Current Status of Marker Genes of Bacteroides and Related Taxa for Identifying Sewage Pollution in Environmental Waters
Abstract
:1. Introduction
2. Assays Targeting Human Fecal Pollution
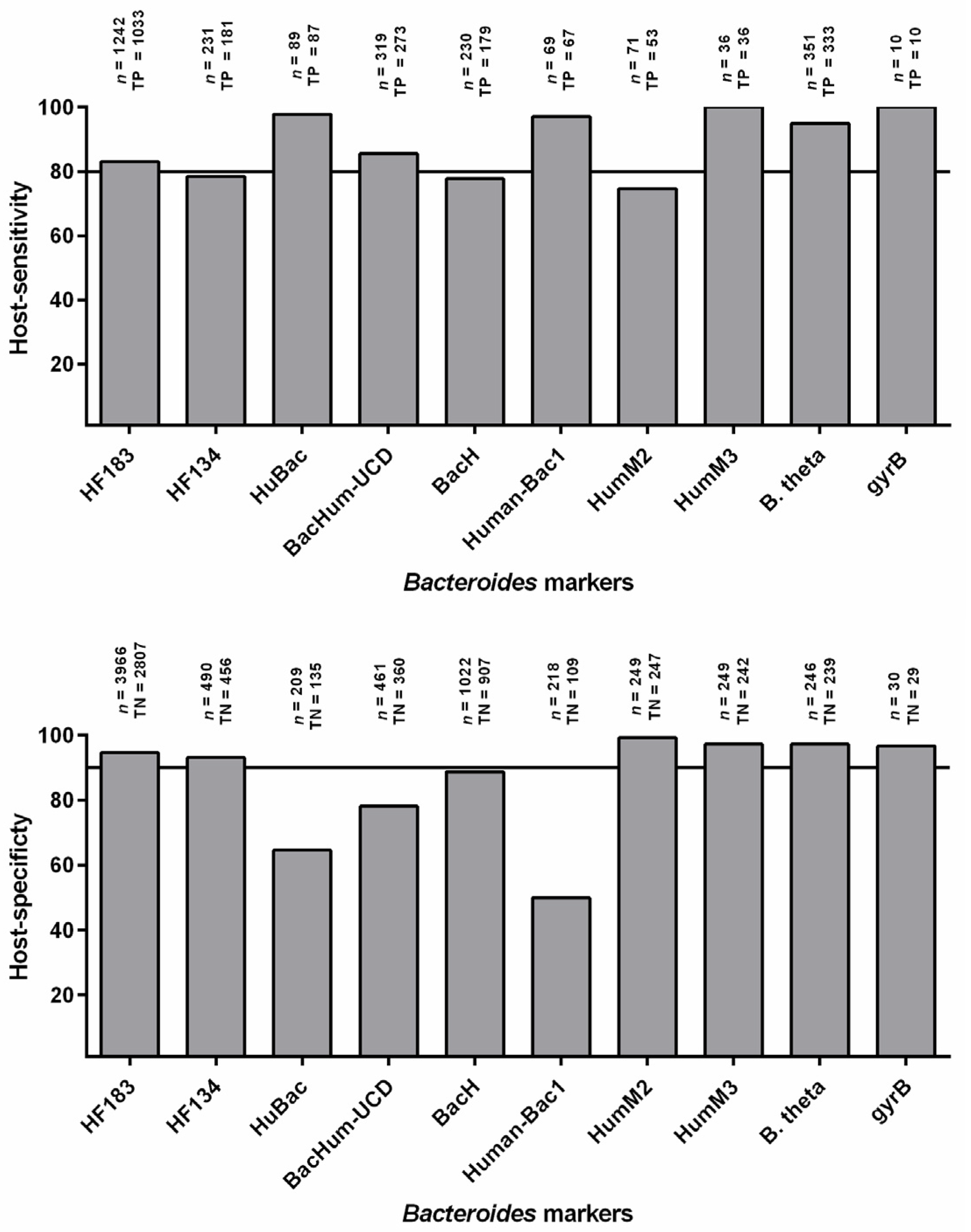
3. Host-Sensitivity and Host-Specificity
3.1. Host Sensitivity
3.2. Host Specificity
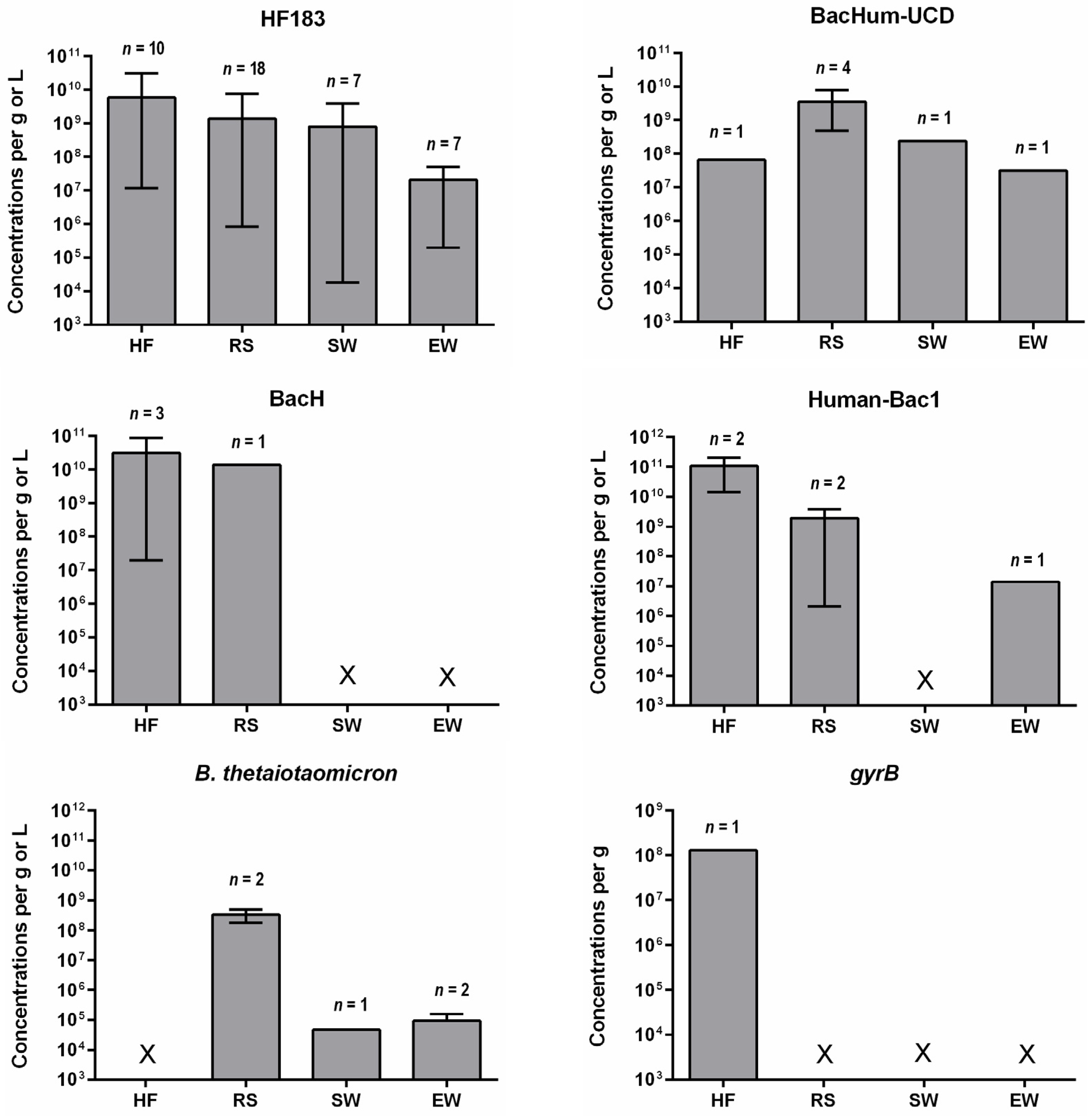
4. Concentrations of Human-Specific Bacteroides Markers in Human Feces, Wastewater and Water Environment
4.1. Human Feces and Wastewater
4.2. Water Environment
5. Decay of Human-Specific Bacteroides Markers in Environmental Matrices
6. Correlation of Human-Specific Bacteroides Markers with Enteric Pathogens in Water
7. Future Directions
8. Conclusions
- Application of human-associated Bacteroides for MST field studies is promising due to their high host specificity and abundance in human wastewater. PCR methods avoid the issue of culture bias but introduce methodological issues such as efficiency of DNA extraction and inhibition of amplification. PCR/qPCR-based methods can be less expensive and are more rapid than culturing bacteria—they can yield results in as little as 4–6 h. However, standardization of assays, units, and the markers’ performance characteristics will be substantial challenges from a regulatory perspective.
- Among the human-specific Bacteroides markers, the performance characteristics and application of the HF183 markers have been more thoroughly evaluated than others. However, BacH, HumM2, and HumM3 have been shown to be useful quantitative tools for tracking sewage pollution in environmental waters. Rapid decay of these markers in the environment makes them very useful markers for tracking recent fecal pollution. However, differential decay rates and a lack of complete host specificity indicate that these Bacteroides markers should be accompanied with more host-specific markers such as human adenoviruses or polyomaviruses in a toolbox format for the accurate and sensitive detection of sewage pollution in environmental waters.
- Little research has been undertaken on the host specificity and sensitivity of the HumM2, HumM3, B. thetaiotaomicron, α-1.6-mannanas, and gyrB markers. Since some animal fecal samples produced false-positive results with these markers due to the presence of the target (or a very closely related) bacteria, further evaluation of performance characteristics should be undertaken to determine the broader applicability of these markers. Non-specific markers may still be useful for source tracking if information is available on the contributing sources and if possible testing should be accompanied with additional markers in a toolbox format.
- Quantitative data on the occurrence of human-specific Bacteroides markers in their hosts is limited other than for HF183. Such data are important to determine the suitability of a marker for detecting fecal pollution in environmental waters. The baseline concentration, which is appropriate to detect fecal pollution and also indicate health risks, needs to be established.
- The high prevalence and levels of human-specific Bacteroides markers in environmental waters in the USA, France, Bangladesh, Canada, Belgium, and several other countries indicate chronic sewage pollution in environmental waters. The application of these tools is encouraged in continents such as Asia and Africa (where possible) where wastewater pollution due to improper sanitation and gastrointestinal diseases is a major concern, but only after assessing the performance of the markers in these regions.
- Regulatory and public health concerns mandate that more studies should be undertaken to gain an understanding of how human-specific Bacteroides markers correlate with FIB, pathogens, and human health risks. The absence of correlations does not necessarily impede the utility of these markers in identifying fecal pollution and the potential for mitigation. Little is known regarding the concentrations of human-specific Bacteroides markers in soils, sediments, and beach sand. These environmental matrices harbor FIB and the limited data indicate that BacHum-UCD markers persist longer in watered beach sand [117]. Bacteroides associated with particular matter in suspension or solid matrices may remain viable for a longer time than if they are dispersed in water. These results have implications for the accuracy of MST tools as regulatory standards for the protection of water quality.
- A significant challenge associated with the field application of these markers is effective, quantitative recovery of DNA from complex environmental samples. Little has been documented on the recovery of these markers from environmental matrices. Quantification of these markers in environmental waters can be difficult due to factors such as dilution, sorption to particulate matters, environmental decay, loss due to recovery and DNA extraction, and the fact that only a small volume of a DNA sample is used for PCR analysis. Recent developments in qPCR technology such as droplet digital PCR may provide more sensitive detection of DNA from environmental waters. Incorporation of quantitative microbial risk assessment (QMRA) with MST science will improve our understanding of the relative public health risks associated with various sources of fecal pollution.
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ahmed, W.; Goonetilleke, A.; Gardner, T. Human and bovine adenoviruses for the detection of source-specific fecal pollution in coastal waters in Australia. Water Res. 2010, 44, 4662–4673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneeberger, C.L.; O’Driscoll, M.; Humphery, C.; Henry, K.; Deal, N.; Seiber, K.; Hill, V.R.; Zarate-Bermudez, M. Fate and transport of enteric microbes from septic systems in a coastal watershed. J. Environ. Health 2015, 77, 22–20. [Google Scholar] [PubMed]
- Sidhu, J.P.S.; Ahmed, W.; Gernjak, W.; Aryal, R.; McCarthy, D.; Palmer, A.; Kolotelo, P.; Toze, S. Sewage pollution in urban stormwater runoff as evident from the widespread presence of multiple microbial and chemical source tracking markers. Sci. Total Environ. 2013, 463–464, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Ryu, H.; Hill, S.; Schoen, M.; Ashbolt, N.; Edge, T.A.; Domingo, J.S. Distribution and potential significance of a gull fecal marker in urban coastal and riverine areas of southern Ontario, Canada. Water Res. 2011, 45, 3960–3968. [Google Scholar] [CrossRef] [PubMed]
- Chase, E.; Hunting, J.; Staley, C.; Harwood, V.J. Microbial source tracking to identify human and ruminant sources of faecal pollution in an ephemeral Florida river. J. Appl. Microbiol. 2012, 113, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Sritharan, T.; Palmer, A.; Sidhu, J.P.S.; Toze, S. Evaluation of bovine feces-associated microbial source tracking markers and their correlations with fecal indicators and zoonotic pathogens in a Brisbane, Australia, Reservoir. Appl. Environ. Microbiol. 2013, 79, 2682–2691. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency. Ambient Water Quality Criteria for Bacteria; Office of Water, EPA 440/5-84-002; U.S. EPA: Washington, DC, USA, 1986.
- U.S. Environmental Protection Agency. Method 1600: Enterococci in Water by Membrane Filtration Using Membrane-Enterococcus indoxyl-d-glucoside agar (mEI); Office of Water, EPA-821-R-02-022; U.S. EPA: Washington, DC, USA, 2002.
- Hörman, A.; Rimhanen-Finne, R.; Maunula, L.; von Bonsdorff, C.H.; Torvela, N.; Heikinheimo, A.; Hänninen, M.L. Campylobacter spp., Giardia spp., Cryptosporidium spp., noroviruses, and indicator organisms in surface water in southwestern Finland, 2000–2001. Appl. Environ. Microbiol. 2004, 70, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Harwood, V.J.; Levine, A.D.; Scott, T.M.; Chivukula, V.; Lukasik, J.; Farrah, S.R.; Rose, J.B. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 2005, 71, 3163–3170. [Google Scholar] [CrossRef] [PubMed]
- Field, K.G.; Samadpour, M. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res. 2007, 41, 3517–3538. [Google Scholar] [CrossRef] [PubMed]
- Byappanahalli, M.N.; Whitman, R.L.; Shively, D.A.; Ting, W.T.; Tseng, C.C.; Nevers, M.B. Seasonal persistence and population characteristics of Escherichia coli and enterococci in deep backshore sand of two freshwater beaches. J. Water Health 2006, 4, 313–320. [Google Scholar] [PubMed]
- Anderson, K.L.; Whitlock, J.E.; Harwood, V.J. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 2005, 71, 3041–3048. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Ksoll, W.B.; Hicks, R.E.; Sadowsky, M.J. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 2006, 72, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Desmarais, T.R.; Solo-Gabriele, H.M.; Palmer, C.J. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 2002, 68, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Whitman, R.L.; Byers, S.E.; Shively, D.A.; Ferguson, D.M.; Byappanahalli, M. Occurrence and growth characteristics of Escherichia coli and enterococci within the accumulated fluid of the northern pitcher plant (Sarracenia purpurea L.). Can. J. Microbiol. 2005, 5, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Harwood, V.J.; Whitlock, J.; Withington, V. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: Use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 2000, 66, 3698–3704. [Google Scholar] [CrossRef] [PubMed]
- Hagedorn, C.; Crozier, J.B.; Mentz, K.A.; Booth, A.M.; Graves, A.K.; Nelson, N.J.; Reneau, R.B., Jr. Carbon source utilization profiles as a method to identify sources of faecal pollution in water. J. Appl. Microbiol. 2003, 94, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Wiggins, B.A.; Cash, P.W.; Creamer, W.S.; Dart, S.E.; Garcia, P.P.; Gerecke, T.M.; Han, J.; Henry, B.L.; Hoover, K.V.; Johnson, E.L.; et al. Use of antibiotic resistance analysis for representativeness testing of multi watershed libraries. Appl. Environ. Microbiol. 2003, 69, 3399–3405. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, A.E.; Field, K.G. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 2000, 66, 4571–4574. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.M.; Jenkins, T.M.; Lukasik, J.; Rose, J.B. Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ. Sci. Technol. 2005, 39, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Ufnar, J.A.; Wang, S.Y.; Christiansen, J.M.; Yampara-Iquise, H.; Carson, C.A.; Ellender, R.D. Detection of the nifH gene of Methanobrevibacter smithii: A potential tool to identify sewage pollution in recreational waters. J. Appl. Microbiol. 2006, 101, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Prystajecky, N.; Huck, P.M.; Schreier, H.; Isaac-Renton, J.L. Assessment of Giardia and Cryptosporidium spp. as a microbial source tracking tool for surface water: Application in a mixed-use watershed. Appl. Environ. Microbiol. 2014, 80, 2328–2336. [Google Scholar] [CrossRef] [PubMed]
- Kirs, M.; Smith, D.C. Multiplex quantitative real-time reverse transcriptase PCR for F+-specific RNA coliphages: A method for use in microbial source tracking. Appl. Environ. Microbiol. 2007, 73, 808–814. [Google Scholar] [CrossRef] [PubMed]
- McQuaig, S.M.; Scott, T.M.; Lukasik, J.O.; Paul, J.H.; Harwood, V.J. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 2009, 75, 3379–3388. [Google Scholar] [CrossRef] [PubMed]
- Harwood, V.J.; Staley, C.; Badgley, B.D.; Borges, K.; Korajkic, A. Microbial source tracking markers for detection of fecal contamination in environmental waters: Relationships between pathogens and human health outcomes. FEMS Microbiol. Rev. 2014, 38, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.F.; Weisberg, S.B.; McGee, C.D. Evaluation of microbial source tracking methods using mixed fecal sources in aqueous test samples. J. Water Health 2003, 1, 141–151. [Google Scholar] [PubMed]
- Hartel, P.G.; Summer, J.D.; Hill, J.L.; Collins, J.V.; Entry, J.A.; Segars, W.I. Geographic variability of Escherichia coli ribotypes from animals in Idaho and Georgia. J. Environ. Qual. 2002, 31, 1273–1278. [Google Scholar] [CrossRef] [PubMed]
- Harwood, V.J.; Wiggins, B.A.; Hagedorn, C.; Ellender, R.D.; Gooch, J.; Kern, J.; Samadpour, M.; Chapman, A.C.H.; Robinson, B.J. Phenotypic library-based microbial source tracking methods: Efficacy in the California collaborative study. J. Water Health 2003, 1, 153–166. [Google Scholar] [PubMed]
- Gordon, D.M.; Bauer, S.; Johnson, J.R. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology. 2002, 148, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Kreader, C.A. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl. Environ. Microbiol. 1995, 66, 2263–2266. [Google Scholar]
- Sghir, A.; Gramet, G.; Suau, A.; Rochet, V.; Pochart, P.; Dore, J. Quantification of bacterial groups within human faecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 2000, 66, 2263–2266. [Google Scholar] [CrossRef] [PubMed]
- Allsop, K.; Stickler, J.D. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. J. Appl. Bacteriol. 1985, 58, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, A.E.; Field, K.G. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 2000, 66, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Reischer, G.H.; Kasper, D.C.; Steinborn, R.; Farnleitner, A.H.; Mach, R.L. A quantitative real-time PCR assay for the highly sensitive and specific detection of human faecal influence in spring water from a large alpine catchment area. Lett. Appl. Microbiol. 2007, 44, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Kildare, B.J.; Leutenegger, C.; McSwain, B.S.; Bambic, D.G.; Rajal, V.B.; Wuertz, S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: A Bayesian approach. Water Res. 2007, 41, 3701–3715. [Google Scholar] [CrossRef] [PubMed]
- Okabe, S.; Okayama, N.; Savichtcheva, O.; Ito, T. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 2007, 74, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Shanks, O.C.; Kelty, C.A.; Sivaganesan, M.; Varma, M.; Haugland, R.A. Quantitative PCR for genetic markers of human fecal pollution. Appl. Environ. Microbiol. 2009, 75, 5507–5513. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Lee, J. Evaluation of new gyrB-based real-time PCR system for the detection of B. fragilis as an indicator of human-specific fecal contamination. J. Microbiol. Methods 2010, 82, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, A.E.; Goyard, T.; Simonich, M.T.; Field, K.G. Application of a rapid method for identifying fecal pollution sources in a multi-use estuary. Water Res. 2003, 37, 909–914. [Google Scholar] [CrossRef]
- Seurinck, S.; Verdievel, M.; Verstraete, W.; Siciliano, S.D. Identification of human fecal pollution sources in a coastal area: A case study at Oostende (Belgium). J. Water Health 2006, 4, 167–175. [Google Scholar] [PubMed]
- Gourmelon, M.; Caparis, M.P.; Segura, R.; Mennec, C.L.; Lozach, S.; Piriou, J.Y.; Rince, R.A. Evaluation of two library-independent microbial source tracking methods to identify sources of fecal contamination in French estuaries. Appl. Environ. Microbiol. 2007, 73, 4857–4866. [Google Scholar] [CrossRef] [PubMed]
- Soller, J.A.; Bartrand, T.; Ashbolt, N.J.; Ravenscroft, J.; Wade, T.J. Estimating the primary aetiologic agents in recreational freshwaters impacted by human sources of faecal contamination. Water Res. 2010, 44, 4736–4747. [Google Scholar] [CrossRef] [PubMed]
- Seurinck, S.; Defoirdt, T.; Verstraete, W.; Siciliano, S.D. Detection and quantification of human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human fecal pollution in freshwater. Environ. Microbiol. 2005, 7, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Layton, A.; McKay, L.; Williams, D.; Garrett, V.; Gentry, R.; Sayler, G. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 2006, 72, 4214–4224. [Google Scholar] [CrossRef] [PubMed]
- Haugland, R.A.; Varma, M.; Kelty, C.A.; Peed, L.; Sivaganesan, M.; Shanks, O.C. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by real-time PCR. Syst. Appl. Microbiol. 2010, 33, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Converse, R.R.; Blackwood, A.D.; Kirs, M.; Griffith, M.; Noble, R.T. Rapid QPCR-based assay for fecal Bacteroides spp. as a tool for assessing fecal contamination in recreational waters. Appl. Environ. Microbiol. Water Res. 2009, 43, 4828–4837. [Google Scholar] [CrossRef] [PubMed]
- Boehm, A.B.; Van De Werfhorst, L.C.; Griffith, J.F.; Holden, P.; Jay, J.; Shanks, O.C.; Wang, D.; Weisberg, S.B. Performance of forty-one microbial source-tracking methods: A twenty seven lab evaluation study. Water Res. 2013, 47, 6812–6828. [Google Scholar] [CrossRef] [PubMed]
- Layton, B.A.; Cao, Y.; Ebentier, D.L.; Hanley, K.; Balleste, E.; Brandao, J.; Byappanahalli, M.; Converse, R.; Farnleitner, A.H.; Gentry-Shields, J.; et al. Performance of human fecal acaerobes-associated PCR-based assays in a multi-laboratory method evaluation study. Water Res. 2013, 47, 6897–6908. [Google Scholar] [CrossRef] [PubMed]
- Green, H.C.; Haugland, R.A.; Verma, M.; Millen, H.T.; Borchardt, M.A.; Field, K.G.; Walters, W.A.; Knight, R.; Kelty, C.A.; Shanks, O.C. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl. Environ. Microbiol. 2014, 80, 3086–3094. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Goonetilleke, A.; Powell, D.; Gardner, T. Evaluation of multiple sewage-associated Bacteroides PCR markers for sewage pollution tracking. Water Res. 2009, 43, 4872–4877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballesté, E.; Bonjoch, X.; Belanche, L.A.; Blanch, A.R. Molecular indicators used in the development of predictive models for microbial source tracking. Appl. Environ. Microbiol. 2010, 76, 1789–1795. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Masters, N.; Toze, S. Consistency in the host specificity and host sensitivity of the Bacteroides HF183 marker for sewage pollution tracking. Lett. Appl. Microbiol. 2012, 55, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Aslan, A.; Rose, J.B. Evaluation of the host-specificity of Bacteroides thetaiotaomicron alpha-1–6, mannanase gene as a sewage marker. Lett. Appl. Microbiol. 2012, 56, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Reischer, G.H.; Edbon, J.E.; Bauer, J.M.; Schuster, N.; Ahmed, W.; Åström, J.; Blanch, A.R.; Blöschl, G.; Byamukama, D.; Coakley, T.; et al. Performance characteristics of qPCR assays targeting human- and ruminant-associated Bacteroidetes for microbial source tracking across sixteen countries on six continents. Environ. Sci. Technol. 2013, 47, 8548–8556. [Google Scholar] [CrossRef] [PubMed]
- Sadowsky, M.J.; Call, D.R.; Santo Domingo, J.W. Application of microbial source tracking studies. In Microbial Source Tracking; Domingo, J.W.S., Sadowsky, M.J., Eds.; ASM Press: Washington, DC, USA, 2007; pp. 235–277. [Google Scholar]
- Carson, C.A.; Christiansen, J.M.; Yampara-Iquise, H.; Benson, V.W.; Baffaut, C.; Davis, J.V.; Broz, R.R.; Kurtz, W.B.; Rogers, W.M.; Fales, W.H. Specificity of a Bacteroides thetaiotaomicron marker for human feces. Appl. Environ. Microbiol. 2005, 71, 4945–4949. [Google Scholar] [CrossRef] [PubMed]
- Yampara-Iquise, H.; Zheng, G.; Jones, J.E.; Carson, C.A. Use of a Bacteroides thetaiotaomicron-specific alpha-1-6, mannanase quantitative PCR to detect human faecal pollution in water. J. Appl. Microbiol. 2008, 105, 1686–1693. [Google Scholar] [CrossRef] [PubMed]
- Shanks, O.C.; Kelty, C.A.; Oshiro, R.; Haugland, R.A.; Madi, T.; Brooks, L.; Field, K.G.; Sivaganesan, M. Data acceptance criteria for standardized human-associated fecal source identification quantitative real-time PCR methods. Appl. Environ. Microbiol. 2016, 82, 2773–2782. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency. Microbial Source Tracking Guide; Document EPA/600/R-05/064; U.S. Environmental Protection Agency: Washington, DC, USA, 2005.
- Gawler, A.H.; Beecher, J.E.; Brandão, J.; Carroll, N.M.; Falcão, L.; Gourmelon, M.; Masterson, B.; Nunes, B.; Porter, J.; Rince, A.; et al. Validation of host-specific Bacteriodales 16S rRNA genes as markers to determine the origin of fecal pollution in Atlantic Rim countries of the European Union. Water Res. 2007, 41, 3780–3784. [Google Scholar] [CrossRef] [PubMed]
- Dorai-raj, S.; O’Grady, J.; Colleran, E. Specificity and sensitivity evaluation of novel and existing Bacteroidales and Bifidobacteria-specific PCR assays on feces and sewage samples and their application for microbial source tracking in Ireland. Water Res. 2009, 43, 4980–4988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odagiri, M.; Schriewer, A.; Hanley, K.; Wuertz, S.; Misra, P.R.; Panigrahi, P.; Jenkins, M.W. Validation of Bacteroidales quantitative PCR assays targeting human and animal fecal contamination in the public and domestic domains in India. Sci. Total Environ. 2015, 502, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Stoeckel, D.M.; Harwood, V.J. Performance, design, and analysis in microbial source tracking studies. Appl. Environ. Microbiol. 2007, 73, 2405–2415. [Google Scholar] [CrossRef] [PubMed]
- Shanks, O.C.; White, K.; Kelty, C.A.; Sivaganesan, M.; Blannon, J.; Meckes, M.; Varma, M.; Haugland, R.A. Performance of PCR-based assays targeting Bacteroides genetic markers of human fecal pollution in sewage and fecal samples. Environ. Sci. Technol. 2010, 44, 6281–6288. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Stewart, J.; Powell, D.; Gardner, T. Evaluation of Bacteroides markers for the detection of human faecal pollution. Lett. Appl. Microbiol. 2008, 46, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, M.W.; Tiwari, S.; Lorente, M.; Gichaba, C.M.; Wuertz, S. Identifying human and livestock sources of fecal contamination in Kenya with host-specific Bacteroidales assays. Water Res. 2009, 43, 4956–4966. [Google Scholar] [CrossRef] [PubMed]
- Edge, T.A.; Hill, S.; Seto, P.; Marsalek, J. Library-dependent and library-independent microbial source tracking to identify spatial variation in faecal contamination source along a Lake Ontario Beach (Ontario, Canada). Water Sci. Technol. 2010, 62, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Fremaux, B.; Gritzfeld, J.; Boa, T.; Yost, C.K. Evaluation of host-specific Bacteroidales 16S rRNA gene markers as a complementary tool for detecting fecal pollution in a prairie watershed. Water Res. 2009, 43, 4838–4849. [Google Scholar] [CrossRef] [PubMed]
- Mieszkin, S.; Furet, J.-P.; Corthier, G.; Gourmelon, M. Estimation of pig fecal contamination in a river catchment by real-time PCR using two pig-specific Bacteroidales 16S rRNA genetic markers. Appl. Environ. Microbiol. 2009, 75, 3045–3054. [Google Scholar] [CrossRef] [PubMed]
- Harwood, V.J.; Brownell, M.; Wang, S.; Lepo, J.; Ellender, R.D.; Ajidahun, A.; Hellein, K.N.; Kennedy, E.; Ye, X.; Flood, C. Validation and field testing of library-independent microbial source tracking methods in the Gulf of Mexico. Water Res. 2009, 43, 4812–4819. [Google Scholar] [CrossRef] [PubMed]
- Stea, E.C.; Hansen, L.T.; Jamieson, R.C.; Yost, C.K. Fecal contamination in the surface waters of a rural and an urban source-watershed. J. Environ. Qual. 2015, 44, 1556–1567. [Google Scholar] [CrossRef] [PubMed]
- Van De Werfhorst, L.; Sercu, B.; Holden, P. Comparison of the host specificities of two Bacteroidales quantitative PCR assays used for tracking human fecal contamination. Appl. Environ. Microbiol. 2011, 77, 6258–6260. [Google Scholar] [CrossRef] [PubMed]
- Tambalo, D.D.; Fremaux, B.; Boa, T.; Yost, C.K. Persistence of host-associated Bacteroidales gene markers and their quantitative detection in an urban and agricultural mixed prairie watershed. Water Res. 2012, 46, 2891–2904. [Google Scholar] [CrossRef] [PubMed]
- Weidhaas, J.L.; Macbeth, T.W.; Olsen, R.L.; Sadowsky, M.J.; Norat, D.; Harwood, V.J. Identification of a Brevibacterium marker gene specific to poultry litter and development of a quantitative PCR assay. J. Appl. Microbiol. 2010, 109, 334–347. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Sidhu, J.P.S.; Smith, K.; Beale, D.; Gyawali, P.; Toze, S. Distributions of fecal markers in wastewater from varying climatic zones for human fecal pollution tracking in Australian surface waters. Appl. Environ. Microbiol. 2016, 82, 1316–1323. [Google Scholar] [CrossRef] [PubMed]
- Kelty, C.A.; Varma, M.; Sivaganesan, M.; Haugland, R.A.; Shanks, O.C. Distribution of genetic marker concentrations for fecal indicator bacteria in sewage and animal feces. Appl. Environ. Microbiol. 2012, 78, 4225–4232. [Google Scholar] [CrossRef] [PubMed]
- Sercu, B.; Van De Werfhorst, L.C.; Murray, J.; Holden, P.A. Storm drains are sources of human fecal pollution during dry weather in three urban southern California watersheds. Environ. Sci. Technol. 2009, 43, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Yusuf, R.; Hasan, I.; Goonetilleke, A.; Gardner, T. Quantitative PCR assay of sewage-associated Bacteroides markers to assess sewage pollution in an urban lake in Dhaka, Bangladesh. Can. J. Microbiol. 2010, 56, 838–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van De Werfhorst, L.C.; Murray, J.L.S.; Reynolds, S.; Reynolds, K.; Holden, P.A. Canine scent detection and microbial source tracking of human waste contamination in storm drains. Water Environ. Res. 2014, 86, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Eichmiller, J.J.; Hicks, R.E.; Sadowsky, M.J. Distribution of genetic markers of fecal pollution on a freshwater sandy shoreline in proximity to wastewater effluent. Environ. Sci. Technol. 2013, 47, 3395–3402. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Gordon, K.V.; Schoen, M.E.; Harwood, V.J. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl. Environ. Microbiol. 2012, 78, 7317–7326. [Google Scholar] [CrossRef] [PubMed]
- Staley, Z.R.; Vogel, L.; Robinson, C.; Edge, T.A. Differential occurrence of Escherichia coli and human Bacteroidales at two great lakes beaches. J. Great Lakes Res. 2015, 41, 530–535. [Google Scholar] [CrossRef]
- Nshimyimana, J.P.; Ekklesia, E.; Shanahan, P.; Chua, L.H.C.; Thomson, J.R. Distribution and abundance of human-specific Bacteroides and relation to traditional indicators in an urban tropical catchment. J. Appl. Microbiol. 2014, 116, 1369–1383. [Google Scholar] [CrossRef] [PubMed]
- Mika, K.B.; Ginsburg, D.W.; Lee, C.M.; Thulsiraj, V.; Jay, J.A. Fecal indicator bacteria levels do not correspond with incidence of human-associated HF183 Bacteroides 16S rRNA gene markers in two urban Southern California. Water Air Soil Pollut. 2014, 225, 1960. [Google Scholar] [CrossRef]
- Liao, H.; Krometis, L.-A.H.; Hession, W.C.; Benitez, R.; Sawyer, R.; Schaberg, E.; von Wagoner, E.; Badgley, B.D. Storm loads of culturable and molecular fecal indicators in an inland urban stream. Sci. Total Environ. 2015, 530–531, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Jardé, E.; Gruau, G.; Pourcher, A.M.; Gourmelon, M.; Jadas-Hécart, A.; Wickmann, A.C.P. Origin of fecal contamination in waters from contrasted areas: Stanols as microbial source tracking markers. Water Res. 2012, 46, 4009–4016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauffret, A.; Caparis, M.P.; Gourmelon, M. Relevance of Bacteroidales and F-specific RNA bacteriophages for efficient fecal contamination tracking at the level of a catchment in France. Appl. Environ. Microbiol. 2012, 78, 5143–5152. [Google Scholar] [CrossRef] [PubMed]
- Gourmelon, M.; Caparis, M.P.; Le Mennec, C.; Mieszkin, S.; Ponthoreau, C.; Gendronneau, M. Application of library-independent microbial source tracking methods for identifying the sources of faecal contamination in coastal waters. Water Sci. Technol. 2010, 61, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Gourmelon, M.; Caparis, M.P.; Mieszkin, S.; Marti, R.; Wery, N.; Jarde, E.; Derrien, M.; Jadas-Hecart, A.; Communal, P.Y.; Jaffrezic, A.; et al. Development of microbial and chemical MST tools to identify the origin of the faecal pollution in bathing and shellfish harvesting waters in France. Water Res. 2010, 44, 4812–4824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marti, R.; Gannon, V.P.J.; Jokinen, C.; Lanthier, M.; Lapen, D.R.; Neuman, N.F.; Ruecker, N.J.; Scott, A.; Wilkes, G.; Zhang, Y.; et al. Quantitative multi-year elucidation of fecal sources of waterborne pathogen contamination in the South Nation River basin using Bacteroidales microbial source tracking markers. Water Res. 2013, 47, 2315–2324. [Google Scholar] [CrossRef] [PubMed]
- Coakley, T.; Brion, G.M.; Fryar, A.E. Prevalence of and relationship between two human-associated DNA biomarkers for Bacteroides in an urban watershed. J. Environ. Qual. 2015, 44, 1694–1698. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.; Hunter, S.; Cyterski, M.; Peed, L.A.; Kelty, C.A.; Sivaganesan, M.; Mooney, T.; Prieto, L.; Shanks, O.C. Factors affecting the presence of human-associated and fecal indicator real-time quantitative PCR genetic markers in urban-impacted recreational beaches. Water Res. 2014, 64, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Sercu, B.; Van De Werfhorst, L.C.; Murray, J.L.S.; Holden, P.A. Sewage exfiltration as a source of storm drain contamination during dry weather in urban watersheds. Environ. Sci. Technol. 2011, 45, 7151–7157. [Google Scholar] [CrossRef] [PubMed]
- Paar, J., III; Doolittle, M.M.; Varma, M.; Siefring, S.; Oshima, K.; Haugland, R.A. Development and evaluation of a culture-independent method for source determination of fecal wastes in surface and storm waters using reverse transcriptase-PCR detection of FRNA coliphage genogroup. J. Microbiol. Methods 2015, 112, 28–35. [Google Scholar] [PubMed]
- Villemur, R.; Imbeau, M.; Vuong, M.N.; Masson, L.; Payment, P. An environmental survey of surface waters using mitochondrial DNA from human, bovine and porcine origin as fecal source tracking markers. Water Res. 2015, 69, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Gentry-Shields, J.; Rowny, J.G.; Stewart, J.R. HuBac and nifH source tracking markers display a relationship to land use but not rainfall. Water Res. 2012, 46, 6163–6174. [Google Scholar] [CrossRef] [PubMed]
- Sauer, E.P.; VandeWalle, J.L.; Bootsma, M.J.; McLellan, S.L. Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment. Water Res. 2011, 45, 4081–4091. [Google Scholar] [CrossRef] [PubMed]
- Bambic, D.G.; Hildare-Hann, B.J.; Rajal, V.B.; Sturm, B.S.M.; Minton, C.B.; Schriewer, A.; Wuertz, S. Spatial and hydrologic variation of Bacteroidales, adenovirus and enterovirus in a semi-arid wastewater effluent-impacted watershed. Water Res. 2015, 75, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Knappett, P.S.K.; Escamilla, V.; Layton, A.; McKay, D.; Emch, M.; Williams, D.E.; Huq, R.; Alam, J.; Farhana, L.; Mailloux, B.J.; et al. Impact of population and latrines on fecal contamination of ponds in rural Bangladesh. Sci. Total Environ. 2011, 409, 3174–3182. [Google Scholar] [CrossRef] [PubMed]
- Ohad, S.; Vaizel-Ohayon, D.; Rom, M.; Guttman, J.; Berger, D.; Kravitz, V.; Pilo, S.; Huberman, Z.; Kashi, Y.; Rorman, E. Microbial source tracking in adjacent Karst Springs. Appl. Environ. Microbiol. 2015, 81, 5037–5047. [Google Scholar] [CrossRef] [PubMed]
- Savichtcheva, O.; Okayama, N.; Okabe, S. Relationships between Bacteroides 16S rRNA genetic markers and presence of bacterial enteric pathogens and conventional fecal indicators. Water Res. 2007, 41, 3615–3628. [Google Scholar] [CrossRef] [PubMed]
- Walters, S.P.; Yamahara, K.H.; Boehm, A.B. Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: Implications for their use in assessing risk in recreational waters. Water Res. 2009, 43, 4929–4939. [Google Scholar] [CrossRef] [PubMed]
- Sinton, L.W.; hall, C.H.; Lynch, P.A.; Davies-Colley, R.J. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl. Environ. Microbiol. 2002, 68, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Dick, L.K.; Stelzer, E.A.; Bertke, E.E.; Fong, D.L.; Stoeckel, D.M. Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Appl. Environ. Microbiol. 2010, 76, 3255–3262. [Google Scholar] [CrossRef] [PubMed]
- Green, H.C.; Shanks, O.C.; Sivaganesan, M.; Haugland, R.A.; Field, K.G. Differential decay of human faecal Bacteroides in marine and freshwater. Environ. Microbiol. 2011, 13, 3235–3249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okabe, S.; Shimazu, Y. Persistence of host-specific Bacteroides-Prevotellla 16S rRNA genetic markers in environmental waters: Effects of temperature and salinity. Appl. Microbiol. Biotechnol. 2007, 76, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Wuertz, S. Rapid decay of host-specific fecal Bacteroidales cells in seawater as measured by quantitative PCR with propidium monoazide. Water Res. 2009, 43, 4850–4859. [Google Scholar] [CrossRef] [PubMed]
- Brooks, Y.; Aslan, A.; Tamarkar, S.; Murali, B.; Mitchell, J.; Rose, J.B. Analysis of the persistence of enteric markers in sewage polluted water on a solid matrix and in liquid suspension. Water Res. 2015, 76, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Gilpin, B.J.; Devane, M.; Robson, B.; Nourozi, F.; Scholes, P.; Lin, S.; Wood, D.R.; Sinton, L.W. Sunlight inactivation of human polymerases chain reaction markers and cultured fecal indicators in river and saline waters. Water Environ. Res. 2013, 85, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Wuertz, S. Decay of host-associated Bacteroidales cells and DNA in continuous flow freshwater and seawater microcosms of identical experimental design and temperature as measured by PMA-qPCR and qPCR. Water Res. 2015, 70, 2015–213. [Google Scholar] [CrossRef] [PubMed]
- Jeanneau, L.; Solecki, O.; Wéry, N.; Jardé, E.; Gourmelon, M.; Communal, P.Y.; Jadas-Hécart, A.; Caparis, M.P.; Gruau, G.; Pourcher, A.M. Relative decay of fecal indicator bacteria and human-associated markers: A microcosm study simulating wastewater input into seawater and freshwater. Environ. Sci. Technol. 2012, 46, 2375–2382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Z.; He, Z.; Zhou, X.; Powell, C.A.; Yang, Y.; Roberts, M.G.; Stoffella, P.J. High diversity and differential decay persistence of fecal Bacteroidales population spiked into freshwater microcosm. Water Res. 2012, 46, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Eichmiller, J.J.; Borchert, A.J.; Sadowsky, M.J. Decay of genetic markers for fecal bacterial indicators and pathogens in sand from lake superior. Water Res. 2014, 59, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Gyawali, P.; Sidhu, J.P.S.; Toze, S. Relative inactivation of faecal indicator bacteria and sewage markers in freshwater and seawater microcosms. Lett. Appl. Microbiol. 2014, 59, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Walters, S.P.; Field, K.G. Survival and persistence of human and ruminant-specific faecal Bacteroidales in freshwater microcosms. Environ. Microbiol. 2009, 11, 1410–1421. [Google Scholar] [CrossRef] [PubMed]
- Yamahara, K.M.; Sassoubre, L.M.; Goodwin, K.D.; Boehm, A.B. Occurrence and persistence of bacterial pathogens and indicator organisms in beach sand along the California Coast. App. Environ. Microbiol. 2012, 78, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Wuertz, S. Survival of host-associated Bacteroides cells and their relationship with Enterococcus spp., Campylobacter jejuni, Salmonella enterica serovar Typhimurium, and adenovirus in freshwater microcosms as measured by propidium monoazide-quantitative PCR. Appl. Environ. Microbiol. 2012, 78, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Powell, D.; Goonetilleke, A.; Gardner, T. Detection and source identification of faecal pollution in non-sewered catchment by means of host-specific molecular markers. Water Sci. Technol. 2008, 58, 579–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, K.V.; Brownell, M.; Wang, S.Y.; Lepo, J.E.; Mott, J.; Nathaniel, R.; Kilgen, M.; Hellein, K.N.; Kennedy, E.; Harwood, V.J. Relationship of human-associated microbial source tracking markers with enterococci in Gulf of Mexico waters. Water Res. 2013, 47, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Bonkosky, M.; Hernandez-Delgado, E.A.; Sandoz, B.; Robledo, I.E.; Norat-Ramirez, J.; Mattei, H. Detection of spatial fluctuations of non-point source fecal pollution in coral reef surrounding waters in southwestern Puerto Rico using PCR-based assays. Mar. Pollut. Bull. 2009, 58, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Staley, C.; Sadowsky, M.J.; Gyawali, P.; Sidhu, J.P.S.; Palmer, A.; Beale, D.J.; Toze, S. Toolbox approaches using molecular markers and 16S rRNA gene amplicon data sets for identification of fecal pollution in surface water. Appl. Environ. Microbiol. 2015, 81, 7067–7077. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Sidhu, J.P.S.; Toze, S. Evaluation of the nifH gene marker of Methanobrevibacter smithii for the detection of sewage pollution in environmental waters in Southeast Queensland, Australia. Environ. Sci. Technol. 2012, 46, 543–550. [Google Scholar] [CrossRef] [PubMed]
- McQuaig, S.; Griffith, J.; Harwood, V.J. The association of fecal indicator bacteria with human viruses and microbial source tracking markers at coastal beaches impacted by nonpoint source pollution. Appl. Environ. Microbiol. 2012, 78, 6423–6432. [Google Scholar] [CrossRef] [PubMed]
- Walters, S.P.; Gannon, V.P.J.; Field, K.G. Detection of Bacteroidales fecal indicators and the zoonotic pathogens E. coli O157:H7, Salmonella, and Campylobacter in river water. Environ. Sci. Technol. 2007, 41, 1856–1862. [Google Scholar] [CrossRef] [PubMed]
- Viau, E.J.; Boehm, A.B. Quantitative PCR-based detection of pathogenic Leptospira in Hawai’ian coastal streams. J. Water Health 2011, 9, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Boehm, A.B.; Fuhrman, J.A.; Mrse, R.D.; Grant, S.B. Tiered approach for identification of a human fecal pollution source at a recreational beach: Case study at Avalon Cay, Catalina Island, California. Environ. Sci. Technol. 2003, 37, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Noble, R.T.; Griffith, J.F.; Blackwood, A.D.; Fuhrman, J.A.; Gregory, J.B.; Hernandez, X.; Liang, X.; Bera, A.A.; Schiff, K. Multitiered approach using quantitative PCR to track sources of fecal pollution affecting Santa Monica Bay, California. Appl. Environ. Microbiol. 2006, 72, 1604–1612. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Reckhow, K.H.; Lukasik, J.; Harwood, V.J. Assessment of sources of human pathogens and fecal contamination in a Florida freshwater lake. Water Res. 2012, 46, 5799–5812. [Google Scholar] [CrossRef] [PubMed]
- Schriewer, A.; Miller, W.A.; Byrne, B.A.; Miller, M.A.; Oates, S.; Conrad, P.A.; Hardin, D.; Yang, H.H.; Chouicha, N.; Melli, A.; et al. Presence of Bacteroidales as a predictor of pathogens in surface waters of the central California Coast. Appl. Environ. Microbiol. 2010, 76, 5802–5814. [Google Scholar] [CrossRef] [PubMed]
- Boehm, A.B.; Yamahara, K.M.; Love, D.C.; Peterson, B.M.; McNeill, K.; Nelson, K.L. Covariation and photoinactivation of traditional and novel indicator organisms and human viruses at a sewage-impacted marine beach. Environ. Sci. Technol. 2009, 43, 8046–8052. [Google Scholar] [CrossRef] [PubMed]
- Viau, E.J.; Lee, D.; Boehm, A.B. Swimmer risk of gastrointestinal illness from exposure to tropical coastal waters impacted by terrestrial dry-weather runoff. Environ. Sci. Technol. 2011, 45, 7158–7165. [Google Scholar] [CrossRef] [PubMed]
- McLellan, S.L.; Eren, A.M. Discovering new indicators of fecal pollution. Trends Microbiol. 2014, 22, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Harwood, V.J.; Nayak, B.; Staley, C.; Sadowsky, M.J. A novel microbial source tracking microarray for pathogen detection and fecal source identification in environmental systems. Environ. Sci. Technol. 2015, 49, 7319–7329. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Harwood, V.J.; Nayak, B.; Weidhaas, J.L. Ultrafiltration and microarray for detection of microbial source tracking marker and pathogen genes in riverine and marine systems. Appl. Environ. Microbiol. 2016, 82, 1625–1635. [Google Scholar] [CrossRef] [PubMed]
- Unno, T.; Jang, J.; Han, D.; Kim, J.H.; Sadowsky, M.J.; Kim, O.S.; Chun, J.; Hur, H.-G. Use of barcoded pyrosequencing and shared OTUs to determine sources of fecal bacteria in watersheds. Environ. Sci. Technol. 2010, 44, 7777–7782. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.C.; Eren, M.A.; Green, H.C.; Shanks, O.C.; Morrison, H.G.; Vineis, J.H.; Sogin, M.L.; McLellan, S.L. Comparison of sewage and animal fecal microbiomes using oligotyping reveals potential human fecal indicators in multiple taxonomic groups. Appl. Environ. Microbiol. 2015, 81, 7023–7033. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Maignien, L.; Sul, W.J.; Murphy, L.G.; Grim, S.L.; Morrison, H.G.; Sogin, M.L. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 2013, 4, 1111–1119. [Google Scholar] [CrossRef] [PubMed]

| Assays | Primer Sequences | Primer and Probe Concentrations | Annealing Temperature (°C) | References |
|---|---|---|---|---|
| 16S rRNA-based assays | ||||
| HF183 | HF183 F:ATCATGAGTTCACATGTCCG Bac708 R:CAATCGGAGTTCTTCGTG | 10 μM 10 μM | 63 | [34] |
| HF183 F:ATCATGAGTTCACATGTCCG SSHBac R:TACCCCGCCTACTATCTAATG | 250 nM 250 nM | 53 | [44] | |
| HF183 F:ATCATGAGTTCACATGTCCG BFDRev:CGTAGGAGTTTGGACCGTGT BFDFam:FAM-CTGAGAGGAAGGTCCCCCACATTGGA-TAMRA | 1 μM 1 μM 80 nM | 60 | [34,46] | |
| HF183 F:ATCATGAGTTCACATGTCCG BacR287:CTTCCTCTCAGAACCCCTATCC BacP234:FAM-CTAATGGAACGCATCCC-MGB | 1 μM 1 μM 80 nM | 60 | [50] | |
| HF134 | HF134 F:GCCGTCTACTCTTGGCCA Bac708 R:CAATCGGAGTTCTTCGTG | 10 μM 10 μM | 63 | [34] |
| HuBac | HuBac566F:GGGTTTAAAGGGAGCGTAGG HuBac692R:CTACACCACGAATTCCGCCT HuBac594P:FAM-TAAGTCAGTTGTGAAAGTTTGCGGCTC-BHQ1 | 15 pmol 15 pmol 5 pmol | 60 | [45] |
| BacHum-UCD | BacHum160F:TGAGTTCACATGTCCGCATGA BacHum241R:CGTTACCCCGCCTACTATCTAATG BacHum1930P:FAM-TCCGGTAGACGATGGGGATGCGTT-TAMRA | 400 nM 400 nM 80 nM | 60 | [36] |
| BacH | BacHF:CTTGGCCAGCCTTCTGAAAG BacHR:CCCCATCGTCTACCGAAAATAC BacHPC:MGBNFQ-GTCCTACCCTAGTACT-FAM BacHPT:MGBNFQ-GTTCTACCGTAGTACT-FAM | 200 nM 200 nM 100 nM 100 nM | 61 | [35] |
| Human-Bac1 | qHS601F:GTTGTGAAAGTTTGCGGCTCA qBac725R:CAATCGGAGTTCTTCGTGATATCTA qHS624P:FAM-CGTAAAATTGCAGTTGA-MGB | 900 nM 900 nM 200 nM | 62 | [37] |
| Non-16S rRNA-based assays | ||||
| HumM2 | Hum2F:CGTCAGGTTTGTTTCGGTATTG Hum2R:TCATCACGTAACTTATTTATATGCATTAGC HumM2P:FAM-TATCGAAAATCTCACGGATTAACTCTTGTGTACGC-TAMRA | 1 μM 1 μM 80 nM | 60 | [38] |
| HumM3 | Hum3F:GTAATTCGCGTTCTTCCTCACAT Hum3R:GGAGGAAACAAGTATGAAGATAGAAGAATTAA HumM2P:FAM-AGGTCTGTCCTTCGAAATAGCGGT-TAMRA | 1 μM 1 μM 80 nM | 60 | [38] |
| B. thetaiotaomicron, α-1.6-mannanase | BtHF:CATCGTTCGTCAGCAGTAACA BtHR:CCAAGAAAAAGGGACAGTGG BtHP:FAM-ACCTGCTG-NFQ | 900 nM 900 nM 250 nM | 60 | [58] |
| gyrB | Bf904F:GGCGGTCTTCCGGGTAAA Bf958R:CACACTTCTGCGGGTCTTTGT Bf923P:FAM-TGGCCGACTGCTC-MGB | 500 nM 500 nM 250 nM | 60 | [39] |
| Assays | Country | Sample Types | No. of Samples Tested (% of Sample Positive) | Concentrations of Gene Copies | References |
|---|---|---|---|---|---|
| HF183 a | USA | Surface water | 10 (30) | 3.2 × 103–1.4 × 104 per L | [82] |
| HF183 a | USA | Creek and storm drain | 90 (44) | Approx. 1.0 × 103–1.9 × 108 per L | [78] |
| HF183 a | Canada | Beach water, stormwater, creek water | 203 (79) | 3.2 × 102–4.0 × 103 per L | [83] |
| HF183 a | USA | Storm drains, creek water | 26 (27) | 4.1 × 102–1.5 × 107 per L | [80] |
| HF183 a | Singapore | Reservoir and catchment water | 54 (93) | 2.5 × 104–6.0 × 105 per L | [84] |
| HF183 a | USA | Coastal water | 230 (29) | 1.2 × 104–1.5 × 104 per L | [85] |
| HF183 a | USA | Stormwater | 6 (100) | 1.0 × 104–3.0 × 106 per L | [86] |
| HF183 a | USA | Harbor water | 10 (100) | 1.3 × 105 per L | [81] |
| HF183 a | France | River water | 14 (7) | 7.3 × 106 per L | [87] |
| HF183 a | France | Catchment water | 240 (17) | 1.6 × 101–2.5 × 102 per L | [88] |
| HF183 a | France | Surface water | 63 (43) | 1.6 × 104–1.0 × 106 per L | [89] |
| HF183 a | France | River water | 23 (43) | 2.3 × 104–9.6 × 105 per L | [90] |
| HF183 a | France | River water | 24 (58) | 3.2 × 104–1.3 × 106 per L | [70] |
| HF183 a | Canada | River water | 1095 (10) | 7.1 × 104 per L | [91] |
| HF183 b | USA | Surface water | 189 (NM) | 5.6 × 103 per μL of extract | [92] |
| HF183 b | USA | Surface water | 29 (76) | 5.1 × 102–2.9 × 104 per L | [93] |
| HF183 a | USA | Stormwater | 14 (43) | 3.9 × 103–6.3 × 106 per L | [94] |
| HF183 c | USA | Stormwater, surface water | 32 (100) | 3.5 × 103–1.4 × 106 per L | [95] |
| HF183 a | Bangladesh | Lake water | 20 (70) | 3.9 × 105–6.3 × 108 per L | [79] |
| HF183 a | Kenya | River water | 18 (11) | 9.5 × 103–4.5 ×104 per L | [67] |
| HF183 a | Canada | Surface water | 374 (10) | 6.3 × 104–6.3 × 106 per L | [72] |
| HF183 a | Canada | Surface water | 184 (72) | 1.0 × 105 per L | [96] |
| HF183 a | USA | River water | 35 (34) | 1.3 × 103–2.4 × 104 per L | [5] |
| HF183 a | Belgium | Coastal water | 80 (76) | Approx. 1.0 × 105–1.0 × 107 per L | [41] |
| HuBac | USA | Surface water and stormwater | 249 (98) | 4.3 × 104–7.8 × 106 per L | [97] |
| HuBac | USA | Creek water | 7 (100) | 0.6 × 100–2.4 × 102 mg of feces per L | [45] |
| BacHum-UCD | USA | Stormwater | 828 (57) | 3.0 × 103–4.1 × 106 per L | [98] |
| BacHum-UCD | USA | Surface water and stormwater | 73 (88) | 3.0 × 105–5.0 × 105 per L | [99] |
| BacHum-UCD | Kenya | River water | 18 (11) | 1.3 × 105–1.6 × 105 per L | [67] |
| BacH | Canada | River water | 130 (88) | 8.0 × 101–1.0 × 103 per L | [74] |
| BacH | Austria | Spring water, river water, watering brook | 6 (50) | 6.5 × 102–1.1 × 106 per L | [35] |
| HuBac | Bangladesh | Pond water | 43 (84) | 1.3 × 105–6.8 × 108 per L | [100] |
| BacH | Israel | Spring water | 46 (100) | 1.1 × 104–3.2 × 105 per L | [101] |
| Human-Bac1 | Japan | River water | 30 (100) | 2.7 × 103–6.5 × 104 per L | [102] |
| HumM2 | USA | Surface water | 29 (76) | 7.6 × 102–2.1 × 103 per L | [91] |
| HumM3 | Israel | Spring water | 46 (65) | 1.1 × 103–9.6 × 103 per L | [101] |
| B. thetaiotaomicron | USA | Surface water | 29 (76) | 6.7 × 102–1.2 × 106 per L | [58] |
| Assays | Matrices | Temperature (°C) | Salinity (ppt) | Conditions | T90 (Days) | Reference |
|---|---|---|---|---|---|---|
| HF183 | Freshwater | 4 | >24.0 a | [44] | ||
| 12 | 10.0 a | |||||
| 28 | 3.00 a | |||||
| HF183 | Canal water | 6–26 | Sunlight | 2.72 a | [113] | |
| 2.08 b | ||||||
| HF183 | Seawater | 20 | 33 | Dark | 2.30 a | [112] |
| Freshwater | 1.70 a | |||||
| HF183 | Beach sand (14% moisture) | 25 | Dark | 1.50 a | [114] | |
| Beach sand (28% moisture) | 4.90 a | |||||
| HF183 | Freshwater | 13 | Light | 0.72 a | [106] | |
| Dark | 0.75 a | |||||
| Marine water | Light | 3.98 a | ||||
| Dark | 0.49 a | |||||
| HF183 | Freshwater | 13 | Light | 0.41 b | [106] | |
| Dark | 1.03 b | |||||
| Marine water | Light | 5.50 b | ||||
| Dark | 1.40 b | |||||
| HF183 | Freshwater | 14–21 | Sunlight | 3.44 a | [115] | |
| Seawater | 2.70 a | |||||
| HF183 | River water | 25 | Artificial sunlight | 0.85 a | [105] | |
| 15 | 1.26 a | |||||
| Sediment | 25 | 1.44 a | ||||
| HF183 | Freshwater | 14 | Sunlight | 1.37 a | [110] | |
| Dark | 2.59 a | |||||
| Seawater | Sunlight | 1.45 a | ||||
| Dark | 1.65 a | |||||
| HF183 | River water | 13 | Sunlight | 0.59 a | [116] | |
| 0.71 b | ||||||
| Dark | 0.83 a | |||||
| 0.91 b | ||||||
| HF134 | River water | 13 | Sunlight | 0.71 a | [116] | |
| 0.66 b | ||||||
| Dark | 0.83 a | |||||
| 0.91 b | ||||||
| BacHum-UCD | Watered Beach sand | 22 | Dark | 14.5 a | [117] | |
| Control beach sand | 36.2 a | |||||
| BacHum-UCD | Seawater | 17 | 34.2 | Sunlight | 1.79 a | [116] |
| Dark | 8.72 a | |||||
| BacHum-UCD | Seawater | 14 | 31.5 | Sunlight | 3.75 a | [108] |
| Dark | 3.58 a | |||||
| Sunlight | 0.56 c | |||||
| Dark | 0.71 c | |||||
| BacHum-UCD | Freshwater | 22 | Sunlight | 0.52 c | [118] | |
| Dark | 1.29 c | |||||
| Sunlight | 2.14 a | |||||
| Sunlight | 1.29 a | |||||
| BacHum-UCD | Freshwater | 22 | Sunlight | 2.44 a | [111] | |
| Dark | 2.50 a | |||||
| Sunlight | 0.79 c | |||||
| Dark | 1.98 c | |||||
| BacHum-UCD | River water | 25 | Artificial sunlight | 0.77 a | [105] | |
| 15 | 1.17 a | |||||
| Sediment | 25 | 1.04 a | ||||
| BacH | Creek water | 20–25 | 0.75 a | [74] | ||
| 3.85 a | ||||||
| Human-Bac1 | River water | 4 | Dark | 14.3 a | [107] | |
| 10 | 3.57 a | |||||
| 20 | 1.96 a | |||||
| 30 | 1.66 a | |||||
| 10 | 0 | 3.33 a | ||||
| 10 | 3.33 a | |||||
| Seawater | 20 | 3.70 a | ||||
| 20 | 3.70 a | |||||
| HumM2 | Freshwater | 13 | Light | 1.10 a | [106] | |
| Dark | 0.18 a | |||||
| Marine water | Light | 4.23 a | ||||
| Dark | 6.95 a | |||||
| B. thetaiotaomicron, α-1.6-mannanase | River water (solid matrix) | 4 | Dark | 27.0 a | [109] | |
| River water | 9.60 a | |||||
| River water (solid matrix) | 27 | 18.0 a | ||||
| River water | 1.80 a | |||||
| River water (solid matrix) | 37 | 3.20 a | ||||
| River water | 1.00 a |
| Analytes | Studies | |
|---|---|---|
| Weak to Strong Positive Correlation | Negative to No Correlation | |
| HF183 vs. Fecal coliform | [96] | [91,121] |
| HF183 vs. E. coli | [5,84,88,98,119,122] | [42,70,86,91,123] |
| HF183 vs. Enterococcus spp. | [81,98,119,120,122] | [86,88,91,121,123] |
| HF183 vs. C. perfringens | - | [91] |
| HF183 vs. Adenoviruses | [124] | - |
| HF183 vs. Campylobacter spp. | [125,126] | [69,72] |
| HF183 vs. Enteroviruses | [127,128] | [129] |
| HF183 vs. Salmonella spp. | [72,91] | [69,129] |
| HF183 vs. Cryptosporidium spp. | - | [91,129] |
| HF183 vs. Giardia spp. | - | [91,129] |
| HF183 vs. E. coli O157:H7 | - | [69,72,91] |
| HF183 vs. Shiga-toxin producing E. coli | - | [69] |
| HF134 vs. E. coli | [119] | - |
| HF134 vs. Enterococcus spp. | [119] | - |
| HuBac vs. E. coli | [100] | - |
| BacHum-UCD vs. Campylobacter spp. | [126,130] | - |
| BacHum-UCD vs. Cryptosporidium spp. | [130] | - |
| BacHum-UCD vs. E. coli O157:H7 | [130] | - |
| BacHum-UCD vs. Leptospira spp. | [126,130] | - |
| BacHum-UCD vs. Salmonella spp. | [130] | - |
| BacHum-UCD vs. Enteroviruses | [131] | - |
| BacHum-UCD vs. Adenoviruses | - | [132] |
| BacHum-UCD vs. noroviruses | - | [132] |
| Human-Bac1 vs. Fecal coliform | - | [37] |
| Human-Bac1 vs. C. perfringens | - | [102] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, W.; Hughes, B.; Harwood, V.J. Current Status of Marker Genes of Bacteroides and Related Taxa for Identifying Sewage Pollution in Environmental Waters. Water 2016, 8, 231. https://doi.org/10.3390/w8060231
Ahmed W, Hughes B, Harwood VJ. Current Status of Marker Genes of Bacteroides and Related Taxa for Identifying Sewage Pollution in Environmental Waters. Water. 2016; 8(6):231. https://doi.org/10.3390/w8060231
Chicago/Turabian StyleAhmed, Warish, Bridie Hughes, and Valerie J. Harwood. 2016. "Current Status of Marker Genes of Bacteroides and Related Taxa for Identifying Sewage Pollution in Environmental Waters" Water 8, no. 6: 231. https://doi.org/10.3390/w8060231






