsenics as bioenergetic substrates. Biochim. Biophys. Acta 2013, 1827, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, E.; Brugna, M.; Baymann, F.; Muller, D.; Lièvremont, D.; Lett, M.-C.; Nitschke, W. Arsenite oxidase, an ancient bioenergetic enzyme. Mol. Biol. Evol. 2003, 20, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Lett, M.-C.; Muller, D.; Lièvremont, D.; Silver, S.; Santini, J. Unified nomenclature for genes involved in prokaryotic aerobic arsenite oxidation. J. Bacteriol. 2012, 194, 207–208. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M. Growth in sulfidic mineral environments: Metal resistance mechanisms in acidophilic micro-organisms. Microbiology 2003, 149, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Salzano, A.M.; Febbraio, F.; Farias, T.; Cetrangolo, G.P.; Nucci, R.; Scaloni, A.; Manco, G. Redox stress proteins are involved in adaptation response of the hyperthermoacidophilic archaeon Sulfolobus solfataricus to nickel challenge. Microb. Cell Fact. 2007, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Lagorce, A.; Fourçans, A.; Dutertre, M.; Bouyssiere, B.; Zivanovic, Y.; Confalonieri, F. Genome-wide transcriptional response of the archaeon Thermococcus gammatolerans to cadmium. PLoS ONE 2012, 7, e41935. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wu, N.; Li, J.; Liu, Y.; Guo, J.; Yao, B.; Fan, Y. Nickel-resistant determinant from Leptospirillum ferriphilum. Appl. Environ. Microbiol. 2007, 73, 2364–2368. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Tian, J.; Zhang, S.; Wu, N.; Fan, Y. Identification of the transcriptional regulator NcrB in the nickel resistance determinant of Leptospirillum ferriphilum UBK03. PLoS ONE 2011, 6, e17367. [Google Scholar] [CrossRef] [PubMed]
- Merroun, M.L.; Selenska-Pobell, S. Bacterial interactions with uranium: An environmental perspective. J. Contam. Hydrol. 2008, 102, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Cologgi, D.L.; Lampa-Pastirk, S.; Speers, A.M.; Kelly, S.D.; Reguera, G. Extracellular reduction of uranium via Geobacter conductive pili as a protective cellular mechanism. Proc. Natl. Acad. Sci. USA 2011, 108, 15248–15252. [Google Scholar] [CrossRef] [PubMed]
- Martinez, R.J.; Beazley, M.J.; Taillefert, M.; Arakaki, A.K.; Skolnick, J.; Sobecky, P.A. Aerobic uranium (VI) bioprecipitation by metal-resistant bacteria isolated from radionuclide- and metal-contaminated subsurface soils. Environ. Microbiol. 2007, 9, 3122–3133. [Google Scholar] [CrossRef] [PubMed]
- Renninger, N.; Knopp, R.; Nitsche, H.; Clark, D.S.; Keasling, J.D. Uranyl precipitation by Pseudomonas aeruginosa via controlled polyphosphate metabolism. Appl. Environ. Microbiol. 2004, 70, 7404–7412. [Google Scholar] [CrossRef] [PubMed]
- Merroun, M.L.; Nedelkova, M.; Ojeda, J.J.; Reitz, T.; Fernández, M.L.; Arias, J.M.; Romero-González, M.; Selenska-Pobell, S. Bio-precipitation of uranium by two bacterial isolates recovered from extreme environments as estimated by potentiometric titration, TEM and X-ray absorption spectroscopic analyses. J. Hazard. Mater. 2011, 197, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Reitz, T.; Merroun, M.L.; Rossberg, A.; Steudtner, R.; Selenska-Pobell, S. Bioaccumulation of U(VI) by Sulfolobus acidocaldarius under moderate acidic conditions. Radiochim. Acta 2011, 99, 543–554. [Google Scholar] [CrossRef]
- Kashefi, K.; Lovley, D.R. Reduction of Fe(III), Mn(IV), and Toxic Metals at 100 C by Pyrobaculum islandicum. Appl. Environ. Microbiol. 2000, 66, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.J.; Gillow, J.B.; Dodge, C.J.; Harris, R.; Beveridge, T.J.; Papenguth, H.W. Uranium association with halophilic and non-halophilic bacteria and archaea. Radiochim. Acta 2004, 92, 481–488. [Google Scholar] [CrossRef]
- Reitz, T.; Merroun, M.; Rossberg, A.; Selenska-Pobell, S. Interactions of Sulfolobus acidocaldarius with uranium. Radiochim. Acta 2010, 98, 249–257. [Google Scholar] [CrossRef]
- Beazley, M.J.; Martinez, R.J.; Sobecky, P.A.; Webb, S.M.; Taillefert, M. Uranium biomineralization as a result of bacterial phosphatase activity: Insights from bacterial isolates from a contaminated subsurface. Environ. Sci. Technol. 2007, 41, 5701–5707. [Google Scholar] [CrossRef] [PubMed]
- Selenska-pobell, S.; Merroun, M. Accumulation of heavy metals by microorganisms: Bio-mineralization and nanocluster formation. In Prokaryotic Cell Wall Compounds; König, H., Claus, H., Varma, A., Eds.; Springer: Berlin, Germany, 2010; pp. 483–500. [Google Scholar]
- Merroun, M.; Hennig, C.; Rossberg, A.; Reich, T.; Selenska-Pobell, S. Characterization of U(VI)-Acidithiobacillus ferrooxidans complexes using EXAFS, transmission electron microscopy, and energy-dispersive X-ray analysis. Radiochim. Acta 2003, 91, 583–592. [Google Scholar] [CrossRef]
- Malekzadeh, F.; Latifi, A.M.; Shahamat, M.; Levin, M.; Colwell, R.R. Effects of selected physical and chemical parameters on uranium uptake by the bacterium Chryseomonas MGF-48. World J. Microbiol. Biotechnol. 2002, 18, 599–602. [Google Scholar] [CrossRef]
- Panak, P.J.; Knopp, R.; Booth, C.H.; Nitsche, H. Spectroscopic studies on the interaction of U(VI) with Bacillus sphaericus. Radiochim. Acta 2002, 90, 779–783. [Google Scholar] [CrossRef]
- Bäuerlein, E. Biomineralization of unicellular organisms: An unusual membrane biochemistry for the production of inorganic nano- and microstructures. Angew. Chem. Int. Ed. 2003, 42, 614–641. [Google Scholar] [CrossRef] [PubMed]
- Eichler, J. Facing extremes: Archaeal surface-layer (glyco)proteins. Microbiology 2003, 149, 3347–3351. [Google Scholar] [CrossRef] [PubMed]
- Dispirito, A.A.; Dugan, P.R.; Tuovinen, O.H. Sorption of Thiobacillus ferrooxidans to particulate material. Biotechnol. Bioeng. 1983, 25, 1163–1168. [Google Scholar] [CrossRef] [PubMed]
- Gehrke, T.; Telegdi, J.; Thierry, D.; Sand, W. Importance of Extracellular Polymeric Substances from Thiobacillus ferrooxidans for Bioleaching. Appl. Environ. Microbiol. 1998, 64, 2743–2747. [Google Scholar] [PubMed]
- Harneit, K.; Göksel, A.; Kock, D.; Klock, J.H.; Gehrke, T.; Sand, W. Adhesion to metal sulfide surfaces by cells of Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans and Leptospirillum ferrooxidans. Hydrometallurgy 2006, 83, 245–254. [Google Scholar] [CrossRef]
- Africa, C.J.; van Hille, R.P.; Sand, W.; Harrison, S.T.L. Investigation and in situ visualisation of interfacial interactions of thermophilic microorganisms with metal-sulphides in a simulated heap environment. Miner. Eng. 2013, 48, 100–107. [Google Scholar] [CrossRef]
- Zhang, R.; Bellenberg, S.; Castro, L.; Neu, T.R.; Sand, W.; Vera, M. Colonization and biofilm formation of the extremely acidophilic archaeon Ferroplasma acidiphilum. Hydrometallurgy 2014, 150, 245–252. [Google Scholar] [CrossRef]
- Noël, N.; Florian, B.; Sand, W. AFM & EFM study on attachment of acidophilic leaching organisms. Hydrometallurgy 2010, 104, 370–375. [Google Scholar]
- Rodriguez-Leiva, M.; Tributsch, H. Morphology of bacterial leaching patterns by Thiobacillus ferrooxidans on synthetic pyrite. Arch. Microbiol. 1988, 149, 401–405. [Google Scholar] [CrossRef]
- Sand, W.; Gerke, T.; Hallmann, R.; Schippers, A. Sulfur chemistry, biofilm, and the (in)direct attack—A critical evaluation of bacterial leaching. Appl. Microbiol. Biotechnol. 1995, 43, 961–966. [Google Scholar] [CrossRef]
- Sand, W.; Gehrke, T.; Jozsa, P.G.; Schippers, A. (Bio)chemistry of bacterial leaching—Direct vs. indirect bioleaching. Hydrometallurgy 2001, 59, 159–175. [Google Scholar] [CrossRef]
- Sand, W.; Gehrke, T. Extracellular polymeric substances mediate bioleaching/biocorrosion via interfacial processes involving iron(III) ions and acidophilic bacteria. Res. Microbiol. 2006, 157, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Florian, B.; Noël, N.; Thyssen, C.; Felschau, I.; Sand, W. Some quantitative data on bacterial attachment to pyrite. Miner. Eng. 2011, 24, 1132–1138. [Google Scholar] [CrossRef]
- Vera, M.; Schippers, A.; Sand, W. Progress in bioleaching: Fundamentals and mechanisms of bacterial metal sulfide oxidation-part A. Appl. Microbiol. Biotechnol. 2013, 97, 7529–7541. [Google Scholar] [CrossRef] [PubMed]
- Schippers, A.; Hedrich, S.; Vasters, J.; Drobe, M.; Sand, W.; Willscher, S. Biomining : Metal Recovery from Ores with Microorganisms. Adv. Biochem. Eng. Biotechnol. 2013, 141, 1–47. [Google Scholar]
- Vandevivere, P.; Kirchman, D.L. Attachment stimulates exopolysaccharide synthesis by a bacterium. Appl. Environ. Microbiol. 1993, 59, 3280–3286. [Google Scholar] [PubMed]
- Gehrke, T.; Hallmann, R.; Kinzler, K.; Sand, W. The EPS of Acidithiobacillus ferrooxidans—A model for structure-function relationships of attached bacteria and their physiology. Water Sci. Technol. 2001, 43, 159–167. [Google Scholar] [PubMed]
- Solari, J.A.; Huerta, G.; Escobar, B.; Vargas, T.; Badilla-Ohlbaum, R.; Rubio, J. Interfacial phenomena affecting the adhesion of Thiobacillus ferrooxidans to sulphide mineral surface. Colloids Surfaces 1992, 69, 159–166. [Google Scholar] [CrossRef]
- Blake, R.C.; Shute, E.A.; Howard, G.T. Solubilization of minerals by bacteria: Electrophoretic mobility of Thiobacillus ferrooxidans in the presence of iron, pyrite, and sulfur. Appl. Environ. Microbiol. 1994, 60, 3349–3357. [Google Scholar] [PubMed]
- Vilinska, A.; Rao, K.H. Surface Thermodynamics and Extended DLVO Theory of Acidithiobacillus ferrooxidans Cells Adhesion on Pyrite and Chalcopyrite. Open Colloid Sci. J. 2009, 2, 1–14. [Google Scholar] [CrossRef]
- Sampson, M.I.; Phillips, C.V.; Blake, R.C. Influence of the attachment of acidophilic bacteria during the oxidation of mineral sulfides. Miner. Eng. 2000, 13, 373–389. [Google Scholar] [CrossRef]
- Zhu, J.; Li, Q.; Jiao, W.; Jiang, H.; Sand, W.; Xia, J.; Liu, X.; Qin, W.; Qiu, G.; Hu, Y.; Chai, L. Adhesion forces between cells of Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans or Leptospirillum ferrooxidans and chalcopyrite. Colloids Surfaces B Biointerfaces 2012, 94, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Andrews, G.F. The selective adsorption of Thiobacilli to dislocation sites on pyrite surfaces. Biotechnol. Bioeng. 1988, 31, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.J.; Rutenberg, A.D. Microbial response to surface microtopography: The role of metabolism in localized mineral dissolution. Chem. Geol. 2001, 180, 19–32. [Google Scholar] [CrossRef]
- Ohmura, N.; Kitamura, K.; Saiki, H. Selective adhesion of Thiobacillus ferrooxidans to pyrite. Appl. Environ. Microbiol. 1993, 59, 4044–4050. [Google Scholar] [PubMed]
- Sanhueza, A.; Ferrer, I.J.; Vargas, T.; Amils, R.; Sánchez, C. Attachment of Thiobacillus ferrooxidans on synthetic pyrite of varying structural and electronic properties. Hydrometallurgy 1999, 51, 115–129. [Google Scholar] [CrossRef]
- Shrihari; Modak, J.M.; Kumar, R.; Gandhi, K.S. Dissolution of particles of pyrite mineral by direct attachment of Thiobacillus ferrooxidans. Hydrometallurgy 1995, 38, 175–187. [Google Scholar] [CrossRef]
- Li, Y.-Q.; Wan, D.-S.; Huang, S.-S.; Leng, F.-F.; Yan, L.; Ni, Y.-Q.; Li, H.-Y. Type IV pili of Acidithiobacillus ferrooxidans are necessary for sliding, twitching motility, and adherence. Curr. Microbiol. 2010, 60, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Pohlschroder, M.; Ghosh, A.; Tripepi, M.; Albers, S.-V. Archaeal type IV pilus-like structures—Evolutionarily conserved prokaryotic surface organelles. Curr. Opin. Microbiol. 2011, 14, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, T.J.; Pouwels, P.H.; Sára, M.; Kotiranta, A.; Lounatmaa, K.; Kari, K.; Kerosuo, E.; Haapasalo, M.; Egelseer, E.M.; Schocher, I.; et al. Functions of S-layers. FEMS Microbiol. Rev. 2006, 20, 99–149. [Google Scholar] [CrossRef]
- Acuña, J.; Rojas, J.; Amaro, A.M.; Toledo, H.; Jerez, C.A. Chemotaxis of Leptospirillum ferrooxidans and other acidophilic chemolithotrophs: Comparison with the Escherichia coli chemosensory system. FEMS Microbiol. Lett. 1992, 75, 37–42. [Google Scholar] [CrossRef]
- Meyer, G.; Schneider-Merk, T.; Böhme, S.; Sand, W. A simple method for investigations on the chemotaxis of Acidithiobacillus ferrooxidans and Desulfovibrio vulgaris. Acta Biotechnol. 2002, 22, 391–399. [Google Scholar] [CrossRef]
- Banderas, A.; Guiliani, N. Bioinformatic prediction of gene functions regulated by quorum sensing in the bioleaching bacterium Acidithiobacillus ferrooxidans. Int. J. Mol. Sci. 2013, 14, 16901–16916. [Google Scholar] [CrossRef] [PubMed]
- Bellenberg, S.; Díaz, M.; Noël, N.; Sand, W.; Poetsch, A.; Guiliani, N.; Vera, M. Biofilm formation, communication and interactions of leaching bacteria during colonization of pyrite and sulfur surfaces. Res. Microbiol. 2014, 165, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.M.; Castro, M.; Barriga, A.; Jerez, C.A.; Guiliani, N. The extremophile Acidithiobacillus ferrooxidans possesses a c-di-GMP signalling pathway that could play a significant role during bioleaching of minerals. Lett. Appl. Microbiol. 2012, 54, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.; Deane, S.M.; Ruiz, L.; Rawlings, D.E.; Guiliani, N. Diguanylate Cyclase Null Mutant Reveals that C-Di-GMP Pathway Regulates the Motility and Adherence of the Extremophile Bacterium Acidithiobacillus caldus. PLoS ONE 2015, 10, e0116399. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Bellenberg, S.; Mamani, S.; Ruiz, L.; Echeverría, A.; Soulère, L.; Doutheau, A.; Demergasso, C.; Sand, W.; Queneau, Y.; et al. AHL signaling molecules with a large acyl chain enhance biofilm formation on sulfur and metal sulfides by the bioleaching bacterium Acidithiobacillus ferrooxidans. Appl. Microbiol. Biotechnol. 2013, 97, 3729–3737. [Google Scholar] [CrossRef] [PubMed]
- Farah, C.; Vera, M.; Morin, D.; Haras, D.; Jerez, C.A.; Guiliani, N. Evidence for a functional quorum-sensing type AI-1 system in the extremophilic bacterium Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2005, 71, 7033–7040. [Google Scholar] [CrossRef] [PubMed]
- Bellenberg, S.; Leon-Morales, C.-F.; Sand, W.; Vera, M. Visualization of capsular polysaccharide induction in Acidithiobacillus ferrooxidans. Hydrometallurgy 2012, 129, 82–89. [Google Scholar] [CrossRef]
- Vera, M.A.; Rohwerder, T.; Bellenberg, S.; Sand, W.; Denis, Y.; Bonnefoy, V. Characterization of biofilm formation by the bioleaching acidophilic bacterium acidithiobacillus ferrooxidans by a microarray transcriptome analysis. Adv. Mater. Res. 2009, 71, 175–178. [Google Scholar] [CrossRef]
- Barreto, M.; Jedlicki, E.; Holmes, D.S. Identification of a gene cluster for the formation of extracellular polysaccharide precursors in the chemolithoautotroph Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2005, 71, 2902–2909. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Paz, M.; Gómez, M.J.; Arcas, A.; Parro, V. Environmental transcriptome analysis reveals physiological differences between biofilm and planktonic modes of life of the iron oxidizing bacteria Leptospirillum spp. in their natural microbial community. BMC Genomics 2010, 11, 404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vera, M.; Krok, B.; Bellenberg, S.; Sand, W.; Poetsch, A. Shotgun proteomics study of early biofilm formation process of Acidithiobacillus ferrooxidans ATCC 23270 on pyrite. Proteomics 2013, 13, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.G. Analysis of heat conservation during copper sulphide heap leaching. Hydrometallurgy 2000, 58, 27–41. [Google Scholar] [CrossRef]
- Olson, G.J.; Brierley, J.A.; Brierley, C.L. Bioleaching review part B: Progress in bioleaching: Applications of microbial processes by the minerals industries. Appl. Microbiol. Biotechnol. 2003, 63, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Brierley, C.L.; Brierley, J.A. Progress in bioleaching: Part B: Applications of microbial processes by the minerals industries. Appl. Microbiol. Biotechnol. 2013, 97, 7543–7552. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, Y.; Ballester, A.; Blázquez, M.L.; González, F.; Muñoz, J.A. New information on the chalcopyrite bioleaching mechanism at low and high temperature. Hydrometallurgy 2003, 71, 47–56. [Google Scholar] [CrossRef]
- Mikkelsen, D.; Kappler, U.; Webb, R.I.; Rasch, R.; McEwan, A.G.; Sly, L.I. Visualisation of pyrite leaching by selected thermophilic archaea: Nature of microorganism-ore interactions during bioleaching. Hydrometallurgy 2007, 88, 143–153. [Google Scholar] [CrossRef]
- Gautier, V.; Escobar, B.; Vargas, T. Cooperative action of attached and planktonic cells during bioleaching of chalcopyrite with Sulfolobus metallicus at 70 °C. Hydrometallurgy 2008, 94, 121–126. [Google Scholar] [CrossRef]
- Bromfield, L.; Africa, C.J.; Harrison, S.T.L.; van Hille, R.P. The effect of temperature and culture history on the attachment of Metallosphaera hakonensis to mineral sulfides with application to heap bioleaching. Miner. Eng. 2011, 24, 1157–1165. [Google Scholar] [CrossRef]
- Zolghadr, B.; Kling, A.; Koerdt, A.; Driessen, A.J.M.; Rachel, R.; Albers, S.V. Appendage-mediated surface adherence of Sulfolobus solfataricus. J. Bacteriol. 2010, 192, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Koerdt, A.; Orell, A.; Pham, T.K.; Mukherjee, J.; Wlodkowski, A.; Karunakaran, E.; Biggs, C.A.; Wright, P.C.; Albers, S.-V. Macromolecular fingerprinting of sulfolobus species in biofilm: A transcriptomic and proteomic approach combined with spectroscopic analysis. J. Proteome Res. 2011, 10, 4105–4119. [Google Scholar] [CrossRef] [PubMed]
- Koerdt, A.; Gödeke, J.; Berger, J.; Thormann, K.M.; Albers, S.-V. Crenarchaeal biofilm formation under extreme conditions. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Pohlschroder, M.; Esquivel, R.N. Archaeal type IV pili and their involvement in biofilm formation. Front. Microbiol. 2015, 6, 190. [Google Scholar] [CrossRef] [PubMed]
- Orell, A.; Peeters, E.; Vassen, V.; Jachlewski, S.; Schalles, S.; Siebers, B.; Albers, S.-V. Lrs14 transcriptional regulators influence biofilm formation and cell motility of Crenarchaea. ISME J. 2013, 7, 1886–1898. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Potrykus, J.; Wexler, M.; Bond, P.L.; Dopson, M. Biofilm development in the extremely acidophilic archaeon “Ferroplasma acidarmanus” Fer1. Extremophiles 2010, 14, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.Y.; Neu, T.R.; Bellenberg, S.; Kuhlicke, U.; Sand, W.; Vera, M. Use of lectins to in situ visualize glycoconjugates of extracellular polymeric substances in acidophilic archaeal biofilms. Microb. Biotechnol. 2015, 8, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Poli, A.; di Donato, P.; Abbamondi, G.R.; Nicolaus, B. Synthesis, production, and biotechnological applications of exopolysaccharides and polyhydroxyalkanoates by Archaea. Archaea 2011, 2011, 693253. [Google Scholar] [CrossRef] [PubMed]
- Fröls, S. Archaeal biofilms: Widespread and complex. Biochem. Soc. Trans. 2013, 41, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Jarrell, K.; Ding, Y.; Nair, D.; Siu, S. Surface Appendages of Archaea: Structure, Function, Genetics and Assembly. Life 2013, 3, 86–117. [Google Scholar] [CrossRef] [PubMed]
- Orell, A.; Fröls, S.; Albers, S.-V. Archaeal biofilms: The great unexplored. Annu. Rev. Microbiol. 2013, 67, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Miot, J.; Benzerara, K.; Kappler, A. Investigating Microbe-Mineral Interactions: Recent Advances in X-Ray and Electron Microscopy and Redox-Sensitive Methods. Annu. Rev. Earth Planet. Sci. 2014, 42, 271–289. [Google Scholar] [CrossRef]
- Gralnick, J.A.; Newman, D.K. Extracellular respiration. Mol. Microbiol. 2007, 65, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gray, H.B.; Winkler, J.R. Electron tunneling through proteins. Q. Rev. Biophys. 2003, 36, 341–372. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.J.; Butt, J.N.; Fredrickson, J.K.; Zachara, J.M.; Shi, L.; Edwards, M.J.; White, G.; Baiden, N.; Gates, A.J.; Marritt, S.J.; et al. The “porin-cytochrome” model for microbe-to-mineral electron transfer. Mol. Microbiol. 2012, 85, 201–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, G.F.; Shi, Z.; Shi, L.; Wang, Z.; Dohnalkova, A.C.; Marshall, M.J.; Fredrickson, J.K.; Zachara, J.M.; Butt, J.N.; Richardson, D.J.; et al. A Rapid electron exchange between surface-exposed bacterial cytochromes and Fe(III) minerals. Proc. Natl. Acad. Sci. USA 2013, 110, 6346–6351. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, M.; Finkel, S. Live Wires. Scientist 2013, 27, 38–43. [Google Scholar]
- Marsili, E.; Baron, D.B.; Shikhare, I.D.; Coursolle, D.; Gralnick, J.A.; Bond, D.R. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. USA 2008, 105, 3968–3973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Canstein, H.; Ogawa, J.; Shimizu, S.; Lloyd, J.R. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 2008, 74, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Brutinel, E.D.; Gralnick, J.A. Shuttling happens: Soluble flavin mediators of extracellular electron transfer in Shewanella. Appl. Microbiol. Biotechnol. 2012, 93, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Reguera, G.; McCarthy, K.D.; Mehta, T.; Nicoll, J.S.; Tuominen, M.T.; Lovley, D.R. Extracellular electron transfer via microbial nanowires. Nature 2005, 435, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Gorby, Y.A.; Yanina, S.; McLean, J.S.; Rosso, K.M.; Moyles, D.; Dohnalkova, A.; Beveridge, T.J.; Chang, I.S.; Kim, B.H.; Kim, K.S.; et al. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. USA 2006, 103, 11358–11363. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, M.Y.; Wanger, G.; Leung, K.M.; Yuzvinsky, T.D.; Southam, G.; Yang, J.; Lau, W.M.; Nealson, K.H.; Gorby, Y.A. Electrical transport along bacterial nanowires from Shewanella oneidensis MR-1. Proc. Natl. Acad. Sci. USA 2010, 107, 18127–18131. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R. Long-range electron transport to Fe(III) oxide via pili with metallic-like conductivity. Biochem. Soc. Trans. 2012, 40, 1186–1190. [Google Scholar] [CrossRef] [PubMed]
- Pirbadian, S.; Barchinger, S.E.; Leung, K.M.; Byun, H.S.; Jangir, Y.; Bouhenni, R.A.; Reed, S.B.; Romine, M.F.; Saffarini, D.A.; Shi, L.; et al. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc. Natl. Acad. Sci. USA 2014, 111, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gorby, Y.; Mclean, J.; Korenevsky, A.; Rosso, K.; El-Naggar, M.Y.; Beveridge, T.J. Redox-reactive membrane vesicles produced by Shewanella. Geobiology 2008, 6, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Snider, R.M.; Strycharz-Glaven, S.M.; Tsoi, S.D.; Erickson, J.S.; Tender, L.M. Long-range electron transport in Geobacter sulfurreducens biofilms is redox gradient-driven. Proc. Natl. Acad. Sci. USA 2012, 109, 15467–15472. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, C.; Larsen, S.; Song, J.; Dong, M.; Besenbacher, F.; Meyer, R.L.; Kjeldsen, K.U.; Schreiber, L.; Gorby, Y.A.; El-Naggar, M.Y.; et al. Filamentous bacteria transport electrons over centimetre distances. Nature 2012, 491, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Malvankar, N.S.; Vargas, M.; Nevin, K.P.; Franks, A.E.; Leang, C.; Kim, B.; Inoue, K.; Mester, T.; Covalla, S.F.; Johnson, J.P.; et al. Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 2011, 6, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Vargas, M.; Malvankar, N.S.; Tremblay, P.L.; Leang, C.; Smith, J.A.; Patel, P.; Synoeyenbos-West, O.; Nevin, K.P.; Lovley, D.R. Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. MBio 2013, 4. [Google Scholar] [CrossRef]
- Pirbadian, S.; El-Naggar, M.Y. Multistep hopping and extracellular charge transfer in microbial redox chains. Phys. Chem. Chem. Phys. 2012, 14, 13802. [Google Scholar] [CrossRef] [PubMed]
- Polizzi, N.F.; Skourtis, S.S.; Beratan, D.N. Physical constraints on charge transport through bacterial nanowires. Faraday Discuss. 2012, 155, 43. [Google Scholar] [CrossRef] [PubMed]
- Strycharz-Glaven, S.M.; Tender, L.M. Reply to the “Comment on ‘On electrical conductivity of microbial nanowires and biofilms’” N.S. Malvankar, M.T. Tuominen and D.R. Lovley. Energy Environ. Sci., 2012, 5, doi:10.1039/c2ee02613a. Energy Environ. Sci. 2012, 5, 6250–6255. [Google Scholar] [CrossRef]
- Li, Y.; Li, H. Type IV pili of Acidithiobacillus ferrooxidans can transfer electrons from extracellular electron donors. J. Basic Microbiol. 2014, 54, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Chapana, J.A.; Tributsch, H. Interfacial activity and leaching patterns of Leptospirillum ferrooxidans on pyrite. FEMS Microbiol. Ecol. 2004, 47, 19–29. [Google Scholar] [CrossRef]
- Medvedev, D.; Stuchebrukhov, A.A. DNA repair mechanism by photolyase: Electron transfer path from the photolyase catalytic cofactor FADH(-) to DNA thymine dimer. J. Theor. Biol. 2001, 210, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.S.; Lower, S.K. Thickness and surface density of extracellular polymers on Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2008, 74, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Chapana, J.A.; Tributsch, H. Bio-leaching of pyrite accelerated by cysteine. Process Biochem. 2000, 35, 815–824. [Google Scholar] [CrossRef]
- Rojas-Chapana, J.A.; Tributsch, H. Biochemistry of sulfur extraction in bio-corrosion of pyrite by Thiobacillus ferrooxidans. Hydrometallurgy 2001, 59, 291–300. [Google Scholar] [CrossRef]
- Blake, R.C.; Sasaki, K.; Ohmura, N. Does aporusticyanin mediate the adhesion of Thiobacillus ferrooxidans to pyrite? Hydrometallurgy 2001, 59, 357–372. [Google Scholar] [CrossRef]
- Kuehn, M.J.; Kesty, N.C. Bacterial outer membrane vesicles and the host—Pathogen interaction. Genes Dev. 2005, 19, 2645–2655. [Google Scholar] [CrossRef] [PubMed]
- Mashburn-Warren, L.; Mclean, R.J.C.; Whiteley, M. Gram-negative outer membrane vesicles: Beyond the cell surface. Geobiology 2008, 6, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Ellen, A.F.; Albers, S.V.; Huibers, W.; Pitcher, A.; Hobel, C.F.V.; Schwarz, H.; Folea, M.; Schouten, S.; Boekema, E.J.; Poolman, B.; et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 2009, 13, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Prangishvili, D.; Holz, I.; Stieger, E.; Nickell, S.; Kristjansson, J.K.; Zillig, W. Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J. Bacteriol. 2000, 182, 2985–2988. [Google Scholar] [CrossRef] [PubMed]
- Ellen, A.F.; Zolghadr, B.; Driessen, A.M.J.; Albers, S.-V. Shaping the archaeal cell envelope. Archaea 2010, 2010, 608243. [Google Scholar] [CrossRef] [PubMed]
- Ellen, A.F.; Albers, S.-V.; Driessen, A.J.M. Comparative study of the extracellular proteome of Sulfolobus species reveals limited secretion. Extremophiles 2010, 14, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Roden, E.E. Microbiological controls on geochemical kinetics 2: Case study on microbial oxidation of metal sulfide minerals and future prospects. In Kinetics of Water-Rock Interaction; Brantley, S.L., Kubicki, J.D., White, A.F., Eds.; Springer: New York, NY, USA, 2008; pp. 417–467. [Google Scholar]
- Johnson, D.B. Biomining-biotechnologies for extracting and recovering metals from ores and waste materials. Curr. Opin. Biotechnol. 2014, 30, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Watling, H.R. The bioleaching of sulphide minerals with emphasis on copper sulphides—A review. Hydrometallurgy 2006, 84, 81–108. [Google Scholar] [CrossRef]
- Watling, H.R. Review of Biohydrometallurgical Metals Extraction from Polymetallic Mineral Resources. Minerals 2014, 5, 1–60. [Google Scholar] [CrossRef]
- Du Plessis, C.A.; Batty, J.D.; Dew, D.W. Commercial Applications of Thermophile Bioleaching. In Biomining; Rawlings, D.E., Johnson, D.B., Eds.; Springer-Verlag: Berlin, Germany, 2007; pp. 57–80. [Google Scholar]
- Neale, J.W.; Robertson, S.W.; Muller, H.H.; Gericke, M. Integrated piloting of a thermophilic bioleaching process for the treatment of a low-grade nickel-copper sulphide concentrate. J. South. African Inst. Min. Metall. 2009, 109, 273–293. [Google Scholar]
- Batty, J.D.; Rorke, G.V. Development and commercial demonstration of the BioCOP™ thermophile process. Hydrometallurgy 2006, 83, 83–89. [Google Scholar] [CrossRef]
- Logan, T.C.; Seal, T.; Brierley, J.A. Whole-ore heap biooxidation of sulfidic gold-bearing ores. In Biomining; Rawlings, D.E., Johnson, D.B., Eds.; Springer-Verlag: Berlin, Germany, 2007; pp. 113–138. [Google Scholar]
- Dew, D.W.; Buuren, C.; van Mcewan, K.; Bowker, C. Bioleaching of base metal sulphide concentrates: A comparison of high and low temperature bioleaching. J. S. Afr. Inst. Min. Metall. 2000, 100, 409–414. [Google Scholar]
- Khoshkhoo, M.; Dopson, M.; Shchukarev, A.; Sandström, Å. Chalcopyrite leaching and bioleaching: An X-ray photoelectron spectroscopic (XPS) investigation on the nature of hindered dissolution. Hydrometallurgy 2014, 149, 220–227. [Google Scholar] [CrossRef]
- Gericke, M.; Govender, Y.; Pinches, A. Tank bioleaching of low-grade chalcopyrite concentrates using redox control. Hydrometallurgy 2010, 104, 414–419. [Google Scholar] [CrossRef]
- Ahonen, L.; Tuovinen, O.H. Silver catalysis of the bacterial leaching of chalcopyrite-containing ore material in column reactors. Miner. Eng. 1990, 3, 437–445. [Google Scholar] [CrossRef]
- Crundwell, F.K. The semiconductor mechanism of dissolution and the pseudo-passivation of chalcopyrite. Can. Metall. Q. 2015. [Google Scholar] [CrossRef]
- Abdollahi, H.; Shafaei, S.Z.; Noaparast, M.; Manafi, Z.; Niemelä, S.I.; Tuovinen, O.H. Mesophilic and thermophilic bioleaching of copper from a chalcopyrite-containing molybdenite concentrate. Int. J. Miner. Process. 2014, 128, 25–32. [Google Scholar] [CrossRef]
- Romano, P.; Blázquez, M.L.; Alguacil, F.J.; Muñoz, J.A.; Ballester, A.; González, F. Comparative study on the selective chalcopyrite bioleaching of a molybdenite concentrate with mesophilic and thermophilic bacteria. FEMS Microbiol. Lett. 2001, 196, 71–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takatsugi, K.; Sasaki, K.; Hirajima, T. Mechanism of the enhancement of bioleaching of copper from enargite by thermophilic iron-oxidizing archaea with the concomitant precipitation of arsenic. Hydrometallurgy 2011, 109, 90–96. [Google Scholar] [CrossRef]
- D’Hughes, P.D.; Foucher, S. HIOX® Project: A bioleaching process for the treatment of chalcopyrite concentrates using extreme thermophilesthermophiles. In Biohydrometallurgy: Fundamentals, Technology and Sustainable Development; Ciminelli, V.S.T., Garcia, O., Eds.; Elsevier: Amsterdam, The Netherlands, 2001; pp. 75–83. [Google Scholar]
- Marsh, R.M.; Norris, P.R. The isolation of some thermophilic, autotrophic, iron- and sulphur-oxidizing bacteria. FEMS Microbiol. Lett. 1983, 17, 311–315. [Google Scholar] [CrossRef]
- Le Roux, N.W.; Wakerley, W. Leaching of Chalcopyrite (CuFeS2) at 70 °C using Sulfolobus. In Proceedings of Biohydrometallurgy 87’; Norris, P.R., Kelly, D.P., Eds.; Science and Technology Letters: Surrey, UK, 1998; pp. 305–317. [Google Scholar]
- Lindstrom, E.B.; Gunneriusson, L. Thermophilic bioleaching of arsenopyrite using Sulfolobus and a semi-continuous laboratory procedure. J. Ind. Microbiol. 1990, 5, 375–382. [Google Scholar] [CrossRef]
- Gericke, M.; Pinches, A.; van Rooyen, J. Bioleaching of a chalcopyrite concentrate using an extremely thermophilic culture. Int. J. Miner. Process. 2001, 62, 243–255. [Google Scholar] [CrossRef]
- Konishi, Y.; Yoshida, S.; Asai, S. Bioleaching of pyrite by acidophilic thermophile Acidianus brierleyi. Biotechnol. Bioeng. 1995, 48, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Sandström, Å.; Petersson, S. Bioleaching of a complex sulphide ore with moderate thermophilic and extreme thermophilic microorganisms. Hydrometallurgy 1997, 46, 181–190. [Google Scholar]
- Harvey, T.J.; Holder, N.; Stanek, T. Thermophilic Bioheap Leaching of Chalcopyrite Concentrates. Eur. J. Miner. Process. Environ. Prot. 2002, 2, 253–263. [Google Scholar]
- Hita, R.; Wang, H.; Bigham, J.M.; Torrent, J.; Tuovinen, O.H. Bioleaching of a pyritic sludge from the Aznalcóllar (Spain) mine spillage at ambient and elevated temperatures. Hydrometallurgy 2008, 93, 76–79. [Google Scholar] [CrossRef]
- Bharadwaj, A.; Ting, Y.P. Bioleaching of spent hydrotreating catalyst by acidophilic thermophile Acidianus brierleyi: Leaching mechanism and effect of decoking. Bioresour. Technol. 2013, 130, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Vukovic, M.; Strbac, N.; Sokic, M.; Grekulovic, V.; Cvetkovski, V. Bioleaching of pollymetallic sulphide concentrate using thermophilic bacteria. Hem. Ind. 2014, 68, 575–583. [Google Scholar] [CrossRef] [Green Version]
- Clark, M.E.; Batty, J.D.; van Buuren, C.B.; Dew, D.W.; Eamon, M.A. Biotechnology in minerals processing: Technological breakthroughs creating value. Hydrometallurgy 2006, 83, 3–9. [Google Scholar] [CrossRef]
- Van Staden, P.J.; Gericke, M.; Craven, P.M. Minerals biotechnology: Trends, opportunities and challenges. In Hydrometallurgy 2008 : Proceedings of the Sixth International Symposium; Society for Mining Metallurgy and Exploration: Littleton, CO, USA, 2008; pp. 6–13. [Google Scholar]
- Petersen, J.; Dixon, D.G. Competitive bioleaching of pyrite and chalcopyrite. Hydrometallurgy 2006, 83, 40–49. [Google Scholar] [CrossRef]
- Javadi Nooshabadi, A.; Hanumantha Rao, K. Formation of hydrogen peroxide by sulphide minerals. Hydrometallurgy 2014, 141, 82–88. [Google Scholar] [CrossRef]
- Jones, G.C.; van Hille, R.P.; Harrison, S.T.L. Reactive oxygen species generated in the presence of fine pyrite particles and its implication in thermophilic mineral bioleaching. Appl. Microbiol. Biotechnol. 2013, 97, 2735–2742. [Google Scholar] [CrossRef] [PubMed]
- Langwaldt, J. Bioleaching of multimetal black shale by thermophilic micro-organisms. Adv. Mater. Res. 2007, 20, 167. [Google Scholar] [CrossRef]
- Mishra, D.; Rhee, Y.H. Microbial leaching of metals from solid industrial wastes. J. Microbiol. 2014, 52, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Pandey, B.D. Bio-processing of solid wastes and secondary resources for metal extraction—A review. Waste Manag. 2012, 32, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Reed, C. Earth microbe prefers living on meteorites. Science 2015. [Google Scholar] [CrossRef]
- Takahashi, Y.; Châtellier, X.; Hattori, K.H.; Kato, K.; Fortin, D. Adsorption of rare earth elements onto bacterial cell walls and its implication for REE sorption onto natural microbial mats. Chem. Geol. 2005, 219, 53–67. [Google Scholar] [CrossRef]
- Tsuruta, T. Accumulation of Rare Earth Elements in Various Microorganisms. J. Rare Earths 2007, 25, 526–532. [Google Scholar] [CrossRef]
- Emmanuel, E.S.C.; Ananthi, T.; Anandkumar, B.; Maruthamuthu, S. Accumulation of rare earth elements by siderophore-forming Arthrobacter luteolus isolated from rare earth environment of Chavara, India. J. Biosci. 2012, 37, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Horiike, T.; Yamashita, M. A new fungal isolate, Penidiella sp. Strain T9, accumulates the rare earth element dysprosium. Appl. Environ. Microbiol. 2015, 81, 3062–3068. [Google Scholar] [CrossRef] [PubMed]
- Pol, A.; Barends, T.R.M.; Dietl, A.; Khadem, A.F.; Eygensteyn, J.; Jetten, M.S.M.; Op den Camp, H.J.M. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. 2014, 16, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, A.; Menon, A.L.; Thorgersen, M.P.; Scott, J.W.; Poole, F.L.; Jenney, F.E.; Lancaster, W.A.; Praissman, J.L.; Shanmukh, S.; Vaccaro, B.J.; et al. Microbial metalloproteomes are largely uncharacterized. Nature 2010, 466, 779–782. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wheaton, G.; Counts, J.; Mukherjee, A.; Kruh, J.; Kelly, R. The Confluence of Heavy Metal Biooxidation and Heavy Metal Resistance: Implications for Bioleaching by Extreme Thermoacidophiles. Minerals 2015, 5, 397-451. https://doi.org/10.3390/min5030397
Wheaton G, Counts J, Mukherjee A, Kruh J, Kelly R. The Confluence of Heavy Metal Biooxidation and Heavy Metal Resistance: Implications for Bioleaching by Extreme Thermoacidophiles. Minerals. 2015; 5(3):397-451. https://doi.org/10.3390/min5030397
Chicago/Turabian StyleWheaton, Garrett, James Counts, Arpan Mukherjee, Jessica Kruh, and Robert Kelly. 2015. "The Confluence of Heavy Metal Biooxidation and Heavy Metal Resistance: Implications for Bioleaching by Extreme Thermoacidophiles" Minerals 5, no. 3: 397-451. https://doi.org/10.3390/min5030397





