Selecting the Best: Evolutionary Engineering of Chemical Production in Microbes
Abstract
:1. Introduction
2. Theoretical Frameworks for Coupling Target Metabolite Production to Growth
3. Creating Diversity: From Gene Level to Population Level
4. Practical Implementation of Adaptive Laboratory Evolution Experiments
5. Deciphering Genetic Basis for Evolved Phenotypes
6. Case Studies of Experimental Growth-Coupling Strategies
7. Challenges in Growth Selection-Based Evolutionary Engineering
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nielsen, J.; Keasling, J.D. Engineering cellular metabolism. Cell 2016, 164, 1185–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgard, A.; Burk, M.J.; Osterhout, R.; Van Dien, S.; Yim, H. Development of a commercial scale process for production of 1,4-butanediol from sugar. Curr. Opin. Biotechnol. 2016, 42, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Chubukov, V.; Mukhopadhyay, A.; Petzold, C.J.; Keasling, J.D.; Martín, H.G. Synthetic and systems biology for microbial production of commodity chemicals. NPJ Syst. Biol. Appl. 2016, 2, 16009. [Google Scholar] [CrossRef] [PubMed]
- Wegner, A.; Meiser, J.; Weindl, D.; Hiller, K. How metabolites modulate metabolic flux. Curr. Opin. Biotechnol. 2015, 34, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Piazza, I.; Kochanowski, K.; Cappelletti, V.; Fuhrer, T.; Noor, E.; Sauer, U.; Picotti, P. A map of protein-metabolite interactions reveals principles of chemical communication. Cell 2018, 172, 358–372.e23. [Google Scholar] [CrossRef] [PubMed]
- Conrad, T.M.; Lewis, N.E.; Palsson, B.Ø. Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol. 2011, 7, 509. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.F.; Hughes, B.S. Microbial experimental evolution. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R17–R25. [Google Scholar] [CrossRef] [PubMed]
- Dragosits, M.; Mattanovich, D. Adaptive laboratory evolution—Principles and applications for biotechnology. Microb. Cell Fact. 2013, 12, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauer, U. Evolutionary engineering of industrially important microbial phenotypes. Adv. Biochem. Eng. Biotechnol. 2001, 73, 129–169. [Google Scholar] [PubMed]
- Mans, R.; Daran, J.-M.G.; Pronk, J.T. Under pressure: Evolutionary engineering of yeast strains for improved performance in fuels and chemicals production. Curr. Opin. Biotechnol. 2017, 50, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Good, B.H.; McDonald, M.J.; Barrick, J.E.; Lenski, R.E.; Desai, M.M. The dynamics of molecular evolution over 60,000 generations. Nature 2017, 551, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Herring, C.D.; Raghunathan, A.; Honisch, C.; Patel, T.; Applebee, M.K.; Joyce, A.R.; Albert, T.J.; Blattner, F.R.; van den Boom, D.; Cantor, C.R.; et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 2006, 38, 1406–1412. [Google Scholar] [CrossRef] [PubMed]
- Wong, B.G.; Mancuso, C.P.; Kiriakov, S.; Bashor, C.J.; Khalil, A.S. A generalizable experimental framework for automated cell growth and laboratory evolution. BioRxiv 2018. [Google Scholar] [CrossRef]
- Feist, A.M.; Zielinski, D.C.; Orth, J.D.; Schellenberger, J.; Herrgard, M.J.; Palsson, B.Ø. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab. Eng. 2010, 12, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Lenski, R.E.; Mongold, J.A.; Sniegowski, P.D.; Travisano, M.; Vasi, F.; Gerrish, P.J.; Schmidt, T.M. Evolution of competitive fitness in experimental populations of E. coli: What makes one genotype a better competitor than another? Antonie Leeuwenhoek 1998, 73, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, T.E.; Lloyd, C.J.; Palsson, B.O.; Feist, A.M. Laboratory evolution to alternating substrate environments yields distinct phenotypic and genetic adaptive strategies. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Caspeta, L.; Chen, Y.; Ghiaci, P.; Feizi, A.; Buskov, S.; Hallström, B.M.; Petranovic, D.; Nielsen, J. Biofuels. Altered sterol composition renders yeast thermotolerant. Science 2014, 346, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, E.T.; Wang, S.; Lennen, R.M.; Herrgård, M.J.; Simmons, B.A.; Singer, S.W.; Feist, A.M. Generation of a platform strain for ionic liquid tolerance using adaptive laboratory evolution. Microb. Cell Fact. 2017, 16, 204. [Google Scholar] [CrossRef] [PubMed]
- Strucko, T.; Zirngibl, K.; Pereira, F.; Kafkia, E.; Mohamed, E.T.; Rettel, M.; Stein, F.; Feist, A.M.; Jouhten, P.; Patil, K.R.; et al. Laboratory evolution reveals regulatory and metabolic trade-offs of glycerol utilization in Saccharomyces cerevisiae. Metab. Eng. 2018, 47, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.; Rogers, J.K.; Taylor, N.D.; Church, G.M. Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. USA 2014, 111, 17803–17808. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.J.; Monk, J.M.; Palsson, B.O. Using genome-scale models to predict biological capabilities. Cell 2015, 161, 971–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monk, J.M.; Lloyd, C.J.; Brunk, E.; Mih, N.; Sastry, A.; King, Z.; Takeuchi, R.; Nomura, W.; Zhang, Z.; Mori, H.; et al. iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 2017, 35, 904–908. [Google Scholar] [CrossRef] [PubMed]
- Burgard, A.P.; Pharkya, P.; Maranas, C.D. Optknock: A bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 2003, 84, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Machado, D.; Herrgård, M.J. Co-evolution of strain design methods based on flux balance and elementary mode analysis. Metab. Eng. Commun. 2015, 2, 85–92. [Google Scholar] [CrossRef] [Green Version]
- Hassanpour, N.; Ullah, E.; Yousofshahi, M.; Nair, N.U.; Hassoun, S. Selection Finder (SelFi): A computational metabolic engineering tool to enable directed evolution of enzymes. Metab. Eng. Commun. 2017, 4, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Klamt, S.; Mahadevan, R. On the feasibility of growth-coupled product synthesis in microbial strains. Metab. Eng. 2015, 30, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Von Kamp, A.; Klamt, S. Growth-coupled overproduction is feasible for almost all metabolites in five major production organisms. Nat. Commun. 2017, 8, 15956. [Google Scholar] [CrossRef] [PubMed]
- Trinh, C.T.; Li, J.; Blanch, H.W.; Clark, D.S. Redesigning Escherichia coli metabolism for anaerobic production of isobutanol. Appl. Environ. Microbiol. 2011, 77, 4894–4904. [Google Scholar] [CrossRef] [PubMed]
- Trinh, C.T.; Srienc, F. Metabolic engineering of Escherichia coli for efficient conversion of glycerol to ethanol. Appl. Environ. Microbiol. 2009, 75, 6696–6705. [Google Scholar] [CrossRef] [PubMed]
- Harder, B.-J.; Bettenbrock, K.; Klamt, S. Model-based metabolic engineering enables high yield itaconic acid production by Escherichia coli. Metab. Eng. 2016, 38, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Cirino, P.C.; Mayer, K.M.; Umeno, D. Generating mutant libraries using error-prone PCR. Methods Mol. Biol. 2003, 231, 3–9. [Google Scholar] [PubMed]
- Li, A.; Acevedo-Rocha, C.G.; Sun, Z.; Cox, T.; Xu, J.L.; Reetz, M.T. Beating bias in the directed evolution of proteins: Combining high-fidelity on-chip solid-phase gene synthesis with efficient gene assembly for combinatorial library construction. Chembiochem 2018, 19, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Liu, R.; Garst, A.D.; Lee, T.; Nogué, V.S.I.; Beckham, G.T.; Gill, R.T. CRISPR EnAbled Trackable genome Engineering for isopropanol production in Escherichia coli. Metab. Eng. 2017, 41, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Garst, A.D.; Bassalo, M.C.; Pines, G.; Lynch, S.A.; Halweg-Edwards, A.L.; Liu, R.; Liang, L.; Wang, Z.; Zeitoun, R.; Alexander, W.G.; et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 2017, 35, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Crook, N.; Abatemarco, J.; Sun, J.; Wagner, J.M.; Schmitz, A.; Alper, H.S. In vivo continuous evolution of genes and pathways in yeast. Nat. Commun. 2016, 7, 13051. [Google Scholar] [CrossRef] [PubMed]
- Esvelt, K.M.; Carlson, J.C.; Liu, D.R. A system for the continuous directed evolution of biomolecules. Nature 2011, 472, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Pu, J.; Zinkus-Boltz, J.; Dickinson, B.C. Evolution of a split RNA polymerase as a versatile biosensor platform. Nat. Chem. Biol. 2017, 13, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Bryson, D.I.; Fan, C.; Guo, L.-T.; Miller, C.; Söll, D.; Liu, D.R. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 2017, 13, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Badran, A.H.; Guzov, V.M.; Huai, Q.; Kemp, M.M.; Vishwanath, P.; Kain, W.; Nance, A.M.; Evdokimov, A.; Moshiri, F.; Turner, K.H.; et al. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature 2016, 533, 58–63. [Google Scholar] [CrossRef] [PubMed]
- D’Oelsnitz, S.; Ellington, A. Continuous directed evolution for strain and protein engineering. Curr. Opin. Biotechnol. 2018, 53, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Bratulic, S.; Badran, A.H. Modern methods for laboratory diversification of biomolecules. Curr. Opin. Chem. Biol. 2017, 41, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Gresham, D.; Hong, J. The functional basis of adaptive evolution in chemostats. FEMS Microbiol. Rev. 2015, 39, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, C.N.; Miller, A.W.; Ekness, F.; Dunham, M.J.; Klavins, E. A low cost, customizable turbidostat for use in synthetic circuit characterization. ACS Synth. Biol. 2015, 4, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Gresham, D.; Dunham, M.J. The enduring utility of continuous culturing in experimental evolution. Genomics 2014, 104, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-H.; Feist, A.M.; Barrett, C.L.; Palsson, B.Ø. Cumulative number of cell divisions as a meaningful timescale for adaptive laboratory evolution of Escherichia coli. PLoS One 2011, 6, e26172. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Balasubramanian, S.; Church, G.M.; Gilbert, W.; Rogers, J.; Schloss, J.A.; Waterston, R.H. DNA sequencing at 40: Past, present and future. Nature 2017, 550, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Zhou, H.; Claesen, J.; Shen, B.; Fischbach, M.A.; Voigt, C.A. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2016, 14, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Long, A.; Liti, G.; Luptak, A.; Tenaillon, O. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat. Rev. Genet. 2015, 16, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.S.L.; Lennen, R.M.; Sonnenschein, N.; Herrgård, M.J. Systems biology solutions for biochemical production challenges. Curr. Opin. Biotechnol. 2017, 45, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Jakočiūnas, T.; Jensen, M.K.; Keasling, J.D. System-level perturbations of cell metabolism using CRISPR/Cas9. Curr. Opin. Biotechnol. 2017, 46, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Mougiakos, I.; Bosma, E.F.; Ganguly, J.; van der Oost, J.; van Kranenburg, R. Hijacking CRISPR-Cas for high-throughput bacterial metabolic engineering: Advances and prospects. Curr. Opin. Biotechnol. 2018, 50, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Shanmugam, K.T.; Ingram, L.O. Functional replacement of the Escherichia coli d-(−)-lactate dehydrogenase gene (ldhA) with the l-(+)-lactate dehydrogenase gene (ldhL) from Pediococcus acidilactici. Appl. Environ. Microbiol. 2003, 69, 2237–2244. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Yomano, L.P.; Shanmugam, K.T.; Ingram, L.O. Fermentation of 10% (w/v) sugar to d(−)-lactate by engineered Escherichia coli B. Biotechnol. Lett. 2005, 27, 1891–1896. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tian, T.; Zhao, J.; Wang, J.; Yan, T.; Xu, L.; Liu, Z.; Garza, E.; Iverson, A.; Manow, R.; et al. Homofermentative production of d-lactic acid from sucrose by a metabolically engineered Escherichia coli. Biotechnol. Lett. 2012, 34, 2069–2075. [Google Scholar] [CrossRef] [PubMed]
- Grabar, T.B.; Zhou, S.; Shanmugam, K.T.; Yomano, L.P.; Ingram, L.O. Methylglyoxal bypass identified as source of chiral contamination in l(+) and d(−)-lactate fermentations by recombinant Escherichia coli. Biotechnol. Lett. 2006, 28, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Fong, S.S.; Burgard, A.P.; Herring, C.D.; Knight, E.M.; Blattner, F.R.; Maranas, C.D.; Palsson, B.O. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol. Bioeng. 2005, 91, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jantama, K.; Moore, J.C.; Shanmugam, K.T.; Ingram, L.O. Production of l-alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2007, 77, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.R.; Lan, E.I.; Dekishima, Y.; Baez, A.; Cho, K.M.; Liao, J.C. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2905–2915. [Google Scholar] [CrossRef] [PubMed]
- Machado, H.B.; Dekishima, Y.; Luo, H.; Lan, E.I.; Liao, J.C. A selection platform for carbon chain elongation using the CoA-dependent pathway to produce linear higher alcohols. Metab. Eng. 2012, 14, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Jantama, K.; Haupt, M.J.; Svoronos, S.A.; Zhang, X.; Moore, J.C.; Shanmugam, K.T.; Ingram, L.O. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 2008, 99, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Tokuyama, K.; Toya, Y.; Horinouchi, T.; Furusawa, C.; Matsuda, F.; Shimizu, H. Application of adaptive laboratory evolution to overcome a flux limitation in an Escherichia coli production strain. Biotechnol. Bioeng. 2018, 115, 1542–1551. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.-S.; Xiong, M.; Jambunathan, P.; Wang, J.; Wang, J.; Stapleton, C.; Zhang, K. Engineering nonphosphorylative metabolism to generate lignocellulose-derived products. Nat. Chem. Biol. 2016, 12, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Otero, J.M.; Cimini, D.; Patil, K.R.; Poulsen, S.G.; Olsson, L.; Nielsen, J. Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS One 2013, 8, e54144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes, L.H.; Gomez, J.M.; Kao, K.C. Improving carotenoids production in yeast via adaptive laboratory evolution. Metab. Eng. 2014, 21, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Liao, J.C. An evolutionary strategy for isobutanol production strain development in Escherichia coli. Metab. Eng. 2011, 13, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Kast, P.; Asif-Ullah, M.; Jiang, N.; Hilvert, D. Exploring the active site of chorismate mutase by combinatorial mutagenesis and selection: The importance of electrostatic catalysis. Proc. Natl. Acad. Sci. USA 1996, 93, 5043–5048. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.M.; Kries, H.; Csuhai, E.; Kast, P.; Hilvert, D. Design, selection, and characterization of a split chorismate mutase. Protein Sci. 2010, 19, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, S.V.; Samsonova, N.N.; Kotliarova, V.A.; Rushkevich, N.Y.; Beznoschenko, O.S.; Bachina, T.A.; Imabayashi, Y.; Sugiyama, M.; Suzuki, S. Bacterium Producing a Product of a Reaction Catalyzed by a Protein Having 2-Oxoglutarate-Dependent Enzyme Activity and a Method for Manufacturing the Product. U.S. Patent 8,524,476 B2, 3 September 2013. [Google Scholar]
- Loenarz, C.; Schofield, C.J. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 2008, 4, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Hansen, A.S.L. Method of Improving Methyltransferase Activity. Patent No. WO2018037098, 1 March 2018. [Google Scholar]
- Notebaart, R.A.; Kintses, B.; Feist, A.M.; Papp, B. Underground metabolism: Network-level perspective and biotechnological potential. Curr. Opin. Biotechnol. 2018, 49, 108–114. [Google Scholar] [CrossRef] [PubMed]
- LaCroix, R.A.; Palsson, B.O.; Feist, A.M. A model for designing adaptive laboratory evolution experiments. Appl. Environ. Microbiol. 2017, 83, e03115-16. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.C.; Pretorius, I.S.; Paulsen, I.T. Synthetic evolution of metabolic productivity using biosensors. Trends Biotechnol. 2016, 34, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.K.; Taylor, N.D.; Church, G.M. Biosensor-based engineering of biosynthetic pathways. Curr. Opin. Biotechnol. 2016, 42, 84–91. [Google Scholar] [CrossRef] [PubMed]




| Product | Mechanism of Coupling | Outcome | Citation | Genetic Design | Experimental setup |
| Prephenate | Restoring of amino acids production via production of prephenate | N/A; Evolved strain | (Kast et al., 1996) [53] | Deletions: pheA, tyrA, aroF | M9 medium |
| Insertions: aroH from Bacillus subtilis, tyrA from Erwinia herbicola, pheC from Pseudomonas aeruginosa | |||||
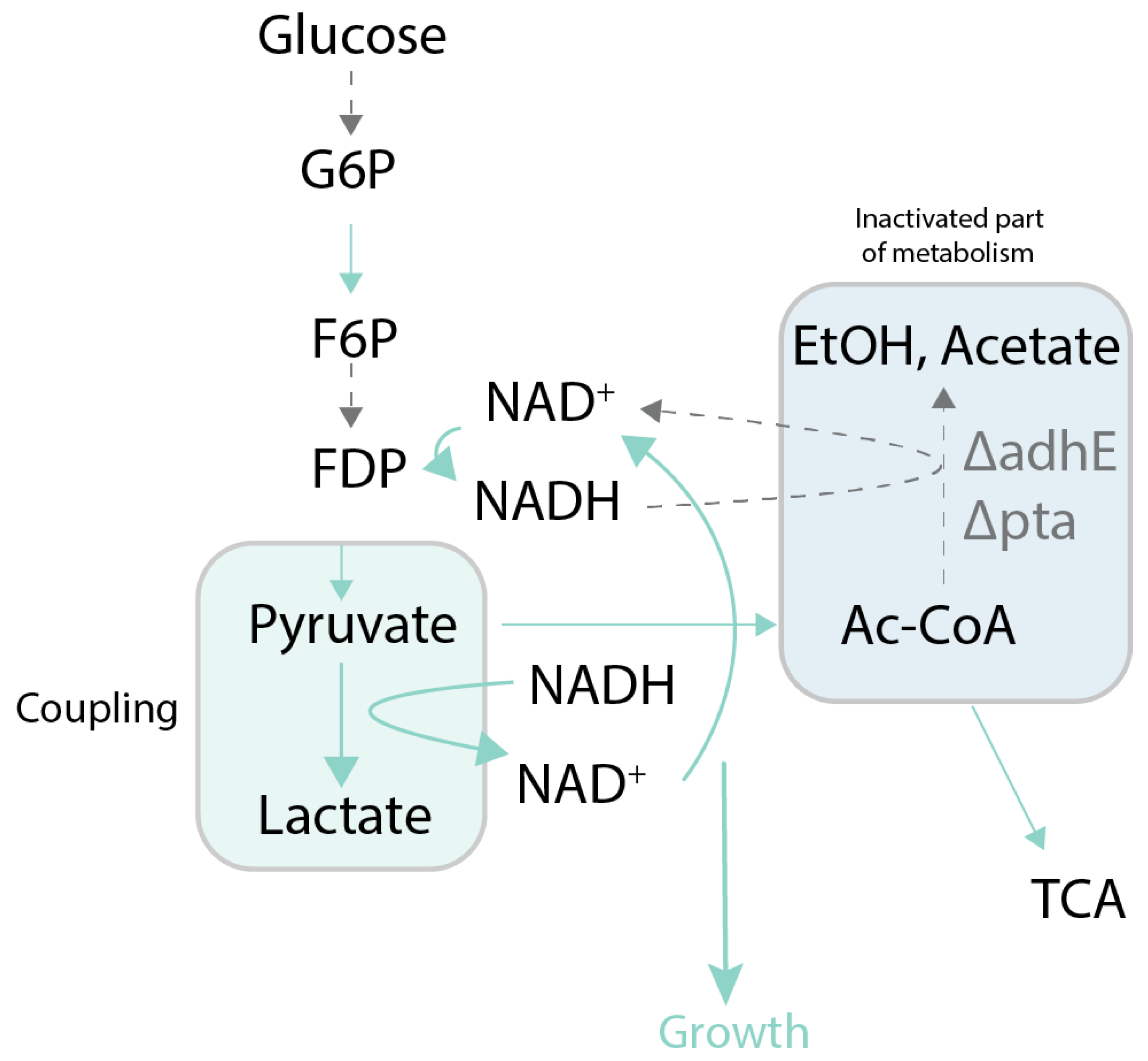
| l-Lactic acid | Redox balance | Yield 93%–95% on glucose and xylose; Evolved strain | (Zhou et al., 2003) [54] | Deletion: focA, pflB, frdB, frdC, adhE, ackA | M9 medium |
| Insertion: ldhA | |||||
| d-Lactic acid | Redox balance | Yield 88%–95% from sugar substrates; Evolved strain | (Zhou et al., 2005) [55] | Deletions: frdA, pflB, adhE, ackA | LB medium |
| d-Lactic acid | Redox balance | 0.87 g/g glucose; Evolved strain | (Fong et al., 2005) [56] | Deletions: adhE, pta, pfk, glk | M9 medium |
| l-Lactic acid | Redox balance | >95% theoretical mass yield from glucose; Evolved strain | (Grabar et al., 2006) [57] | Deletions: frdA, pflB, adhE, ackA, ldhA, mgsA | NBS medium |
| l-Alanine | Redox balance | 95% mass yield from glucose; Evolved strain | (Zhang et al., 2007) [58] | Deletions: frdA, pflB, adhE, ackA, ldhA, mgsA | NBS medium |
| Succinate and malate | Redox balance | Succinate 0.78 g/g glucose yield Malate 1.0 g/g glucose yield; Evolved strains | (Jantama et al., 2008) [59] | Deletions (succinate): ldhA, adhE, ackA, focA, pflB, mgsA, poxB | NBS medium |
| Deletions (malate): ldhA, adhE, ackA, focA, pflB, mgsA | |||||
| 1-butanol | Redox balance | 70%–88% of maximum theoretical yield; Identification of mutation in Ter protein | (Shen et al., 2011) [60] | Deletions: adhE, ldhA, frd, pta | LB medium |
| Insertions: ter from Treponema denticola, atoB from E. coli | |||||
| Isobutanol | Feeding with norvaline thus increasing production of valine | 0.3 g/g glucose (76% of maximum theoretical yield); Set of mutations | (Smith and Liao 2011) [61] | Random mutagenesis + selection | M9 medium with norvaline |
| Higher-chain alcohols | Redox balance | Yield - N/A; Set of mutations | (Machado et al., 2012) [62] | Deletions: adhE, ldhA, frd | LB medium |
| Insertions: atoB from E. coli, adhE2, crt from C. acetobutylicum, hbd from Ralstonia eutropha | |||||
| d-Lactic acid | Redox balance | Product yield of 85%; Evolved strain | (Wang et al., 2012) [63] | Deletions: adhE, frdA, frdB, frdC, frdD, pta, pflB, aldA, cscR | NBS medium |
| Succinate | Production of glycine and serine for biomass coupled to succinate production | 0.02 g succinate/g glucose; Evolved strain | (Otero et al., 2013) [64] | Deletions: sdh, ser3, ser33 | Minimal chemically defined medium |
| 4-hydroxy-l-isoleucine (4-HIL) | Production of 4-HIL due to “shunting”of the citric acid cycle by simultaneously oxidizing isoleucine and α-ketoglutarate | N/A | (Smirnov et al., 2013) [65] | Deletions: sucAB, aceAK | N/A |
| Insertion: ido from Bacillus thuringiensis | |||||
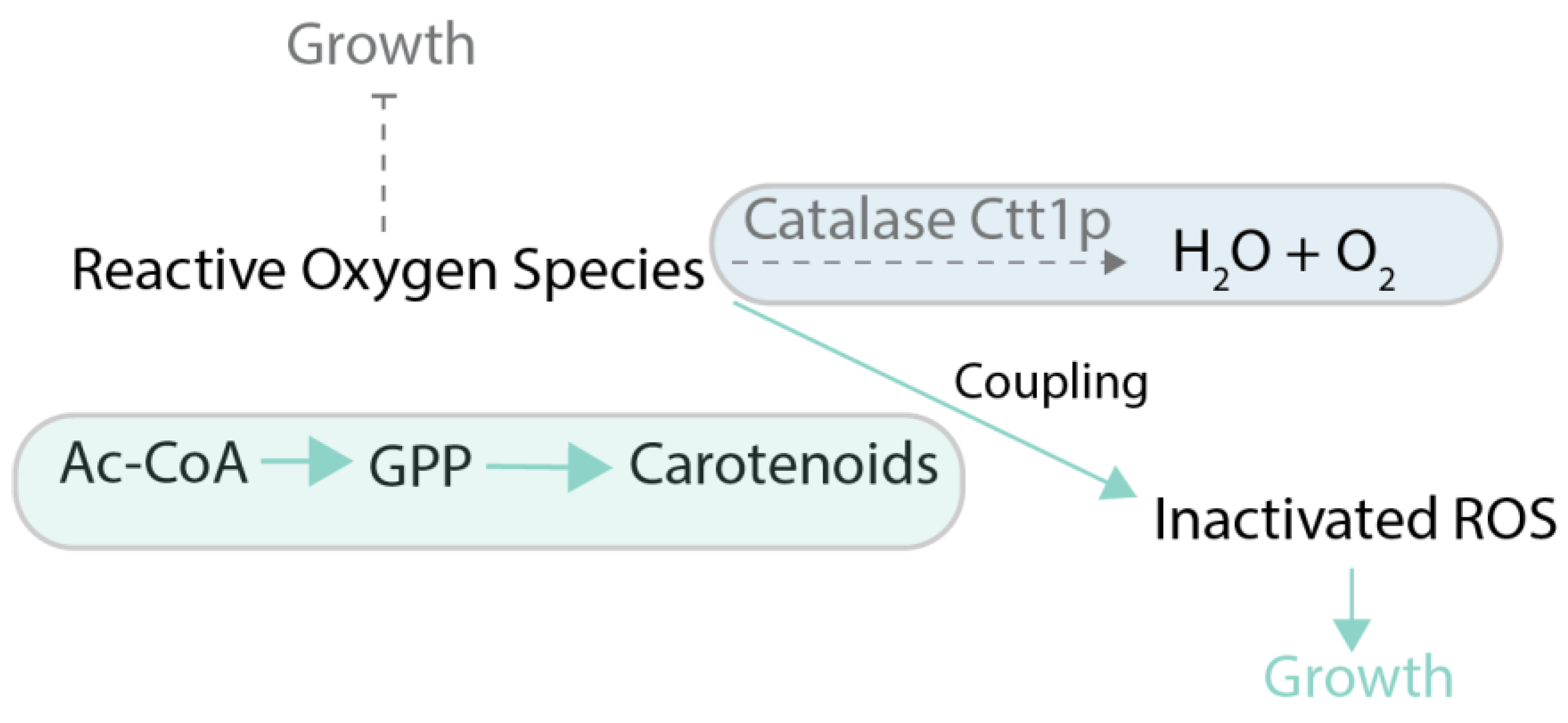
| Carotenoids | Carotenoids production as protection against oxidative stress | 18 mg/g dry cell weight; Evolved strain | (Reyes et al., 2014) [66] | Deletions: CTT1 | Yeast extract Peptone Dextrose medium |
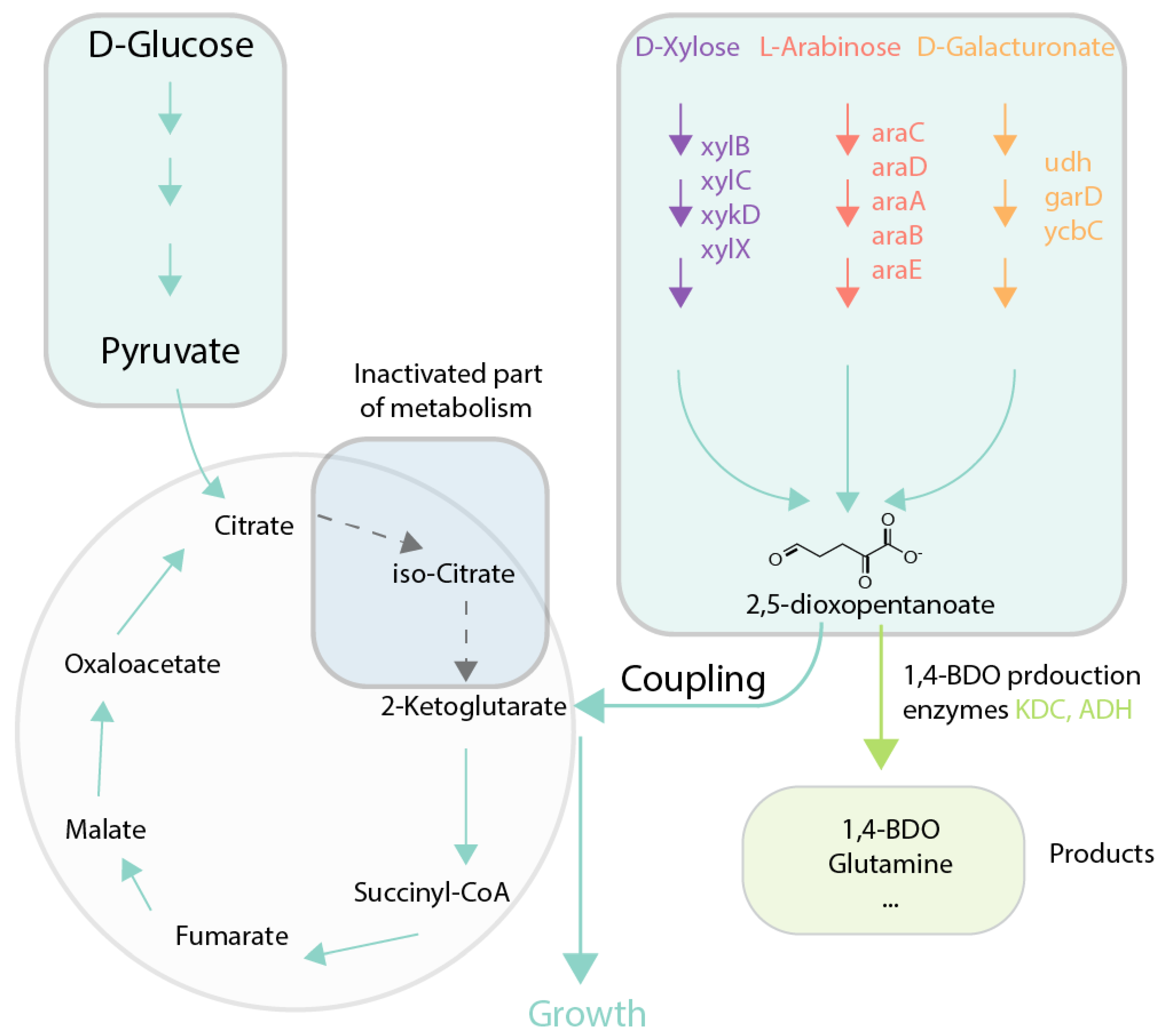
| 1,4-butanediol (1,4-BDO) | Production of 2-ketoglutarate (2-KG) via utilization of xylose and other compounds is the sole source of 2-KG | 1,4-BDO yield of 0.37 g/g d-xylose; Evolved strains and set of mutations | (Tai et al., 2016) [67] | Deletions: icd, xylA, yagE, yjhH Insertions: xylBCDX from C. crescentus | M9 medium with xylose |
| Deletions: icd, araA Insertions: araCDABE from B. multivorans | M9 medium with arabinose | ||||
| Deletions: icd, uxaC, garL Insertions: udh from P. putida, garD from E. coli, and ycbC from Bacillus subtilis | M9 medium with galactose | ||||
| Succinate | Source of succinate on glycerol medium | 0.68 g/g glucose; Set of mutations | (Tokuyama et al., 2018) [68] | Deletions: adhE, pykAF, gldA, pflB | M9 medium |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shepelin, D.; Hansen, A.S.L.; Lennen, R.; Luo, H.; Herrgård, M.J. Selecting the Best: Evolutionary Engineering of Chemical Production in Microbes. Genes 2018, 9, 249. https://doi.org/10.3390/genes9050249
Shepelin D, Hansen ASL, Lennen R, Luo H, Herrgård MJ. Selecting the Best: Evolutionary Engineering of Chemical Production in Microbes. Genes. 2018; 9(5):249. https://doi.org/10.3390/genes9050249
Chicago/Turabian StyleShepelin, Denis, Anne Sofie Lærke Hansen, Rebecca Lennen, Hao Luo, and Markus J. Herrgård. 2018. "Selecting the Best: Evolutionary Engineering of Chemical Production in Microbes" Genes 9, no. 5: 249. https://doi.org/10.3390/genes9050249





