Coliphages as Model Organisms in the Characterization and Management of Water Resources
Abstract
:1. Introduction
2. Coliphages
3. Replication of Coliphages Outside the Gut
4. Are the Methods Cumbersome?
4.1. Somatic Coliphages
4.2. F-Specific Coliphages
4.3. Molecular and Fast Methods
4.4. Concentration Methods
4.5. Additional Methodological Issues
4.5.1. Scale-Up
4.5.2. Reference Material
4.5.3. Costs
5. Which Group Should Be Used? Somatic vs. F-Specific Coliphages
5.1. Abundance in Pollution Sources
5.2. Persistence in Water Environments and Resistance to Treatment
5.3. Resistance to Treatment
5.4. Abundance of Bacteriophages in Surface Water and Groundwater
5.5. General Conclusions for Practical Uses
6. Relationship of Coliphages to the Occurrence of Human Viruses and to Health Risk
6.1. Relationship to Human Viruses
6.2. Relationship to Health Risk
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- AWPRC Study Group on Health Related Water Microbiology. Bacteriophages as model viruses in water quality controlag. Water Res. 1991, 25, 529–545. [Google Scholar]
- Armon, R.; Kott, Y. Bacteriophages as indicators of pollution. Crit. Rev. Environ. Sci. Technol. 1996, 26, 299–335. [Google Scholar] [CrossRef]
- Grabow, W. Bacteriophages: update on application as models for viruses in water. Water SA 2001, 27, 251–268. [Google Scholar] [CrossRef]
- Jofre, J. Indicators of waterborne viruses. In Human Viruses in Water; Bosch, A., Ed.; Perspectives in Medical Virology Volume 17; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Mendez, J.; Jofre, J.; Lucena, F.; Contreras, N.; Mooijman, K.; Araujo, R. Conservation of phage reference materials and water samples containing bacteriophages of enteric bacteria. J. Virol. Methods 2002, 106, 215–224. [Google Scholar] [CrossRef]
- Mooijman, K.A.; Ghameshlou, Z.; Bahar, M.; Jofre, J.; Havelaar, A.H. Enumeration of bacteriophages in water by different laboratories of the European Union in two interlaboratory comparison studies. J. Virol. Methods 2005, 127, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Queensland-Government. Queensland Water Recycling Guidelines; WaterWise; Queensland Environmental Protection Agency: Brisbane, Australia, 2005. [Google Scholar]
- North Carolina Environmental Quality. North Carolina Adm. Code 15A NCAC 2U Reclaimed Water; North Carolina Department of Environment and Natural Resources: Raleigh, NC, USA, 2011. [Google Scholar]
- USEPA. National Primary Drinking Water Regulations: Groundwater rule. Final Rule; 40 CFR Parts 9, 141 and 142, Federal Register, Vol. 71, n. 216; USEPA: Washington, DC, USA, 2006. [Google Scholar]
- Department of Environment and Conservation. Western Australian Guidelines for Biosolids Management; Department of Environment and Conservation: Perth, Australia, 2012. [Google Scholar]
- Criterios para el uso de biosólidos generados en plantas de tratamiento de aguas residuales municipales; Decreto Número 1287; Ministerio de Vivienda, Ciudad y Territorio de Colombia: Bogot, Colombia, 2014.
- USEPA Office of Water. Review of Coliphages as Possible Indicators of Fecal Contamination for Ambient Water Quality. EPA 820-R-15-098; USEPA Office of Water: Washington, DC, USA, 2015. [Google Scholar]
- Rajala-Mustonen, R.L.; Heinonen-Tanski, H. Sensitivity of host strains and host range of coliphages isolated from Finnish and Nicaraguan wastewater. Water Res. 1994, 28, 1811–1815. [Google Scholar] [CrossRef]
- Muniesa, M.; Lucena, F.; Jofre, J. Study of the potential relationship between the morphology of infectious somatic coliphages and their persistence in the environment. J. Appl. Microbiol. 1999, 87, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Fauquet, C.; Mayo, M.A.; Maniloff, J.; Desselberger, U.; Ball, L. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses; Elsevier/Academic Press: London, UK, 2005. [Google Scholar]
- Furuse, K. Distribution of coliphages in the environment: general considerations. In Phage Ecology; Goyal, S.M., Gerba, C.P., Bitton, G., Eds.; John Wiley & Sons, Inc.: New York, NY, USA, 1987; pp. 87–124. [Google Scholar]
- Beekwilder, J.; Nieuwenhuizen, R.; Havelaar, A.H.; van Duin Leiden, J. An oligonucleotide hybridization assay for the identification and enumeration of F-specific RNA phages in surface water. J. Appl. Bacteriol. 1996, 80, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.C.; Shieh, Y.S.; van Duin, J.; Beekwilder, M.J.; Sobsey, M.D. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl. Environ. Microbiol. 1995, 61, 3960–3966. [Google Scholar] [PubMed]
- Muniesa, M.; Lucena, F.; Blanch, A.R.; Payán, A.; Jofre, J. Use of abundance ratios of somatic coliphages and bacteriophages of Bacteroides thetaiotaomicron GA17 for microbial source identification. Water Res. 2012, 46, 6410–6418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Farahbakhsh, K. Removal of native coliphages and coliform bacteria from municipal wastewater by various wastewater treatment processes: Implications to water reuse. Water Res. 2007, 41, 2816–2824. [Google Scholar] [CrossRef] [PubMed]
- Haramoto, E.; Fujino, S.; Otagiri, M. Distinct behaviors of infectious F-specific RNA coliphage genogroups at a wastewater treatment plant. Sci. Total Environ. 2015, 520, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, O.; Casas, S.; Galvañ, C.; Lucena, F.; Bosch, A.; Galofré, B.; Mesa, J.; Jofre, J.; Bernat, X. Direct ultrafiltration performance and membrane integrity monitoring by microbiological analysis. Water Res. 2015, 83, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Rezaeinejad, S.; Vergara, G.G.; Woo, C.H.; Lim, T.T.; Sobsey, M.D.; Gin, K.Y. Surveillance of enteric viruses and coliphages in a tropical urban catchment. Water Res. 2014, 58, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F. Enterobacteriales. In Bergey’s Manual® of Systematic Bacteriology. Volume 2: The Proteobacteria, Part B: The Gammaproteobacteria; Garrity, G., Brenner, D.J., Krieg, N.R., Staley, J.R., Eds.; Springer US: Boston, MA, USA, 2005; pp. 587–850. [Google Scholar]
- Ishii, S.; Sadowsky, M.J. Escherichia coli in the Environment: Implications for Water Quality and Human Health. Microbes Environ. 2008, 23, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, J.M.; Metcalf, T.G. Coliphages as indicators of enteric viruses in shellfish and shellfish raising estuarine waters. Water Res. 1975, 9, 613–616. [Google Scholar] [CrossRef]
- Seeley, N.D.; Primrose, S.B. The effect of temperature on the ecology of aquatic bacteriophages. J. Gen. Virol. 1980, 46, 87–95. [Google Scholar] [CrossRef]
- Grabow, W.O.K.; Burger, J.S.; Nupen, E.P. Evaluation of acid-fast bacteria, Candida albicans, enteric viruses and conventional indicators for monitoring wastewater reclamation systems. Prog. Water Technol 1980, 12, 803–817. [Google Scholar]
- Borrego, J.J.; Córnax, R.; Moriñigo, M.A.; Martínez-Manzanares, E.; Romero, P. Coliphages as an indicator of faecal pollution in water. their survival and productive infectivity in natural aquatic environments. Water Res. 1990, 24, 111–116. [Google Scholar] [CrossRef]
- Wiggins, B.A.; Alexander, M. Minimum bacterial density for bacteriophage replication: Implications for significance of bacteriophages in natural ecosystems. Appl. Environ. Microbiol. 1985, 49, 19–23. [Google Scholar] [PubMed]
- Muniesa, M.; Jofre, J. Factors influencing the replication of somatic coliphages in the water environment. Antonie Van Leeuwenhoek 2004, 86, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Muniesa, M.; Mocé-Llivina, L.; Katayama, H.; Jofre, J. Bacterial host strains that support replication of somatic coliphages. Antonie Van Leeuwenhoek 2003, 83, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Cornax, R.; Moriñigo, M.A.; Balebona, M.C.; Castro, D.; Borrego, J.J. Significance of several bacteriophage groups as indicators of sewage pollution in marine waters. Water Res. 1991, 25, 673–678. [Google Scholar] [CrossRef]
- Raivio, T. Identifying your enemies—Could envelope stress trigger microbial immunity? Mol. Microbiol. 2011, 79, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Perez-Rodriguez, R.; Haitjema, C.; Huang, Q.; Nam, K.H.; Bernardis, S.; Ke, A.; DeLisa, M.P. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol. 2011, 79, 584–599. [Google Scholar] [CrossRef] [PubMed]
- Muniesa, M.; Jofre, J. The contribution of induction of temperate phages to the numbers of free somatic coliphages in waters is not significant. FEMS Microbiol. Lett. 2007, 270, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Delgado, E.A.; Toranzos, G.A. In situ replication studies of somatic and male-specific coliphages in a tropical pristine river. Water Sci. Technol. 1995, 31, 247–250. [Google Scholar] [CrossRef]
- Jofre, J. Is the replication of somatic coliphages in water environments significant? J. Appl. Microbiol. 2009, 106, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Havelaar, A.H.; Pot-Hogeboom, W.M. F-specific RNA-Bacteriophages as model viruses in water hygiene: Ecological aspects. Water Sci. Technol. 1988, 20, 399–407. [Google Scholar]
- Woody, M.A.; Cliver, D.O. Replication of coliphage Q beta as affected by host cell number, nutrition, competition from insusceptible cells and non-FRNA coliphages. J. Appl. Microbiol. 1997, 82, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. Bacteriophages; Interscience Publishers, Inc.: New York, NY, USA, 1959. [Google Scholar]
- USEPA Office of Water. Results of the Interlaboratory Validation of EPA Method 1601 for Presence/Absence of Male-specific (F+) and Somatic Coliphages in Water by Two-Step Enrichment; EPA 821-R-03-015; USEPA Office of Water: Washington, DC, USA, 2003. [Google Scholar]
- USEPA Office of Water. Results of the Interlaboratory Validation of EPA Method 1602 for Enumeration of Male-specific (F+) and Somatic Coliphage in Water by Single Agar Layer (SAL); EPA 821-R-03-016; USEPA Office of Water: Washington, DC, USA, 2003. [Google Scholar]
- International Standardization Organization. Water Quality—Detection and Enumeration of Bacteriophages—Part 2: Enumeration of Somatic Coliphages; ISO-10705-2; International Standardization Organization: Geneva, Switzerland, 2000. [Google Scholar]
- USEPA Office of Water. Method 1601: Male-specific (F+) and Somatic Coliphage in Water by Two-step Enrichment Procedure; EPA 821-R-01-030; USEPA Office of Water: Washington, DC, USA, 2001. [Google Scholar]
- USEPA Office of Water. Method 1602: Male-specific (F+) and Somatic Coliphage in Water by Single Agar Layer (SAL) Procedure; EPA 821-R-01-029; USEPA Office of Water: Washington, DC, USA, 2001. [Google Scholar]
- Rice, E.W.; Baird, R.B.; Eaton, A.D.; Clesceri, L.S. Standard Methods for the Examination of Water and Wastewater; American Public Health Association, American Water Works Association, Water Environment Federation: Washington, DC, USA, 2012. [Google Scholar]
- Grabow, W.O.K.; Holtzhausen, C.S.; de Villiers, J.C. Research on Bacteriophages as Indicators of Water Quality; WRC Report No. 321/1/93; Water Research Commission: Pretoria, South Africa, 1993. [Google Scholar]
- Guzmán, C.; Mocé-Llivina, L.; Lucena, F.; Jofre, J. Evaluation of Escherichia coli host strain CB390 for simultaneous detection of somatic and F-specific coliphages. Appl. Environ. Microbiol. 2008, 74, 531–534. [Google Scholar] [CrossRef] [PubMed]
- Green, J. The APHA standard method for the enumeration of somatic coliphages in water has low efficiency of plating. Water Res. 2000, 34, 759–762. [Google Scholar] [CrossRef]
- Havelaar, A.H.; Hogeboom, W.M. Factors affecting the enumeration of coliphages in sewage and sewage-polluted waters. Antonie Van Leeuwenhoek 1983, 49, 387–397. [Google Scholar] [PubMed]
- USEPA Office of Research and Development. USEPA Manual of Methods for Virology—Chapter 16; EPA 600/4-84/013 (N16); USEPA Office of Research and Development: Washington, DC, USA, 2001. [Google Scholar]
- Debartolomeis, J.; Cabelli, V.J. Evaluation of an Escherichia coli host strain for enumeration of F male-specific bacteriophages. Appl. Environ. Microbiol. 1991, 57, 1301–1305. [Google Scholar] [PubMed]
- Havelaar, A.H.; Hogeboom, W.M. A method for the enumeration of male-specific bacteriophages in sewage. J. Appl. Bacteriol. 1984, 56, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Sobsey, M.D. Comparative survival of indicator viruses and enteric viruses in seawater and sediment. Water Sci. Technol. 1993, 27, 425–428. [Google Scholar]
- Schaper, M.; Jofre, J. Comparison of methods for detecting genotypes of F-specific RNA bacteriophages and fingerprinting the origin of faecal pollution in water samples. J. Virol. Methods 2000, 89, 1–10. [Google Scholar] [CrossRef]
- International Standardization Organization. Water Quality—Detection and Enumeration of Bacteriophages—Part 1: Enumeration of F-specific RNA Bacteriophages; ISO-10705-1; International Standardization Organization: Geneva, Switzerland, 1995; p. 11. [Google Scholar]
- Love, D.C.; Sobsey, M.D. Simple and rapid F+ coliphage culture, latex agglutination, and typing assay to detect and source track fecal contamination. Appl. Environ. Microbiol. 2007, 73, 4110–4118. [Google Scholar] [CrossRef] [PubMed]
- Ogorzaly, L.; Gantzer, C. Development of real-time RT-PCR methods for specific detection of F-specific RNA bacteriophage genogroups: Application to urban raw wastewater. J. Virol. Methods 2006, 138, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Kirs, M.; Smith, D.C. Multiplex quantitative real-time reverse transcriptase PCR for F+-specific RNA coliphages: a method for use in microbial source tracking. Appl Env. Microbiol 2007, 73, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.D.; Cooper, E.M.; Casanova, L.; Sobsey, M.D.; Genthner, F.J. A reverse transcription-PCR assay to distinguish the four genogroups of male-specific (F+) RNA coliphages. J. Virol. Methods 2009, 159, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Hewitt, J.; Greening, G.E. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl. Environ. Microbiol. 2010, 76, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Long, S.C.; El-Khoury, S.S.; Oudejans, S.J.G.; Sobsey, M.D.; Vinjé, J. Assessment of sources and diversity of male-specific coliphages for source tracking. Environ. Eng. Sci. 2005, 22, 367–377. [Google Scholar] [CrossRef]
- Ijzerman, M.M.; Falkinham, J.O.; Hagedorn, C. A liquid, colorimetric presence-absence coliphage detection method. J. Virol. Methods 1993, 45, 229–234. [Google Scholar] [CrossRef]
- Guzmán Luna, C.; Costán-Longares, A.; Lucena, F.; Jofre, J. Detection of somatic coliphages through a bioluminescence assay measuring phage mediated release of adenylate kinase and adenosine 5’-triphosphate. J. Virol. Methods 2009, 161, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Salter, R.S.; Durbin, G.W. Modified USEPA method 1601 to indicate viral contamination of groundwater. J. Am. Water Works Assoc. 2012, 104, 480–488. [Google Scholar] [CrossRef]
- Salter, R.S.; Durbin, G.W.; Conklin, E.; Rosen, J.; Clancy, J. Proposed modifications of Environmental Protection Agency Method 1601 for detection of coliphages in drinking water, with same-day fluorescence-based detection and evaluation by the performance-based measurement system and alternative test protocol valida. Appl. Environ. Microbiol. 2010, 76, 7803–7810. [Google Scholar] [CrossRef] [PubMed]
- Sobsey, M.D.; Schwab, K.J.; Handzel, T.R. A simple membrane filter method to concentrate and enumerate male-specific RNA coliphages. J. Am. Water Work. Assoc. 1990, 82, 52–59. [Google Scholar]
- Méndez, J.; Audicana, A.; Isern, A.; Llaneza, J.; Moreno, B.; Tarancón, M. L.; Jofre, J.; Lucena, F. Standardised evaluation of the performance of a simple membrane filtration-elution method to concentrate bacteriophages from drinking water. J. Virol. Methods 2004, 117, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Schulze, E.; Lenk, J. Concentration of coliphages from drinking water by Mg(OH)2 flocculation. Naturwissenschaften 1983, 70, 612–613. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Coll, N.; Lucena, F.; Mooijman, K.; Havelaar, A.; Pierz, V.; Boque, M.; Gawler, A.; Höller, C.; Lambiri, M.; Mirolo, G.; et al. Occurrence and levels of indicator bacteriophages in bathing waters throughout Europe. Water Res. 2002, 36, 4963–74. [Google Scholar] [CrossRef]
- Gerba, C.P.; Pepper, I.L.; Whitehead, L.F. A risk assessment of emerging pathogens of concern in the land application of biosolids. Water Sci. Technol. 2002, 46, 225–230. [Google Scholar] [PubMed]
- Lodder, W.J.; de Roda Husman, A.M. Presence of noroviruses and other enteric viruses in sewage and surface waters in The Netherlands. Appl. Environ. Microbiol. 2005, 71, 1453–1461. [Google Scholar] [CrossRef] [PubMed]
- Gomila, M.; Solis, J.J.; David, Z.; Ramon, C.; Lalucat, J. Comparative reductions of bacterial indicators, bacteriophage-infecting enteric bacteria and enteroviruses in wastewater tertiary treatments by lagooning and UV-radiation. Water Sci. Technol. 2008, 58, 2223–2233. [Google Scholar] [CrossRef] [PubMed]
- Gantzer, C.; Maul, A.; Audic, J.M.; Schwartzbrod, L. Detection of infectious enteroviruses, enterovirus genomes, somatic coliphages, and Bacteroides fragilis phages in treated wastewater. Appl. Environ. Microbiol. 1998, 64, 4307–4312. [Google Scholar] [PubMed]
- Costán-Longares, A.; Montemayor, M.; Payán, A.; Méndez, J.; Jofre, J.; Mujeriego, R.; Lucena, F. Microbial indicators and pathogens: Removal, relationships and predictive capabilities in water reclamation facilities. Water Res. 2008, 42, 4439–4448. [Google Scholar] [CrossRef] [PubMed]
- Harwood, V.J.; Levine, A.D.; Scott, T.M.; Chivukula, V.; Lukasik, J.; Farrah, S.R.; Rose, J.B. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 2005, 71, 3163–3170. [Google Scholar] [CrossRef] [PubMed]
- Lodder, W.J.; van den Berg, H.H.J.L.; Rutjes, S.A.; de Roda Husman, A.M. Presence of enteric viruses in source waters for drinking water production in The Netherlands. Appl. Environ. Microbiol. 2010, 76, 5965–5971. [Google Scholar] [CrossRef] [PubMed]
- Rubiano, M.-E.; Agulló-Barceló, M.; Casas-Mangas, R.; Jofre, J.; Lucena, F. Assessing the effects of tertiary treated wastewater reuse on a Mediterranean river (Llobregat, NE Spain): Pathogens and indicators. Environ. Sci. Pollut. Res. Int. 2012, 19, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Lipp, E.K.; Farrah, S.A.; Rose, J.B. Assessment and impact of microbial fecal pollution and human enteric pathogens in a coastal community. Mar. Pollut. Bull. 2001, 42, 286–293. [Google Scholar] [CrossRef]
- Guzmán, C.; Jofre, J.; Montemayor, M.; Lucena, F. Occurrence and levels of indicators and selected pathogens in different sludges and biosolids. J. Appl. Microbiol. 2007, 103, 2420–2429. [Google Scholar] [CrossRef] [PubMed]
- Martin-Diaz, J.; Casas-Mangas, R.; Garcia-Aljaro, C.; Blanch, A.R.; Lucena, F. Somatic coliphages as surrogates for enteroviruses in sludge hygienization treatments. Water Sci. Technol. 2016, 73, wst2016066. [Google Scholar] [CrossRef] [PubMed]
- Rutgers, S.A.; Rutjes, S.A.; Lodder, W.J.; Van Leeuwen, A.D.; de Roda Husman, A.M. Detection of infectious rotavirus in naturally contaminated source waters for drinking water production. J. Appl. Microbiol. 2009, 107, 97–105. [Google Scholar]
- Tani, N.; Shimamoto, K.; Ichimura, K.; Nishii, Y.; Tomita, S.; Oda, Y. Enteric virus levels in river water. Water Res. 1992, 26, 45–48. [Google Scholar] [CrossRef]
- Lee, S.-H.; Kim, S.-J. Detection of infectious enteroviruses and adenoviruses in tap water in urban areas in Korea. Water Res. 2002, 36, 248–256. [Google Scholar] [CrossRef]
- Bosch, A.; Pintó, R.M.; Blanch, A.; Jofre, J. Detection of human rotavirus in sewage through two concentration procedures. Water Res. 1988, 22, 343–348. [Google Scholar] [CrossRef]
- Gerba, C.P.; Rose, J.B.; Haas, C.N.; Crabtree, K.D. Waterborne rotavirus: A risk assessment. Water Res. 1996, 30, 2929–2940. [Google Scholar] [CrossRef]
- USEPA Office of Research and Development. Method 1615. Measurement of Enterovirus and Norovirus Occurrence in Water by Culture and RT-qPCR; Version 1.1; National Exposure Research Laboratory, USEPA Office of Research and Development: Cincinnati, OH, USA, 2012. [Google Scholar]
- Havelaar, A.H.; Furuse, K.; Hogeboom, W.M. Bacteriophages and indicator bacteria in human and animal faeces. J. Appl. Bacteriol. 1986, 60, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.O.K.; Neubrech, T.E.; Holtzhausen, C.S.; Jofre, J. Bacteroides fragilis and Escherichia coli bacteriophages: excretion by humans and animals. Water Sci. Technol. 1995, 31, 223–230. [Google Scholar] [CrossRef]
- Lee, J.E.; Lim, M.Y.; Kim, S.Y.; Lee, S.; Lee, H.; Oh, H.-M.; Hur, H.-G.; Ko, G. Molecular characterization of bacteriophages for microbial source tracking in Korea. Appl. Environ. Microbiol. 2009, 75, 7107–7114. [Google Scholar] [CrossRef] [PubMed]
- Havelaar, A.H.; Pot-Hogeboom, W.M.; Furuse, K.; Pot, R.; Hormann, M.P. F-specific RNA bacteriophages and sensitive host strains in faeces and wastewater of human and animal origin. J. Appl. Bacteriol. 1990, 69, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Calci, K.R.; Burkhardt, W.; Watkins, W.D.; Rippey, S.R. Occurrence of male-specific bacteriophage in feral and domestic animal wastes, human feces, and human-associated wastewaters. Appl. Environ. Microbiol. 1998, 64, 5027–5029. [Google Scholar] [PubMed]
- Lucena, F.; Mendez, X.; Moron, A.; Calderon, E.; Campos, C.; Guerrero, A.; Cardenas, M.; Gantzer, C.; Shwartzbrood, L.; Skraber, S.; et al. Occurrence and densities of bacteriophages proposed as indicators and bacterial indicators in river waters from Europe and South America. J. Appl. Microbiol. 2003, 94, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Yahya, M.; Hmaied, F.; Jebri, S.; Jofre, J.; Hamdi, M. Bacteriophages as indicators of human and animal faecal contamination in raw and treated wastewaters from Tunisia. J. Appl. Microbiol. 2015, 118, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.; Sobsey, M. Microbial indicator reductions in alternative treatment systems for swine wastewater. Water Sci. Technol. 1998, 38, 119–122. [Google Scholar] [CrossRef]
- Blanch, A.R.; Belanche-Muñoz, L.; Bonjoch, X.; Ebdon, J.; Gantzer, C.; Lucena, F.; Ottoson, J.; Kourtis, C.; Iversen, A.; Kühn, I.; et al. Integrated analysis of established and novel microbial and chemical methods for microbial source tracking. Appl. Environ. Microbiol. 2006, 72, 5915–5926. [Google Scholar] [CrossRef] [PubMed]
- Aw, T.G.; Gin, K.Y.-H. Environmental surveillance and molecular characterization of human enteric viruses in tropical urban wastewaters. J. Appl. Microbiol. 2010, 109, 716–730. [Google Scholar] [CrossRef] [PubMed]
- Lucena, F.; Duran, A.E.; Morón, A.; Calderón, E.; Campos, C.; Gantzer, C.; Skraber, S.; Jofre, J.; Moron, A.; Calderon, E.; et al. Reduction of bacterial indicators and bacteriophages infecting faecal bacteria in primary and secondary wastewater treatments. J. Appl. Microbiol. 2004, 97, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Alcalde, L.; Oron, G.; Gillerman, L.; Salgot, M.; Manor, Y. Removal of fecal coliforms, somatic coliphages and F-specific bacteriophages in a stabilization pond and reservoir system in arid regions. Water Sci. Technol. Water Supply 2003, 3, 177–184. [Google Scholar]
- Mandilara, G.; Mavridou, A.; Lambiri, M.; Vatopoulos, A.; Rigas, F. The use of bacteriophages for monitoring the microbiological quality of sewage sludge. Environ. Technol. 2006, 27, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Sinton, L.W.; Finlay, R.K.; Lynch, P.A. Sunlight inactivation of fecal bacteriophages and bacteria in sewage-polluted seawater. Appl. Environ. Microbiol. 1999, 65, 3605–3613. [Google Scholar] [PubMed]
- Sinton, L.W.; Hall, C.H.; Lynch, P.A.; Davies-Colley, R.J. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl. Environ. Microbiol. 2002, 68, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Mocé-Llivina, L.; Lucena, F.; Jofre, J. Enteroviruses and bacteriophages in bathing waters. Appl. Environ. Microbiol. 2005, 71, 6838–6844. [Google Scholar] [CrossRef] [PubMed]
- Durán, A.E.; Muniesa, M.; Méndez, X.; Valero, F.; Lucena, F.; Jofre, J. Removal and inactivation of indicator bacteriophages in fresh waters. J. Appl. Microbiol. 2002, 92, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Gessel, P.D.; Hansen, N.C.; Goyal, S.M.; Johnston, L.J.; Webb, J. Persistence of zoonotic pathogens in surface soil treated with different rates of liquid pig manure. Appl. Soil Ecol. 2004, 25, 237–243. [Google Scholar] [CrossRef]
- Agulló-Barceló, M.; Polo-López, M.I.; Lucena, F.; Jofre, J.; Fernández-Ibáñez, P. Solar advanced oxidation processes as disinfection tertiary treatments for real wastewater: Implications for water reclamation. Appl. Catal. B Environ. 2013, 136–137, 341–350. [Google Scholar] [CrossRef]
- Cole, D.; Long, S.C.; Sobsey, M.D. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl. Environ. Microbiol. 2003, 69, 6507–6514. [Google Scholar] [CrossRef] [PubMed]
- Rahman, R.; Alum, A.; Ryu, H.; Abbaszadegan, M. Identification of microbial faecal sources in the New River in the United States-Mexican border region. J. Water Health 2009, 7, 267–275. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, M.R.; Rose, J.B. Application of Bacteroides fragilis phage as an alternative indicator of sewage pollution in Tampa Bay, Florida. Estuaries Coasts 2006, 29, 246–256. [Google Scholar] [CrossRef]
- Allwood, P.B.; Malik, Y.S.; Hedberg, C.W.; Goyal, S.M. Survival of F-specific RNA coliphage, feline calicivirus, and Escherichia coli in water: A comparative study. Appl. Environ. Microbiol. 2003, 69, 5707–5710. [Google Scholar] [CrossRef] [PubMed]
- Nieuwstad, T.J.; Mulder, E.P.; Havelaar, A.H.; Van Olphen, M. Elimination of micro-organisms from wastewater by tertiary precipitation and simultaneous precipitation followed by filtration. Water Res. 1988, 22, 1389–1397. [Google Scholar] [CrossRef]
- Rose, J.B.; Farrah, S.R.; Harwood, V. Reduction of Pathogens, Indicator Bacteria, and Alternative Indicators by Wastewater Treatment and Reclamation Processes; IWA Publishing: Lodon, UK; Water Environment Research Foundation: Alexandria, VA, USA, 2005. [Google Scholar]
- Ottoson, J.; Hansen, A.; Westrell, T.; Johansen, K.; Norder, H.; Stenström, T.A. Removal of noro- and enteroviruses, Giardia cysts, Cryptosporidium oocysts, and fecal indicators at four secondary wastewater treatment plants in Sweden. Water Environ. Res. 2006, 78, 828–834. [Google Scholar] [CrossRef] [PubMed]
- De Luca, G.; Sacchetti, R.; Leoni, E.; Zanetti, F. Removal of indicator bacteriophages from municipal wastewater by a full-scale membrane bioreactor and a conventional activated sludge process: Implications to water reuse. Bioresour. Technol. 2013, 129, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Monclús, H.; Jofre, J.; Rodriguez-Roda, I.; Comas, J.; Balcázar, J.L. Removal of microbial indicators from municipal wastewater by a membrane bioreactor (MBR). Bioresour. Technol. 2011, 102, 5004–5009. [Google Scholar] [CrossRef] [PubMed]
- Montemayor, M.; Costan, A.; Lucena, F.; Jofre, J.; Muñoz, J.; Dalmau, E.; Mujeriego, R.; Sala, L. The combined performance of UV light and chlorine during reclaimed water disinfection. Water Sci. Technol. 2008, 57, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Mandilara, G.D.; Smeti, E.M.; Mavridou, A.T.; Lambiri, M.P.; Vatopoulos, A.C.; Rigas, F.P. Correlation between bacterial indicators and bacteriophages in sewage and sludge. FEMS Microbiol. Lett. 2006, 263, 119–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mignotte-Cadiergues, B.; Gantzer, C.; Schwartzbrod, L. Evaluation of bacteriophages during the treatment of sludge. Water Sci. Technol. 2002, 46, 189–194. [Google Scholar] [PubMed]
- Sinton, L.W.; Finlay, R.K.; Pang, L.; Scott, D.M. Transport of bacteria and bacteriophages in irrigated effluent into and through an alluvial gravel aquifer. Water. Air. Soil Pollut. 1996, 98, 17–42. [Google Scholar] [CrossRef]
- Skraber, S.; Schijven, J.; Italiaander, R.; de Roda Husman, A.M. Accumulation of enteric bacteriophage in fresh water sediments. J. Water Health 2009, 7, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Ibarluzea, J.M.; Santa Marina, L.; Moreno, B.; Serrano, E.; Larburu, K.; Maiztegi, M.J.; Yarzabal, A. Somatic coliphages and bacterial indicators of bathing water quality in the beaches of Gipuzkoa, Spain. J. Water Health 2007, 5, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Noble, R.; Chu, W. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 2001, 67, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Jiang, S.C. Real-time PCR quantification of human adenoviruses in urban rivers indicates genome prevalence but low infectivity. Appl. Environ. Microbiol. 2005, 71, 7426–7433. [Google Scholar] [CrossRef] [PubMed]
- Skraber, S.; Gantzer, C.; Maul, A.; Schwartzbrod, L. Fates of bacteriophages and bacterial indicators in the Moselle river (France). Water Res. 2002, 36, 3629–3637. [Google Scholar] [CrossRef]
- Love, D.C.; Rodriguez, R.A.; Gibbons, C.D.; Griffith, J.F.; Yu, Q.; Stewart, J.R.; Sobsey, M.D. Human viruses and viral indicators in marine water at two recreational beaches in Southern California, USA. J. Water Health 2014, 12, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Locas, A.; Barthe, C.; Barbeau, B.; Carrière, A.; Payment, P. Virus occurrence in municipal groundwater sources in Quebec, Canada. Can. J. Microbiol. 2007, 53, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Lucena, F.; Ribas, F.; Duran, A.E.; Skraber, S.; Gantzer, C.; Campos, C.; Morón, A.; Calderón, E.; Jofre, J. Occurrence of bacterial indicators and bacteriophages infecting enteric bacteria in groundwater in different geographical areas. J. Appl. Microbiol. 2006, 101, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Yoo, C.H.; Koo, E.S.; Kim, H.M.; Na, Y.; Jheong, W.H.; Jeong, Y.S. Occurrence of norovirus and other enteric viruses in untreated groundwaters of Korea. J. Water Health 2011, 9, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Méndez, J.; Audicana, A.; Cancer, M.; Isern, A.; Llaneza, J.; Moreno, B.; Navarro, M.; Tarancón, M.L.; Valero, F.; Ribas, F. Assessment of drinking water quality using indicator bacteria and bacteriophages. J. Water Health 2004, 2, 201–214. [Google Scholar] [PubMed]
- Abbaszadegan, M.; LeChevallier, M.; Gerba, C. Occurrence of viruses in U.S. groundwaters. J. Am. Water Work. Assoc. 2003, 95, 107–120. [Google Scholar]
- Bailey, E. Evaluation of a candidate bacteria host for simultaneous detection and quantification of somatic and male-specific/F+ coliphages in reclaimed water. In Proceedings of the Water Microbiology Conference, University of North Carolina at Chapel Hill, NC, USA, 18–21 May 2015; pp. 75–77.
- Hot, D.; Legeay, O.; Jacques, J.; Gantzer, C.; Caudrelier, Y.; Guyard, K.; Lange, M.; Andréoletti, L. Detection of somatic phages, infectious enteroviruses and enterovirus genomes as indicators of human enteric viral pollution in surface water. Water Res. 2003, 37, 4703–4710. [Google Scholar] [CrossRef]
- Westrell, T.; Teunis, P.; van den Berg, H.; Lodder, W.; Ketelaars, H.; Stenström, T.A.; de Roda Husman, A.M. Short- and long-term variations of norovirus concentrations in the Meuse river during a 2-year study period. Water Res. 2006, 40, 2613–2620. [Google Scholar] [CrossRef] [PubMed]
- Viau, E.J.; Lee, D.; Boehm, A.B. Swimmer risk of gastrointestinal illness from exposure to tropical coastal waters impacted by terrestrial dry-weather runoff. Environ. Sci. Technol. 2011, 45, 7158–7165. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Chu, W.; He, J.-W. Seasonal detection of human viruses and coliphage in Newport Bay, California. Appl. Environ. Microbiol. 2007, 73, 6468–6474. [Google Scholar] [CrossRef] [PubMed]
- Wyer, M.D.; Wyn-Jones, A.P.; Kay, D.; Au-Yeung, H.-K.C.; Gironés, R.; López-Pila, J.; de Roda Husman, A.M.; Rutjes, S.; Schneider, O. Relationships between human adenoviruses and faecal indicator organisms in European recreational waters. Water Res. 2012, 46, 4130–4141. [Google Scholar] [CrossRef] [PubMed]
- Boehm, A.B.; Yamahara, K.M.; Love, D.C.; Peterson, B.M.; McNeill, K.; Nelson, K.L. Covariation and photoinactivation of traditional and novel indicator organisms and human viruses at a sewage-impacted marine beach. Environ. Sci. Technol. 2009, 43, 8046–8052. [Google Scholar] [CrossRef] [PubMed]
- Borchardt, M.A.; Haas, N.L.; Hunt, R.J. Vulnerability of drinking-water wells in La Crosse, Wisconsin, to enteric-virus contamination from surface water contributions. Appl. Environ. Microbiol. 2004, 70, 5937–5946. [Google Scholar] [CrossRef] [PubMed]
- Ballester, N.A.; Fontaine, J.H.; Margolin, A.B. Occurrence and correlations between coliphages and anthropogenic viruses in the Massachusetts Bay using enrichment and ICC-nPCR. J. Water Health 2005, 3, 59–68. [Google Scholar] [PubMed]
- Haramoto, E.; Katayama, H.; Oguma, K.; Ohgaki, S. Application of cation-coated filter method to detection of noroviruses, enteroviruses, adenoviruses, and torque teno viruses in the Tamagawa River in Japan. Appl. Environ. Microbiol. 2005, 71, 2403–2411. [Google Scholar] [CrossRef] [PubMed]
- Skraber, S.; Gassilloud, B.; Gantzer, C. Comparison of coliforms and coliphages as tools for assessment of viral contamination in river water. Appl. Environ. Microbiol. 2004, 70, 3644–3649. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Jeong, Y.S. Comparison of total culturable virus assay and multiplex integrated cell culture-PCR for reliability of waterborne virus detection. Appl. Environ. Microbiol. 2004, 70, 3632–3636. [Google Scholar] [CrossRef] [PubMed]
- Wyn-Jones, A.P.; Carducci, A.; Cook, N.; D’Agostino, M.; Divizia, M.; Fleischer, J.; Gantzer, C.; Gawler, A.; Girones, R.; Höller, C.; et al. Surveillance of adenoviruses and noroviruses in European recreational waters. Water Res. 2011, 45, 1025–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelzaher, A.M.; Wright, M.E.; Ortega, C.; Hasan, A.R.; Shibata, T.; Solo-Gabriele, H.M.; Kish, J.; Withum, K.; He, G.; Elmir, S.M.; et al. Daily measures of microbes and human health at a non-point source marine beach. J. Water Health 2011, 9, 443–457. [Google Scholar] [CrossRef] [PubMed]
- Jofre, J.; Stewart, J.R.; Grabow, W. Phage Methods. In Microbial Source Tracking: Methods, Applications, and Case Studies; Hagedorn, C., Blanch, A.R., Harwood, V.J., Eds.; Springer New York: New York, NY, USA, 2011; pp. 137–156. [Google Scholar]
- Wade, T.J.; Sams, E.; Brenner, K.P.; Haugland, R.; Chern, E.; Beach, M.; Wymer, L.; Rankin, C.C.; Love, D.; Li, Q.; et al. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: A prospective cohort study. Environ. Health 2010, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.V.; Dawson, S.R.; Ward, S.; Surman, S.B.; Neal, K. Bacteriophages are a better indicator of illness rates than bacteria amongst users of a white water course fed by a lowland river. Water Sci. Technol. 1997, 35, 165–170. [Google Scholar] [CrossRef]
- van Asperen, I. Risk of gastroenteritis among triathletes in relation to faecal pollution of fresh waters. Int. J. Epidemiol. 1998, 27, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Wiedenmann, A.; Krüger, P.; Dietz, K.; López-Pila, J.M.; Szewzyk, R.; Botzenhart, K. A randomized controlled trial assessing infectious disease risks from bathing in fresh recreational waters in relation to the concentration of Escherichia coli, intestinal enterococci, Clostridium perfringens, and somatic coliphages. Environ. Health Perspect. 2006, 114, 228–236. [Google Scholar] [CrossRef] [PubMed]
- von Schirnding, Y.E.; Kfir, R.; Cabelli, V.; Franklin, L.; Joubert, G. Morbidity among bathers exposed to polluted seawater. A prospective epidemiological study. S. Afr. Med. J. 1992, 81, 543–546. [Google Scholar] [PubMed]
- Colford, J.M.; Wade, T.J.; Schiff, K.C.; Wright, C.C.; Griffith, J.F.; Sandhu, S.K.; Burns, S.; Sobsey, M.; Lovelace, G.; Weisberg, S.B. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology 2007, 18, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Mookerjee, S.; Batabyal, P.; Halder, M.; Palit, A. Specificity of coliphages in evaluating marker efficacy: A new insight for water quality indicators. J. Virol. Methods 2014, 208, 115–118. [Google Scholar] [CrossRef] [PubMed]
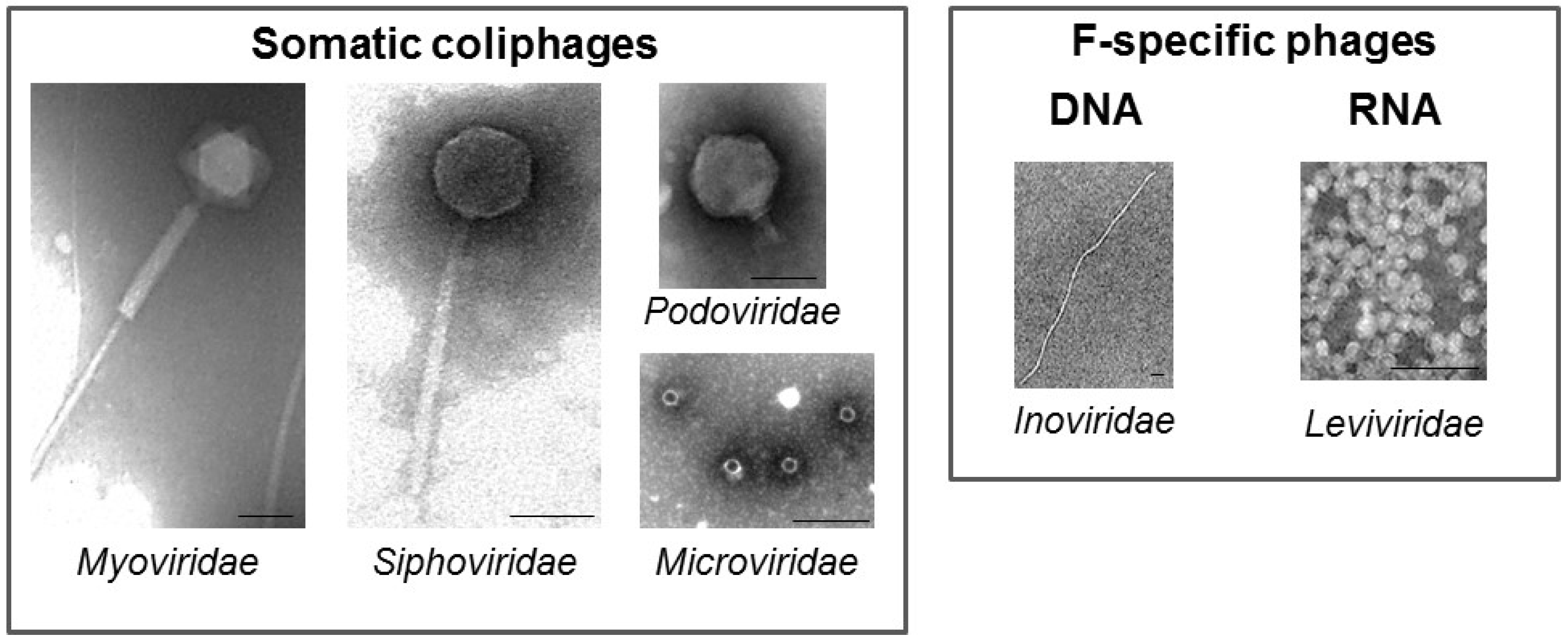
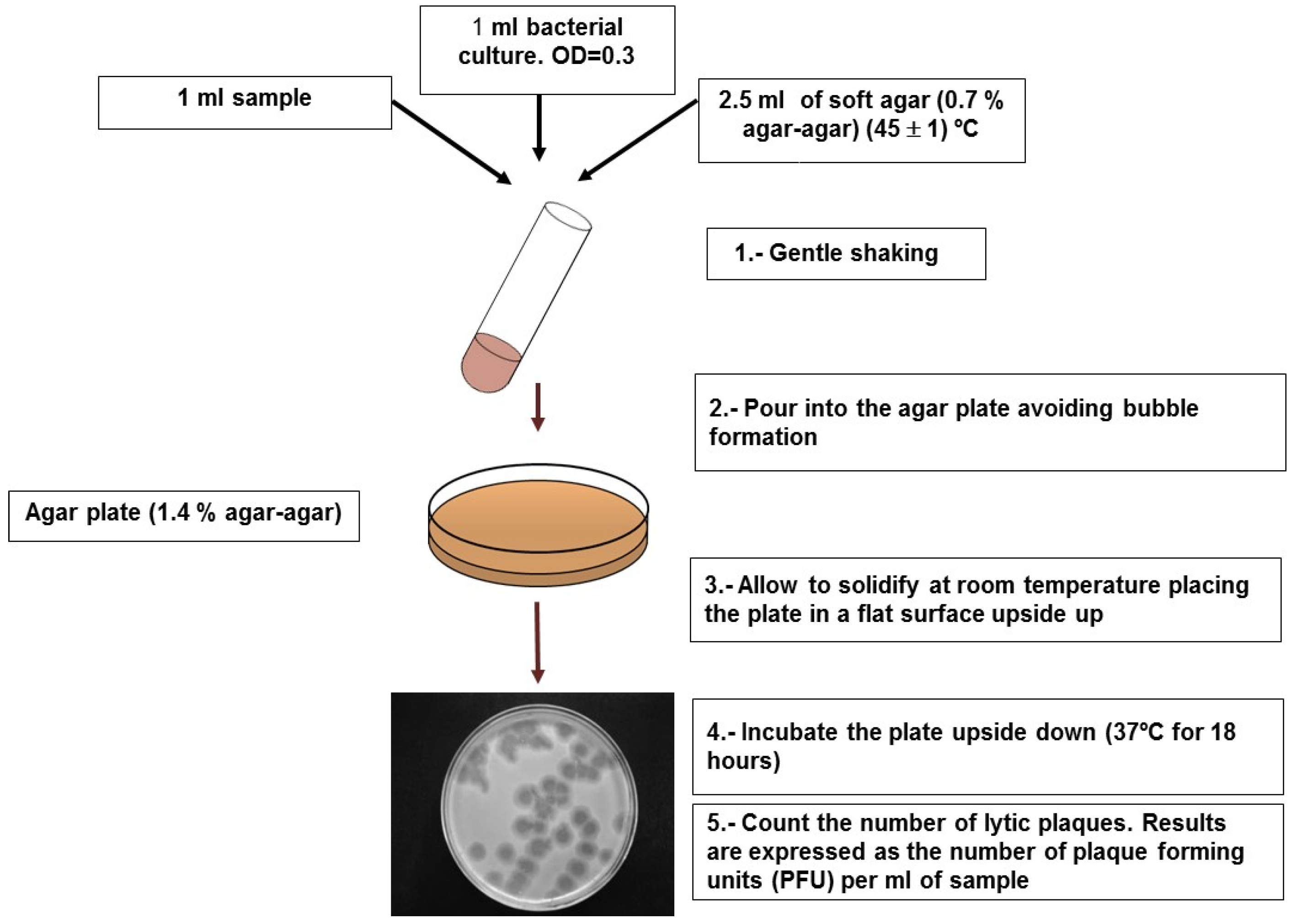
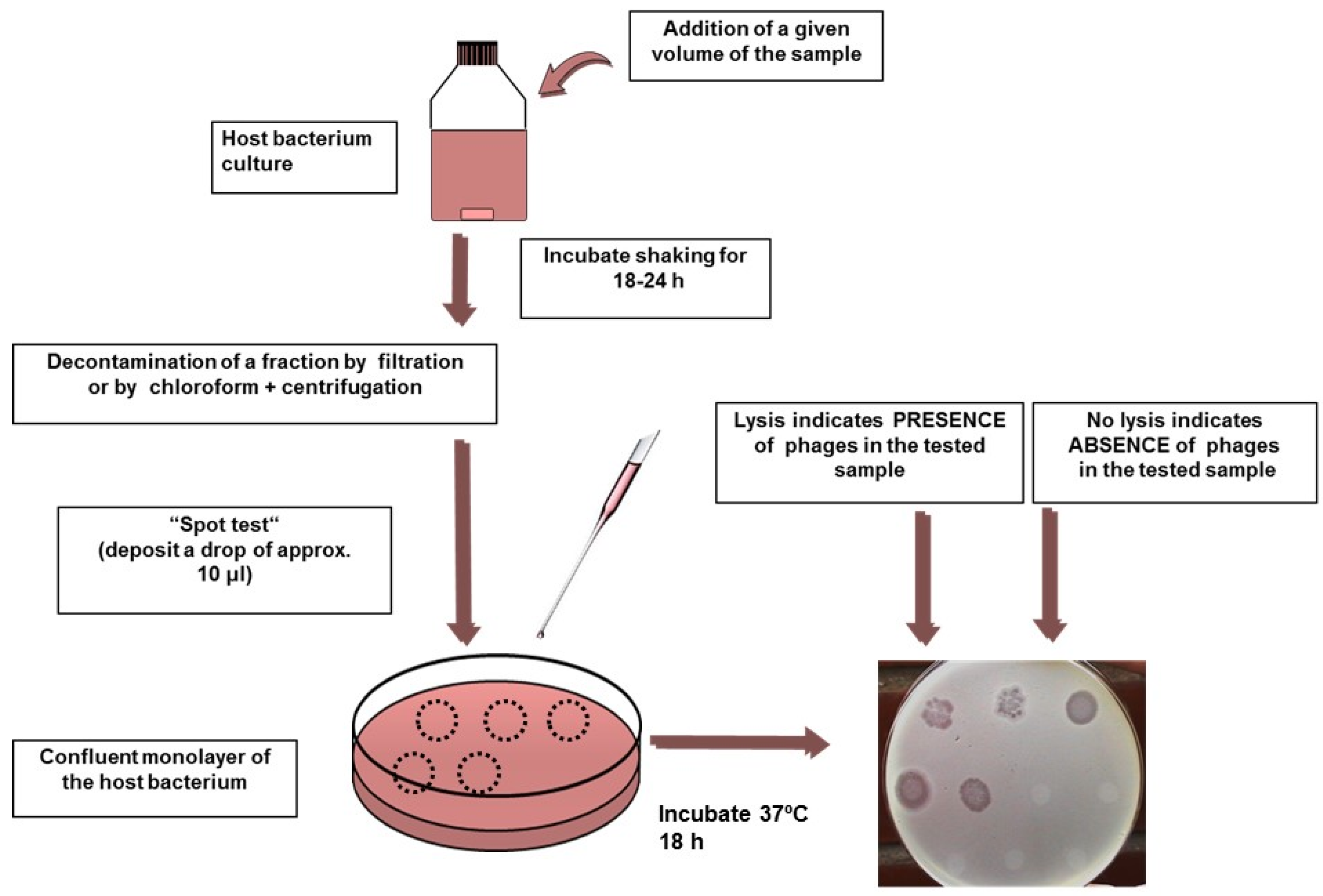
| Samples | Methods | Number of Samples | Geographical Location | Somatic Coliphages(% +) | F-Specific(% +) | RNA F-Specific(% +) | Reference |
|---|---|---|---|---|---|---|---|
| River water | ISO | 392 | Spain, France, Colombia, Argentina | 6.2 × 103 (?) | 5 × 102 (?) | [94] | |
| Freshwater reservoir | USEPA | 65 | Singapore | 2.2 × 102 (98) | 1.1 × 102 (98) | [23] | |
| Sea water | ISO | 806 for somatic and 427 for RNA F-specific phages | Spain | 32.8 (72.6) | 8 (25.5) | [122] | |
| Fresh and sea water | ISO | 139 | 9 European countries, 13 sampling sites | 1.7 × 102 (92) | 12 (50) | [71] | |
| Fresh and sea water | USEPA | 12 | California | 2.0–3.3 × 102 (100) | <0.02–30 (25) | [123] | |
| River water | USEPA | 120 | California | 6.0–103 (?) | 5.0–1.1 × 102 (?) | [124] | |
| River water | ISO | 96 | France | 1.7 × 103 (100) | 2.0 × 102 (92) | [125] | |
| Sea water | ISO | 20 | Spain | <10–1.2 × 104 (95) | 0–84 (15) | [104] | |
| River water | ISO | 75 (10 sites) | The Netherlands | *8.8–4.3 × 102 (100) | 0.04–93.6 (?) | [78] | |
| Sea water | USEPA | 436 | California | 3.1–4.9 (median) | 0.3 (median) | [126] |
| Sampling site | Method | Number of Samples | Geographical Location | Somatic Coliphages | F-Specific Phages | RNA F-Specific Phages | Reference |
|---|---|---|---|---|---|---|---|
| Wells of varied characteristics | USEPA | 160 | Canada | 8.7 | 1.8 | [127] | |
| Variety of wells and springs | ISO | 197 | Argentina, Colombia, France, Spain | 41.7 | 28.8 | [128] | |
| Variety of wells | USEPA | 39 | Korea | 12.5 | 7.5 | [129] | |
| Wells and springs | ISO | 125 | Spain | 53.6 | 36.0 | [130] | |
| Variety of wells | F+ ISO, somatic coliphages C3000 | 444 | USA | 10.8 | 9.5 | [131] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jofre, J.; Lucena, F.; Blanch, A.R.; Muniesa, M. Coliphages as Model Organisms in the Characterization and Management of Water Resources. Water 2016, 8, 199. https://doi.org/10.3390/w8050199
Jofre J, Lucena F, Blanch AR, Muniesa M. Coliphages as Model Organisms in the Characterization and Management of Water Resources. Water. 2016; 8(5):199. https://doi.org/10.3390/w8050199
Chicago/Turabian StyleJofre, Juan, Francisco Lucena, Anicet R. Blanch, and Maite Muniesa. 2016. "Coliphages as Model Organisms in the Characterization and Management of Water Resources" Water 8, no. 5: 199. https://doi.org/10.3390/w8050199






