Genome-Wide Identification of PIFs in Grapes (Vitis vinifera L.) and Their Transcriptional Analysis under Lighting/Shading Conditions
Abstract
:1. Introduction
2. Methods
2.1. Plant Materials and Treatments
2.1.1. Grape Berries in Development
2.1.2. Grape Leaves Used for Lighting/Shading Studies
2.1.3. Grape Berry Samples Used for Lighting/Shading Studies
2.2. Genome-Wide Identification and Annotation of Grape PIFs Genes
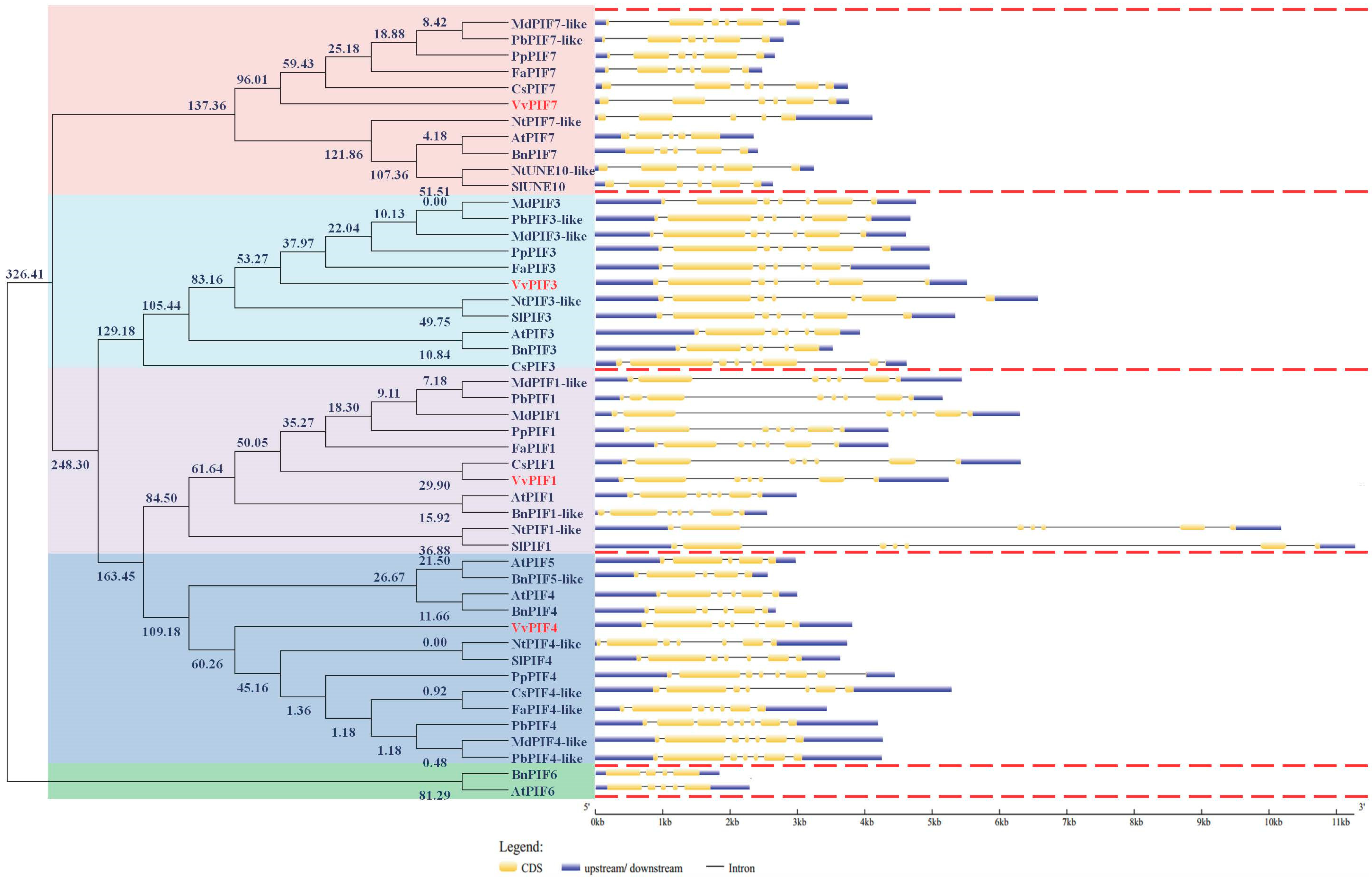
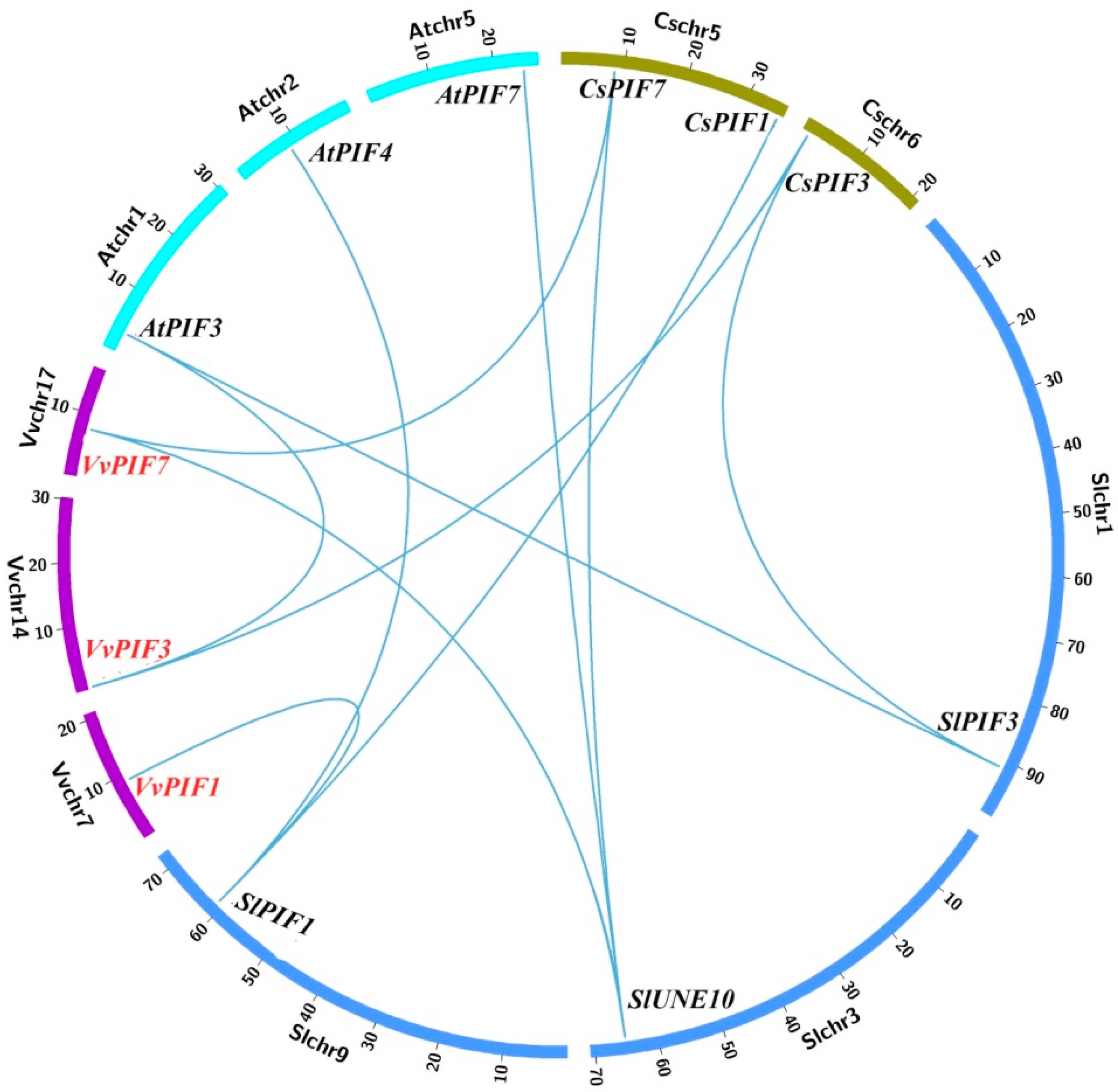
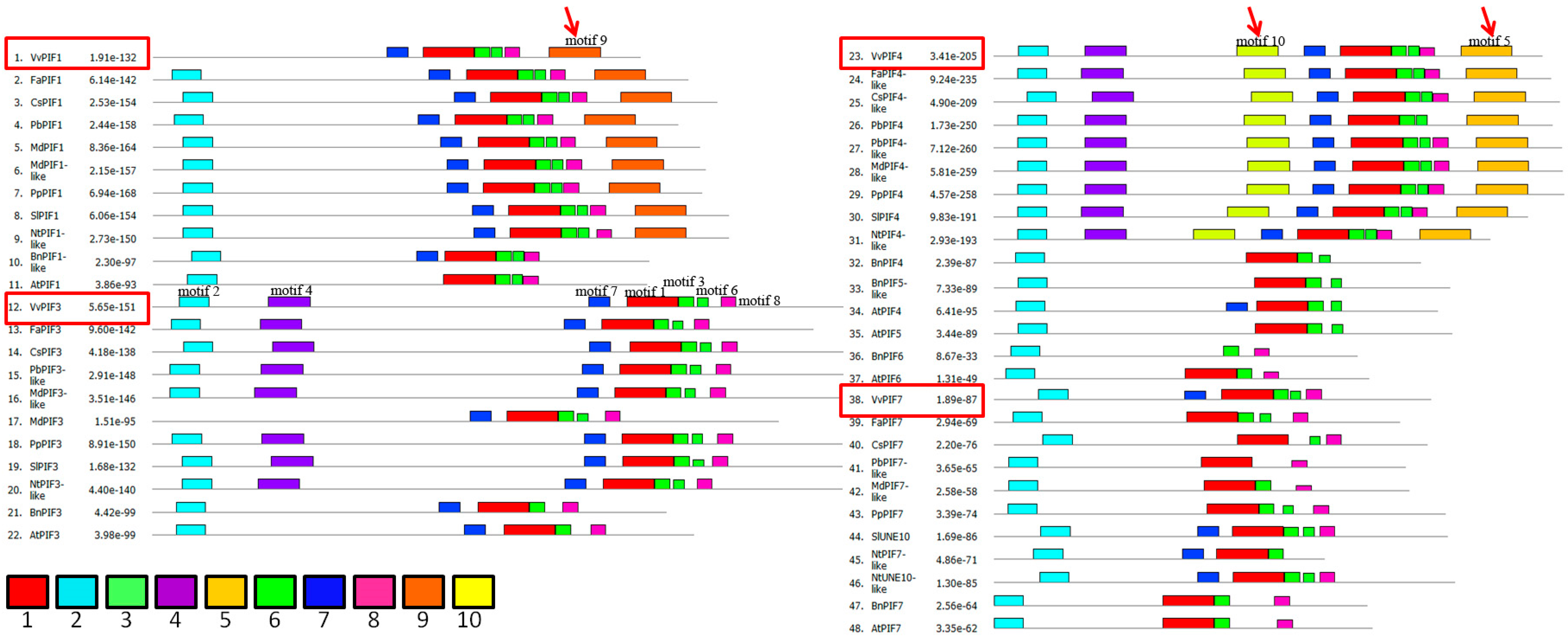
2.3. Gene Structure, Phylogenetic, Conserved Motifs and Syntenic Analysis of PIFs Family
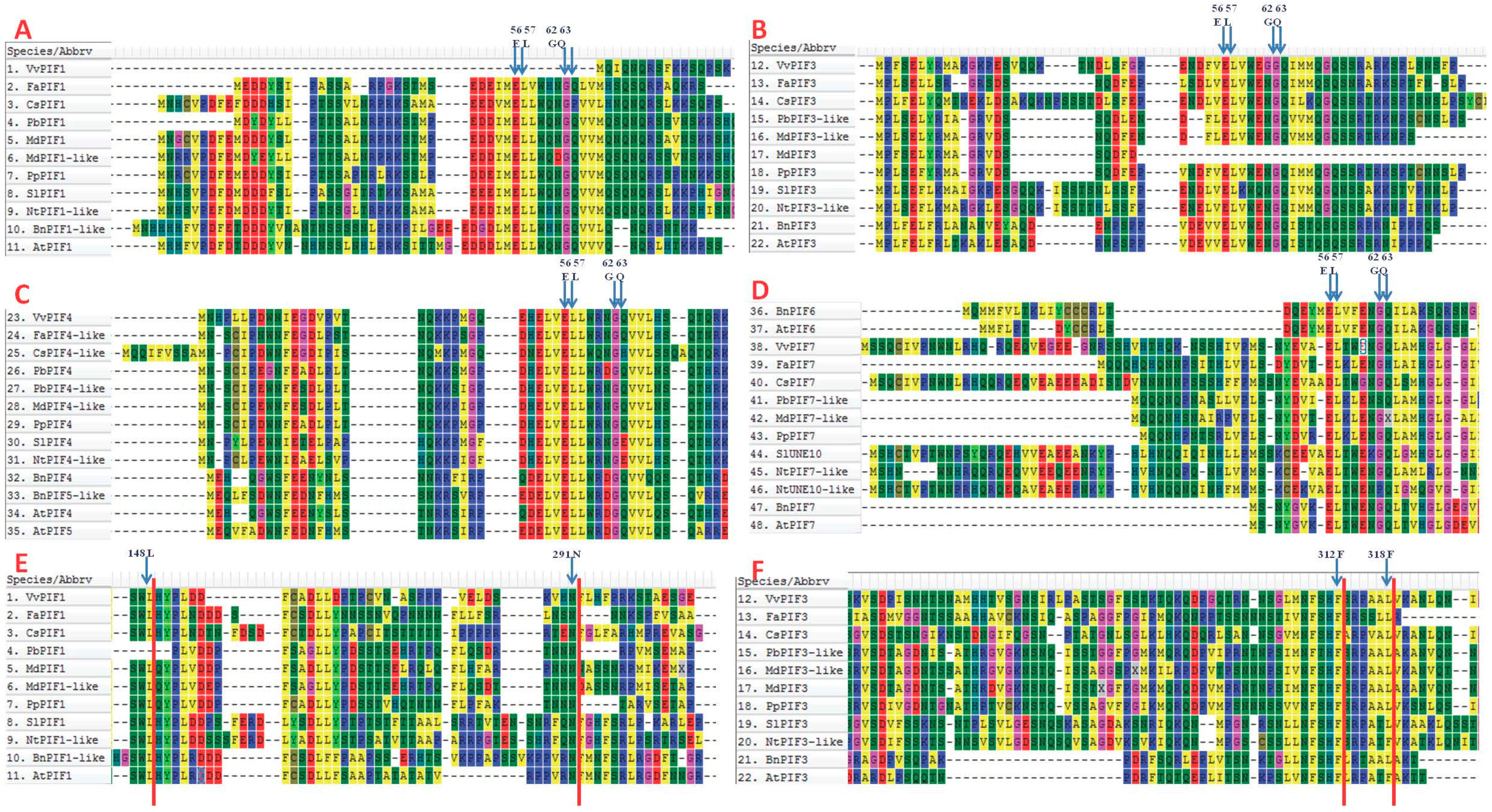
2.4. Multiple Sequence Alignments, Promoter Analysis and Interaction Protein Prediction
2.5. Transcriptome and Microarray Data Acquisition and Analysis
2.6. Total RNA Isolation, cDNA Synthesis and Gene Expression Analysis
2.7. Statistical Analysis
3. Results
3.1. Identification of Grape PIFs
3.2. Phylogenetic, Conserved Structural and Syntenic Analysis of PIFs
3.3. Comparison of Conserved Motifs
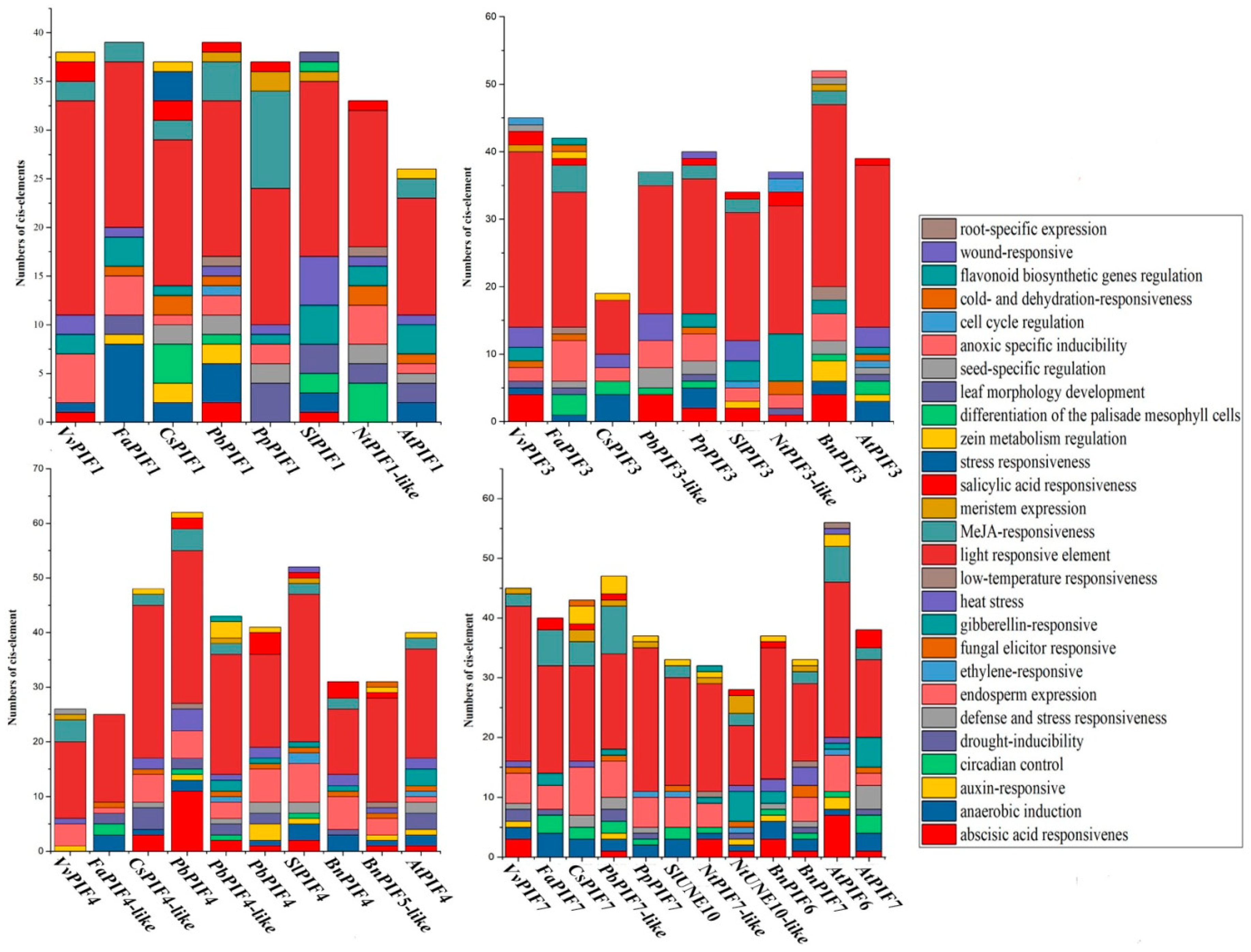
3.4. Cis-Element Analysis in the PIF Gene Promoters and Functional Prediction of PIFs Proteins
3.5. Expression Profiles of PIFs in Different Organs
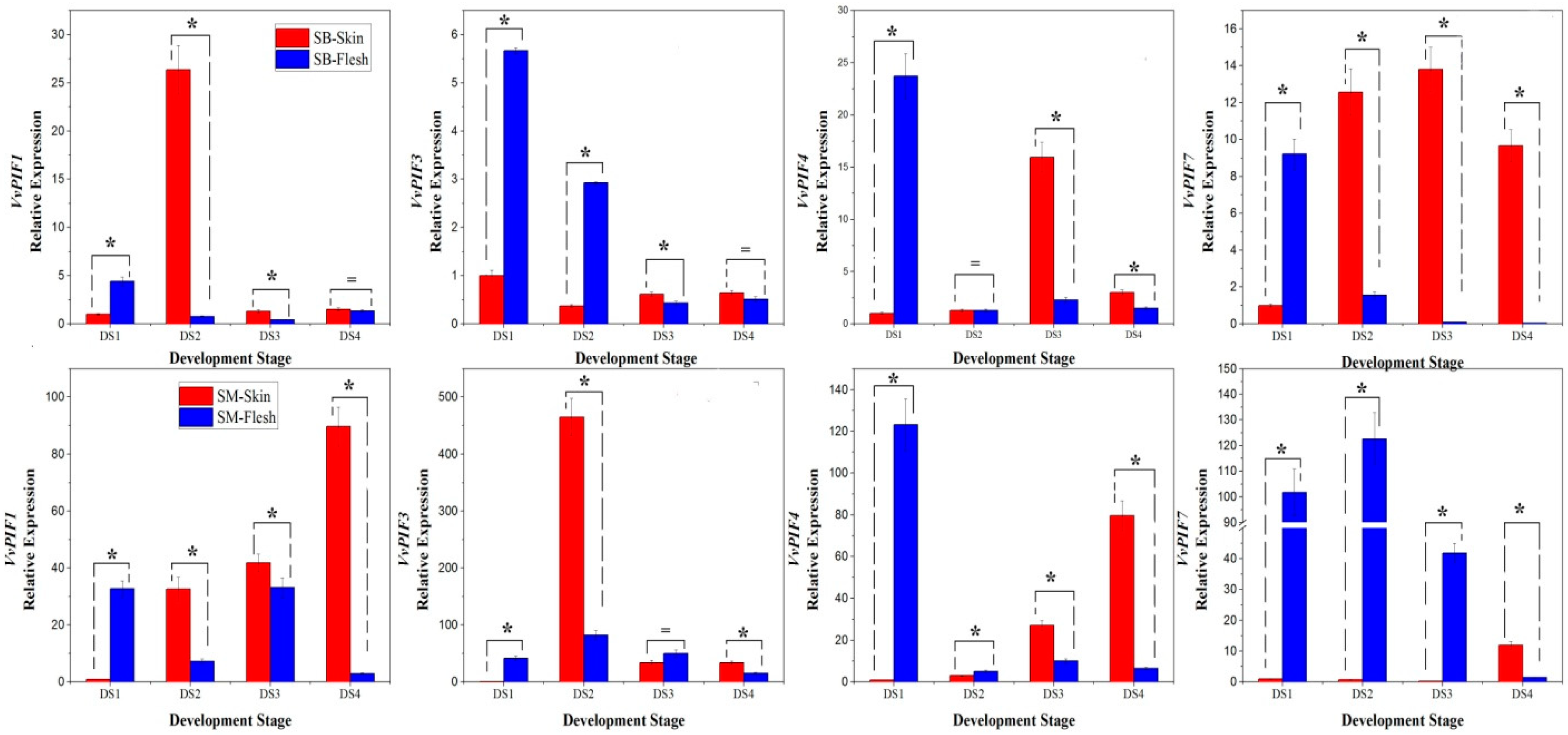
3.6. Expression Profiles of PIFs During Grape Berries Development
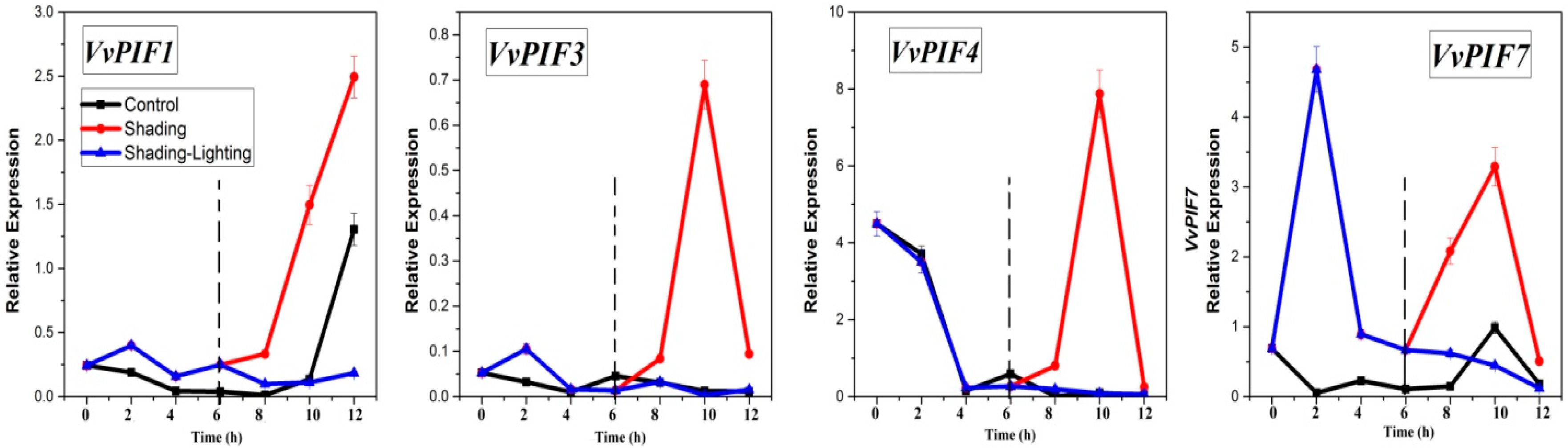
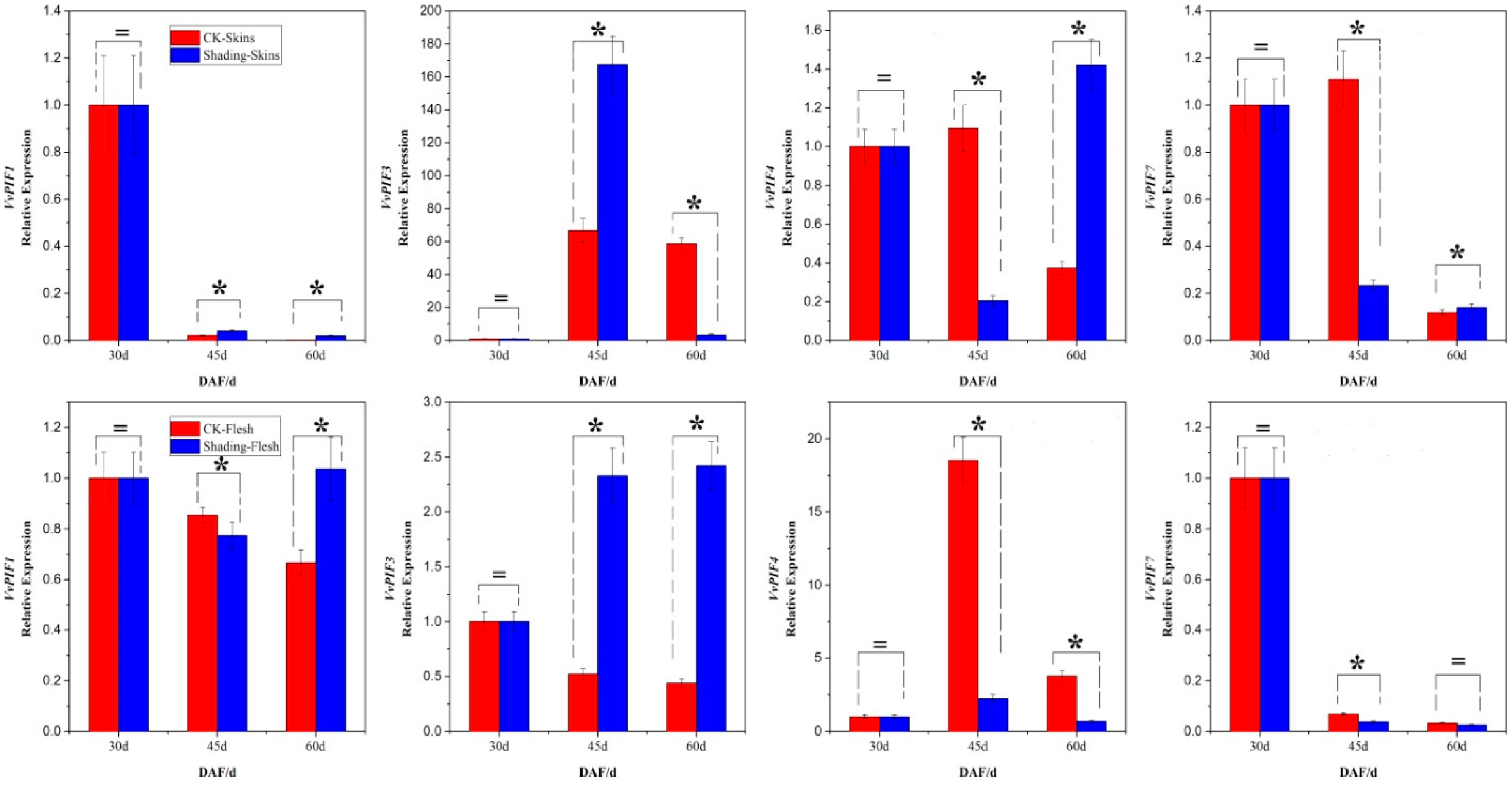
3.7. Expression Profiles of PIFs under Different Treatments
4. Discussion
4.1. PIFs Family and Their Evolutionary Analyses in Grapes
4.2. Function Analysis of the PIFs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and phytochrome interacting factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Bae, G.; Choi, G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Su, Y.S.; Lagarias, J.C. Phytochrome structure and signaling mechanisms. Annu. Rev Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Choi, G. Phytochrome-interacting factor from Arabidopsis to liverwort. Curr. Opin. Plant Biol. 2017, 5, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Possart, A.; Xu, T.; Paik, I.; Hanke, S.; Keim, S.; Hermann, H.M.; Wolf, L.; Hiss, M.; Becker, C.; Huq, E.; et al. Characterization of phytochrome interacting factors from the moss Physcomitrella patens illustrates conservation of phytochrome signaling modules in land plants. Plant Cell 2017, 29, 310–330. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Kim, J.; Park, E.; Kim, J.I.; Kang, C.; Choi, G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004, 16, 3045–3058. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Zhu, L.; Shen, H.; Huq, E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 9433–9438. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Shi, H.; Xue, C.; Wang, L.; Xi, Y.; Li, J.; Quail, P.H.; Deng, X.W.; Guo, H. A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 2012, 22, 1530–1535. [Google Scholar] [CrossRef] [PubMed]
- Monte, E.; Tepperman, J.M.; Al-Sady, B.; Kaczorowski, K.A.; Alonso, J.M.; Ecker, J.R.; Li, X.; Zhang, Y.; Quail, P.H. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 2004, 101, 16091–16098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.; Park, E.; Choi, G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007, 49, 981–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.M.; Thomashow, M.F. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 15054–15059. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Oh, E.; Wang, T.; Wang, Z.Y. TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 2016, 7, 13692. [Google Scholar] [CrossRef] [PubMed]
- Lorrain, S.; Allen, T.; Duek, P.D.; Whitelam, G.C.; Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008, 53, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Wang, J.; Li, P.; Zhao, C.; Chen, Y.; Bi, Y. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci. 2015, 238, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Josse, E.M.; Halliday, K.J. A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol. Biol. 2010, 73, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ljung, K.; Breton, G.; Schmitz, R.J.; Pruneda-Paz, J.; Cowing-Zitron, C.; Cole, B.J.; Ivans, L.J.; Pedmale, U.V.; Jung, H.S.; et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012, 26, 785–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leivar, P.; Monte, E.; Al-Sady, B.; Carle, C.; Storer, A.; Alonso, J.M.; Ecker, J.R.; Quail, P.H. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 2008, 20, 337–352. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Jing, Y.; Jiang, Z.; Lin, R. The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 2014, 26, 2472–2485. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Rodríguez-Concepción, M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheehan, M.J.; Farmer, P.R.; Brutnell, T.P. Structure and expression of maize phytochrome family homeologs. Genetics 2004, 167, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Li, F.W.; Melkonian, M.; Rothfels, C.J.; Villarreal, J.C.; Stevenson, D.W.; Graham, S.W.; Wong, G.K.; Pryer, K.M.; Mathews, S. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat. Commun. 2015, 6, 7852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llorente, B.; D’andrea, L.; Ruiz-Sola, M.A.; Botterweg, E.; Pulido, P.; Andilla, J.; Loza-Alvarez, P.; Rodriguez-Concepcion, M. Tomato fruit carotenoid biosynthesis is adjusted to actual ripening progression by a light-dependent mechanism. Plant J. 2016, 85, 107–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phoenix Bioinformatics Corporation. The Arabidopsis Information Resource (TAIR) Database. Available online: https://www.arabidopsis.org/portals/education/aboutarabidopsis.jsp (accessed on 6 September 2018).
- Genoscope. Grapevine Genome Browser. Available online: http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/ (accessed on 6 September 2018).
- Grape Genome Database, CRIBI, Version 2.1. Available online: http://genomes.cribi.unipd.it/grape/ (accessed on 6 September 2018).
- Zhu, X.; Wang, M.; Li, X.; Jiu, S.; Wang, C.; Fang, J. Genome-wide analysis of the sucrose synthase gene family in grape (Vitis vinifera): Structure, evolution, and expression profiles. Genes 2017, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- ELIXIR. InterProScan. Available online: http://www.ebi.ac.uk/Tools/pfa/iprscan5/ (accessed on 6 September 2018).
- NCBI. The Conserved Domain Database, CDD. Available online: http://www.ncbi.nlm.nih.gov/cdd (accessed on 6 September 2018).
- ExPasy. ProtParam Tool. Available online: http://web.expasy.org/protparam/ (accessed on 6 September 2018).
- Song, J.; Zhou, Y.; Zhang, J.; Zhang, K. Structural, expression and evolutionary analysis of the non-specific phospholipase C gene family in Gossypium hirsutum. BMC Genom. 2017, 18, 979. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, C.; Guo, Y.; Niu, W.; Wang, Y.; Xu, Y. Evolution and expression analysis reveal the potential role of the HD-Zip gene family in regulation of embryo abortion in grapes (Vitis vinifera L.). BMC Genom. 2017, 18, 744. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 2009 19, 1639–1645. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Han, J.; Shangguan, L.; Yang, G.; Kayesh, E.; Zhang, Y.; Leng, X.; Fang, J. Depiction of grapevine phenology by gene expression information and a test of its workability in guiding fertilization. Plant Mol. Biol. 2014, 32, 1070–1084. [Google Scholar] [CrossRef]
- Khanna, R.; Huq, E.; Kikis, E.A..; Al-Sady, B.; Lanzatella, C.; Quail, P.H. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 2004, 16, 3033–3044. [Google Scholar] [CrossRef] [PubMed]
- Al-Sady, B.; Ni, W.; Kircher, S.; Schäfer, E.; Quail, P.H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 2006, 23, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Frugoli, J.A.; McPeek, M.A.; Thomas, T.L.; McClung, C.R. Intron loss and gain during evolution of the catalase gene family in angiosperms. Genetics 1998, 149, 355–365. [Google Scholar] [PubMed]
- Lecharny, A.; Boudet, N.; Gy, I.; Aubourg, S.; Kreis, M. Introns in, introns out in plant gene families: A genomic approach of the dynamics of gene structure. J. Struct. Funct. Genom. 2003, 3, 111–116. [Google Scholar] [CrossRef]
- Zhou, L.J.; Mao, K.; Qiao, Y.; Jiang, H.; Li, Y.Y.; Hao, Y.J. Functional identification of MdPIF1 as a Phytochrome Interacting Factor in Apple. Plant Physiol. Biochem. 2017, 119, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Zhu, L.; Castillon, A.; Majee, M.; Downie, B.; Huq, E. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 2008, 20, 1586–1602. [Google Scholar] [CrossRef] [PubMed]
- Huq, E.; Al-Sady, B.; Hudson, M.; Kim, C.; Apel, K.; Quail, P.H. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 2004, 305, 1937–1941. [Google Scholar] [CrossRef] [PubMed]
- Kumar, I.; Swaminathan, K.; Hudson, K.; Hudson, M.E. Evolutionary divergence of phytochrome protein function in Zea mays PIF3 signaling. J. Exp. Bot. 2016, 67, 4231–4240. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463. [Google Scholar] [PubMed]
- Rosado, D.; Gramegna, G.; Cruz, A.; Lira, B.S.; Freschi, L.; de Setta, N.; Rossi, M. Phytochrome interacting factors (PIFs) in Solanum lycopersicum: Diversity, evolutionary history and expression profiling during different developmental processes. PLoS ONE 2016, 11, e0165929. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Xu, S.L.; González-Grandío, E.; Chalkley, R.J.; Huhmer, A.F.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 2017, 8, 15236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Jeong, J.; Kim, J.; Lee, N.; Kim, M.E.; Lee, S.; Kim, S.C.; Choi, G. PIF1 regulates plastid development by repressing photosynthetic genes in the endodermis. Mol. Plant 2016, 9, 1415–1427. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wu, M.; Zhang, M.; Jiang, W.; Ren, X.; Liang, E.; Zhang, D.; Zhang, C.; Xiao, N.; Li, Y.; et al. A Maize phytochrome-interacting factors Protein ZmPIF1 Enhances Drought Tolerance by Inducing Stomatal Closure and Improves Grain Yield in Oryza sativa. Plant Biotechnol. J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alós, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Johansson, H.; Lee, K.P.; Bou-Torrent, J.; Stewart, K.; Steel, G.; Rodríguez-Concepción, M.; Halliday, K.J. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 2014, 10, e1004416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S.; et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475. [Google Scholar] [CrossRef] [PubMed]
- Nozue, K.; Covington, M.F.; Duek, P.D.; Lorrain, S.; Fankhauser, C.; Harmer, S.L.; Maloof, J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007, 448, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Soy, J.; Leivar, P.; González-Schain, N.; Sentandreu, M.; Prat, S.; Quail, P.H.; Monte, E. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 2012, 71, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Soy, J.; Leivar, P.; Monte, E. PIF1 promotes phytochrome-regulated growth under photoperiodic conditions in Arabidopsis together with PIF3, PIF4, and PIF5. J. Exp. Bot. 2014, 65, 2925–2936. [Google Scholar] [CrossRef] [PubMed]

| Name | Locus Id | Genomic DNA Size (bp) | Number of Amino Acids | Predicted Mw (kDa) | Theoretical pI | Chromosome Location | Functional Domains (Start-End, bp) |
|---|---|---|---|---|---|---|---|
| PIF1 | LOC100264869 | 5238 | 516 | 56.32 | 5.32 | 7 | 308-357/bHLH |
| PIF3 | LOC100247781 | 5463 | 709 | 75.861 | 6.3 | 14 | 462-511/bHLH |
| PIF4 | LOC100262490 | 3798 | 531 | 57.765 | 7.3 | 12 | 338-387/bHLH |
| PIF7 | LOC100262138 | 3617 | 423 | 46.631 | 8.94 | 17 | 223-272/bHLH |
| VvPIF1 | VvPIF3 | VvPIF4 | VvPIF7 | |
|---|---|---|---|---|
| VvPIF1 | - | 48/29 | 55/19 | 51/24 |
| VvPIF3 | 26/34/39 | - | 50/29 | 44/41 |
| VvPIF4 | 33/42/24 | 27/38/28 | - | 49/26 |
| VvPIF7 | 28/41/20 | 22/31/41 | 25/35/34 | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Zheng, T.; Zhu, X.; Jiu, S.; Liu, Z.; Guan, L.; Jia, H.; Fang, J. Genome-Wide Identification of PIFs in Grapes (Vitis vinifera L.) and Their Transcriptional Analysis under Lighting/Shading Conditions. Genes 2018, 9, 451. https://doi.org/10.3390/genes9090451
Zhang K, Zheng T, Zhu X, Jiu S, Liu Z, Guan L, Jia H, Fang J. Genome-Wide Identification of PIFs in Grapes (Vitis vinifera L.) and Their Transcriptional Analysis under Lighting/Shading Conditions. Genes. 2018; 9(9):451. https://doi.org/10.3390/genes9090451
Chicago/Turabian StyleZhang, Kekun, Ting Zheng, Xudong Zhu, Songtao Jiu, Zhongjie Liu, Le Guan, Haifeng Jia, and Jinggui Fang. 2018. "Genome-Wide Identification of PIFs in Grapes (Vitis vinifera L.) and Their Transcriptional Analysis under Lighting/Shading Conditions" Genes 9, no. 9: 451. https://doi.org/10.3390/genes9090451





