Overexpression of a Novel Apple NAC Transcription Factor Gene, MdNAC1, Confers the Dwarf Phenotype in Transgenic Apple (Malus domestica)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth
2.2. Isolation of MdNAC1 from Apple
2.3. Phylogenetic Analysis, Multiple Alignments, and Examination of Gene Structure
2.4. Subcellular Localization and Analysis of Transcriptional Activity for MdNAC1
2.5. Generation of Transgenic Apple Plants
2.6. qRT-PCR Analysis and Promoter Element Analysis
2.7. Growth Parameters
2.8. Photosynthetic Characteristics
2.9. Anatomical Structure Analysis
2.10. Concentrations of Plant Hormones
2.11. Statistical Analysis
3. Results
3.1. Expression, Cloning, and Molecular Characterization of MdNAC1 Gene
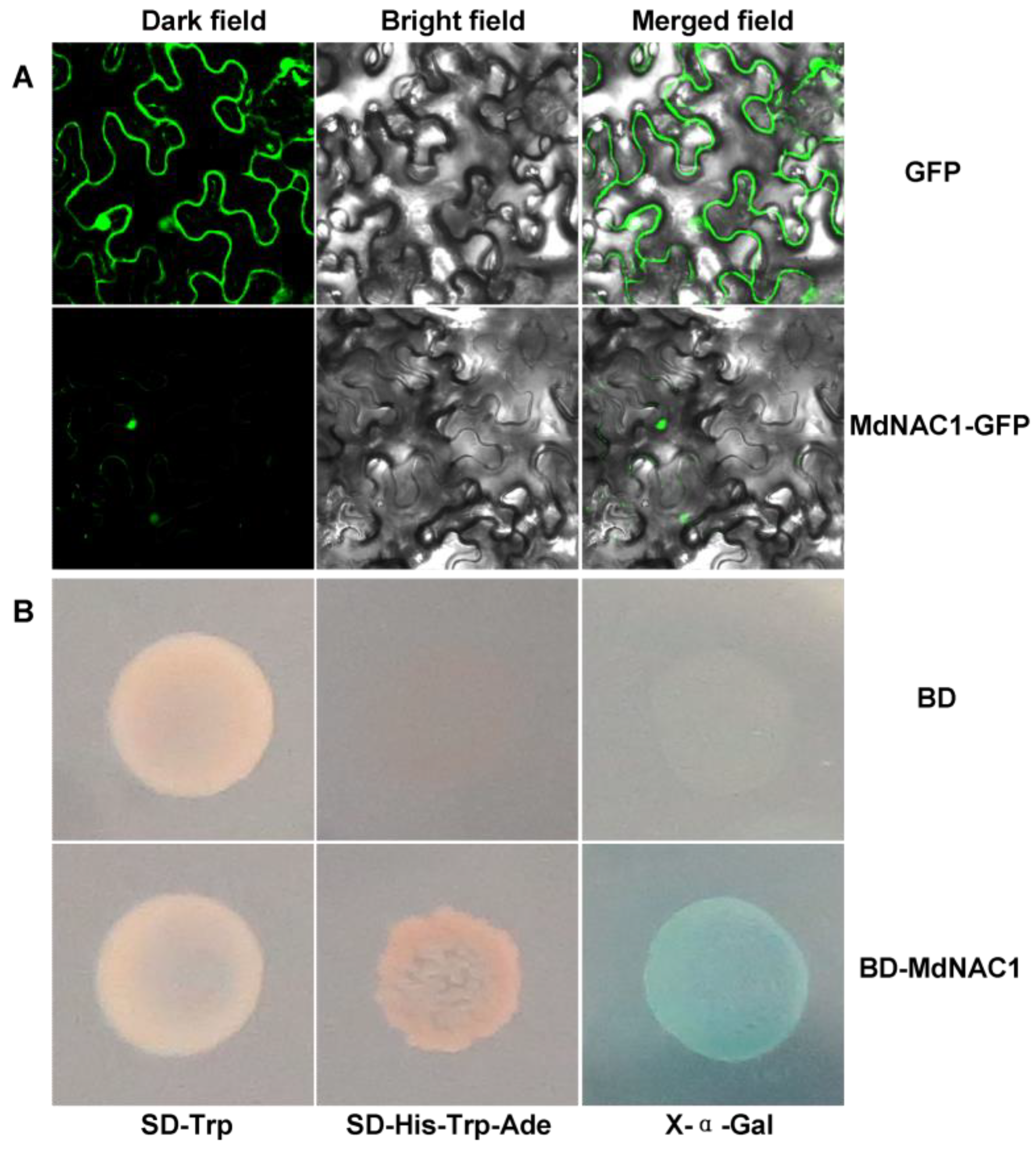
3.2. MdNAC1 Is Mainly Located in the Nucleus and Has Transcriptional Activity
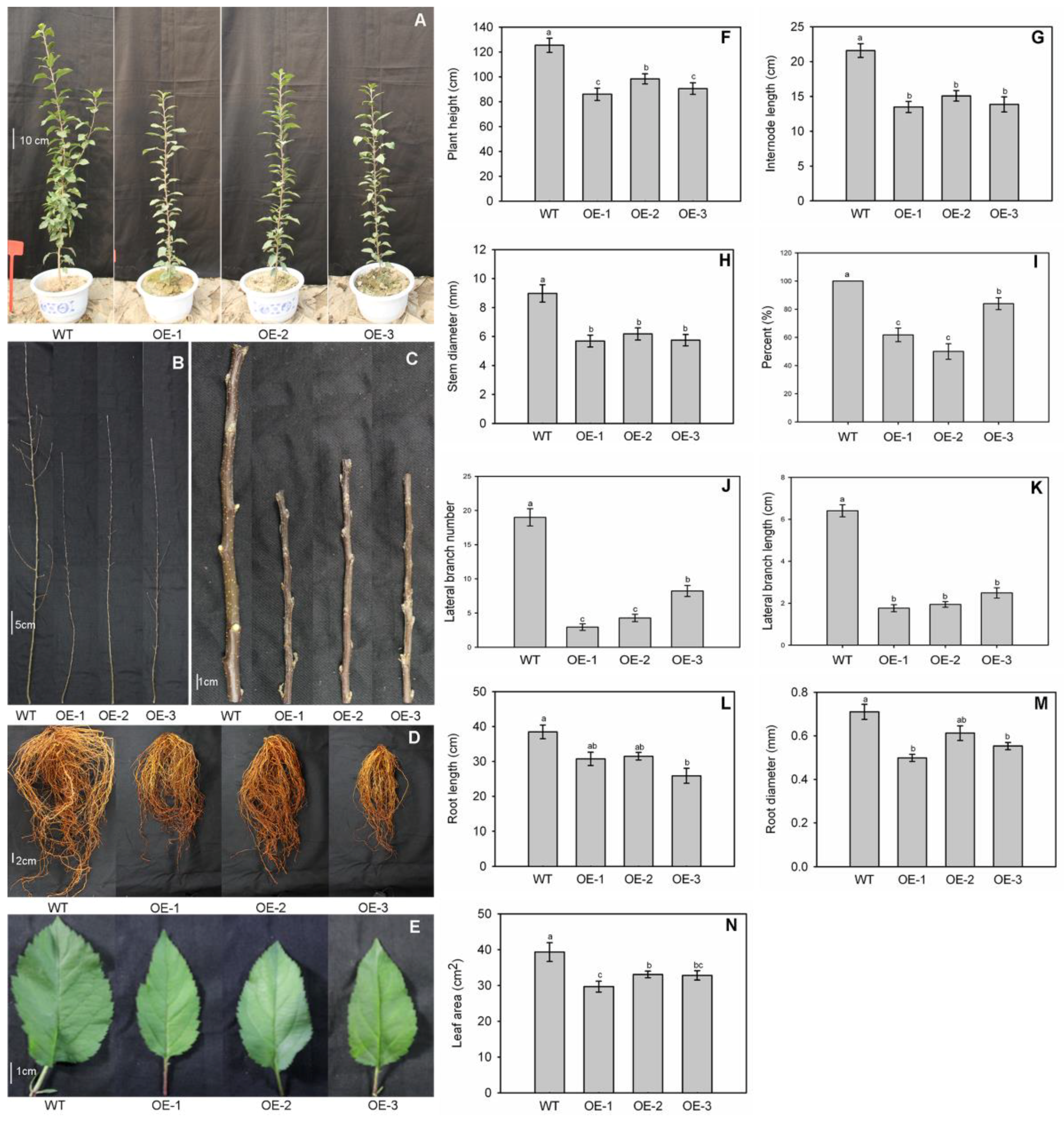
3.3. Dwarf Phenotype of the MdNAC1 Overexpression Apple Lines
3.4. Ultrastructure Analysis of Stems and Leaves
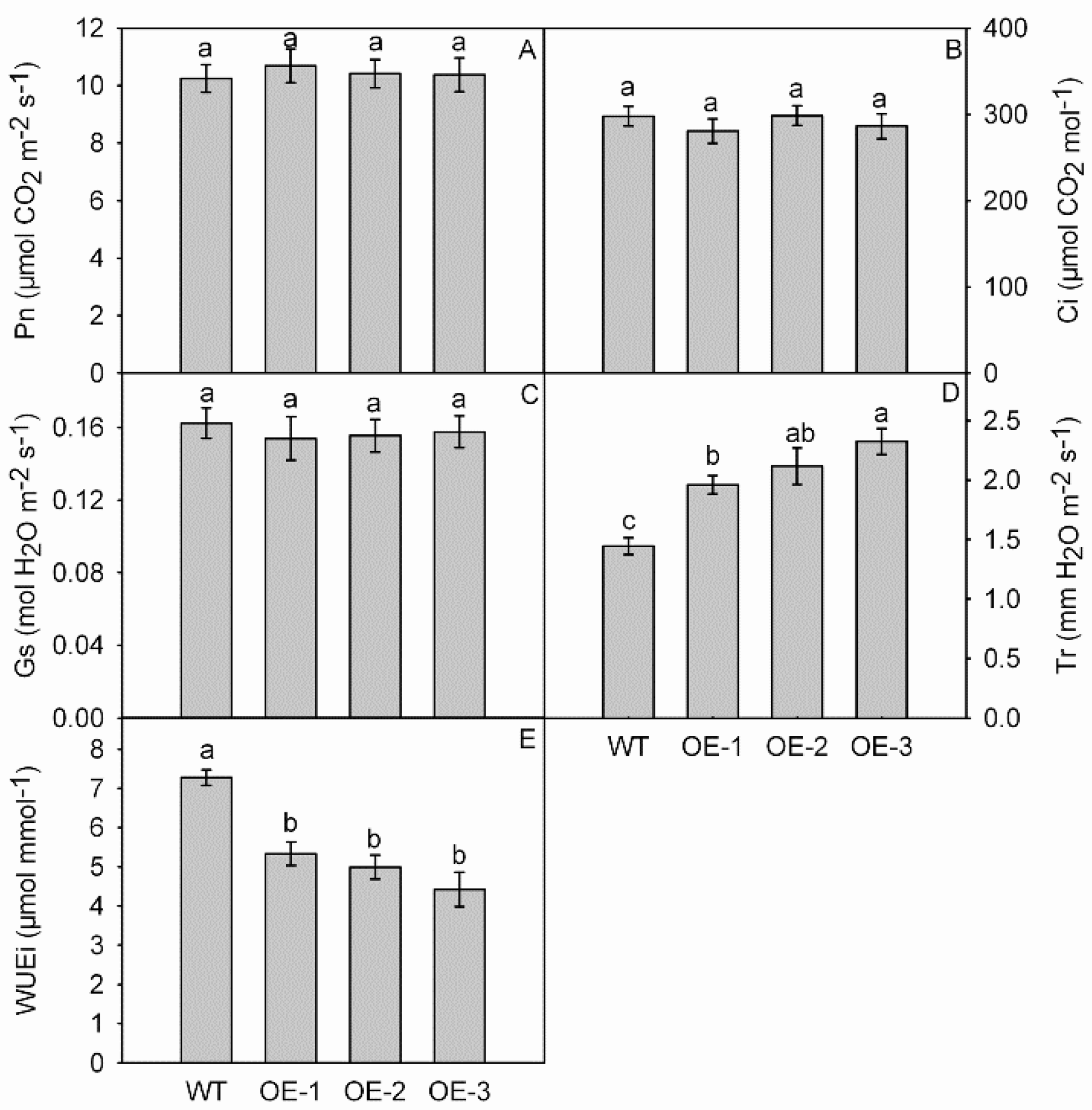
3.5. Photosynthesis and Gas Exchange Parameters
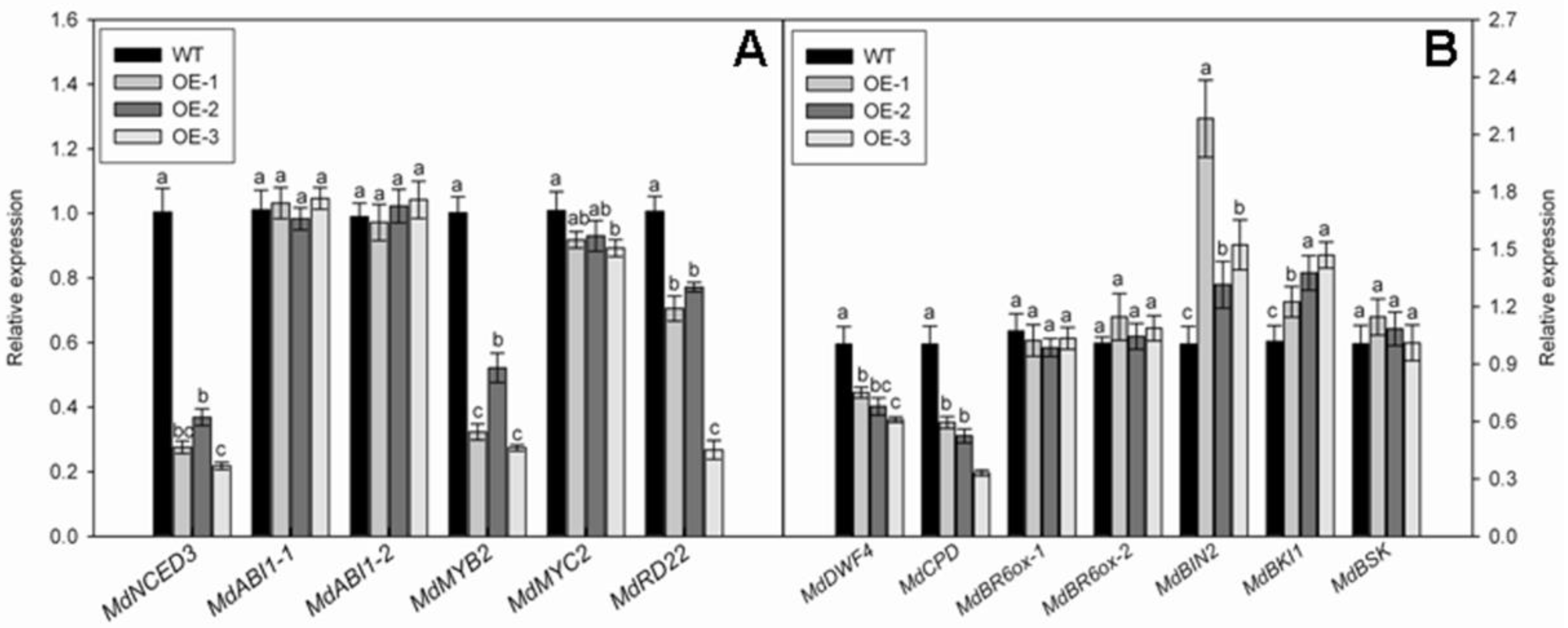
3.6. Overexpression of MdNAC1 Reduces the Biosynthesis of ABA and BR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Cohen, S.; Naor, A.; Bennink, J.; Grava, A.; Tyree, M. Hydraulic resistance components of mature apple trees on rootstocks of different vigours. J. Exp. Bot. 2007, 58, 4213–4224. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.M.; Celton, J.M.; Chagné, D.; Tustin, D.S.; Gardiner, S.E. Two quantitative trait loci, Dw1 and Dw2, are primarily responsible for rootstock–induced dwarfing in apple. Hortic. Res. 2015, 2, 15001. [Google Scholar] [CrossRef] [PubMed]
- Ephritikhine, G.; Fellner, M.; Vannini, C.; Lapous, D.; Barbier–Brygoo, H. The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 1999, 18, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.H.; Li, X.Y.; Welander, M. Overexpression of the Arabidopsis gai gene in apple significantly reduces plant size. Plant Cell Rep. 2008, 27, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Imai, A.; Hanzawa, Y.; Komura, M.; Yamamoto, K.T.; Komeda, Y.; Takahashi, T. The dwarf phenotype of the Arabidopsis acl5 mutant is suppressed by a mutation in an upstream ORF of a bHLH gene. Development 2006, 133, 3575–3585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Xue, H.; Zhang, L.; Zhang, F.; Ou, C.; Wang, F.; Zhang, Z. Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus × domestica). Sci. Rep. 2016, 6, 26719. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhao, Y.; Shan, D.; Shi, K.; Wang, L.; Li, Q.; Wang, N.; Zhou, J.; Yao, J.; Xue, Y.; et al. MdWRKY9 overexpression confers intensive dwarfing in the M26 rootstock of apple by directly inhibiting brassinosteroid synthetase MdDWF4 expression. New Phytol. 2018, 22, 1086–1098. [Google Scholar]
- Sharp, R.E.; LeNoble, M.E.; Else, M.A.; Thorne, E.T.; Gherardi, F. Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: Evidence for an interaction with ethylene. J. Exp. Bot. 2000, 51, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Nagel, O.W.; Konings, H.; Lambers, H. Growth rate, plant development and water relations of the ABA–deficient tomato mutant sitiens. Physiol. Plant. 1994, 92, 102–108. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Rock, C.D. Abscisic Acid Biosynthesis and Response. In The Arabidopsis Book; Somerville, C.R., Meyerowitz, E.M., Eds.; American Society of Plant Biologists: Rockville, MD, USA, 2002; pp. 1–52. [Google Scholar]
- Cheon, J.; Park, S.Y.; Schulz, B.; Choe, S. Arabidopsis brassinosteroid biosynthetic mutant dwarf7–1 exhibits slower rates of cell division and shoot induction. BMC Plant Biol. 2010, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Azpiroz, R.; Wu, Y.; LoCascio, J.C.; Feldmann, K.A. An Arabidopsis brassinosteroid–dependent mutant is blocked in cell elongation. Plant Cell 1998, 10, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.; Jiang, S.; Wang, F.; Tang, C.; Hao, N. An RNA–Seq analysis of the pear (Pyrus communis L.) transcriptome, with a focus on genes associated with dwarf. Plant Gene 2015, 4, 69–77. [Google Scholar] [CrossRef]
- Zhang, D.; Ren, L.; Yue, J.H.; Shi, Y.B.; Zhuo, L.H.; Wang, L.; Shen, X.H. RNA–Seq–based transcriptome analysis of stem development and dwarfing regulation in Agapanthus praecox ssp. orientalis (Leighton) Leighton. Gene 2015, 565, 252–267. [Google Scholar] [PubMed]
- Kato, H.; Motomura, T.; Komeda, Y.; Saito, K.; Kato, A. Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 2010, 167, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Avci, U.; Grant, E.H.; Haigler, C.H.; Beers, E.P. XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J. 2008, 53, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Kubo, M.; Fukuda, H.; Demura, T. Vascular-related NAC–DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008, 55, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tran, L.S.P.; Nguyen, D.V.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi–Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.; Bellizzi, M.; Wan, C.; Cui, Z.; Li, Y.; Wang, G.L. The NAC transcription factor OsSWN1 regulates secondary cell wall development in Oryza sativa. J. Plant Biol. 2015, 58, 44–51. [Google Scholar] [CrossRef]
- Grant, E.H.; Fujino, T.; Beers, E.P.; Brunner, A.M. Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 2010, 232, 337–352. [Google Scholar] [CrossRef] [PubMed]
- BLAST, Basic Local Alignment Search Tool from Genome Database for Rosaceae. Available online: https://www.rosaceae.org/blast (accessed on 19 April 2018).
- Hidden Markov Model (HMM), version v3.1b2. Available online: http://hmmer.org/ (accessed on 19 April 2018).
- Gen Bank Database. Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 19 April 2018).
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 19 April 2018).
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, R.; Qi, M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000, 22, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.S.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration–induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Li, W.; Han, G.; Yang, Y.; Ma, Y.; Li, H.; Zhang, Z. Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci. Hortic. 2013, 164, 202–208. [Google Scholar] [CrossRef]
- Liang, B.; Li, C.; Ma, C.; Wei, Z.; Wang, Q.; Huang, D.; Chen, Qi.; Li, C.; Ma, F. Dopamine alleviates nutrient deficiency-induced stress in Malus hupehensis. Plant Physiol. Biochem. 2017, 119, 346–359. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Cheng, L.; Ma, F.; Zou, Y.; Liang, D. Growth, biomass allocation, and water use efficiency of 31 apple cultivars grown under two water regimes. Agrofor. Syst. 2012, 84, 117–129. [Google Scholar] [CrossRef]
- Ma, Y.; Sawhney, V.K.; Steeves, T.A. Staining of paraffin-embedded plant material in safranin and fast green without prior removal of the paraffin. Can. J. Bot. 1993, 140, 996–999. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J.; Wang, Z.; Zhu, Q.; Wang, W. Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 2001, 127, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Weiler, E.W.; Jourdan, P.S.; Conrad, W. Levels of indole -3-acetic acid in intact and decapitated coleoptiles as determined by a specific and highly sensitive solid-phase enzyme immunoassay. Planta 1981, 153, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Merlot, S.; Gosti, F.G.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; lwasaki, T.; Hosokawa, D.; Shinozak, K. Role of Arabidopsis MYC and MYB homologs in drought and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar] [PubMed]
- Zhao, B.; Li, J. Regulation of Brassinosteroid Biosynthesis and Inactivation. J. Integr. Plant Biol. 2012, 54, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, S.; Hardtke, C.S. Signalling crosstalk in plant growth regulation. Curr. Biol. 2011, 21, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Hibara, K.I.; Takada, S.; Tasaka, M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 2003, 36, 687–696. [Google Scholar] [CrossRef] [PubMed]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Richardson, E.A.; Ye, Z.H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Kim, S.G.; Park, J.E.; Park, H.Y.; Lim, M.H.; Chua, N.H.; Park, C.M. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 2006, 18, 3132–3144. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, C.; Britton, J.H.; Estelle, M. Growth and development of the axrl mutants of Arabidopsis. Plant Cell 1990, 2, 1071–1080. [Google Scholar]
- Silva, T.; Davies, P.J. Elongation rates and endogenous indoleacetic acid levels in roots of pea mutants differing in internode length. Physiol. Plant. 2007, 129, 804–812. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Onckelen, H.V.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Sharp, R.E. Interaction with ethylene: changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ. 2002, 25, 211–222. [Google Scholar] [CrossRef]
- Barrero, J.M.; Piqueras, P.; Gonzán, M.; Serrano, R.; Rodríguez, P.L.; Ponce, M.R.; Micol, J.L. A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J. Exp. Bot. 2005, 56, 2071–2083. [Google Scholar] [CrossRef] [PubMed]
- LeNoble, M.E.; Spollen, W.G.; Sharp, R.E. Maintenance of shoot growth by endogenous ABA: Genetic assessment of the involvement of ethylene suppression. J. Exp. Bot. 2004, 55, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Nitsch, L.; Kohlen, W.; Oplaat, C.; Charnikhova, T.; Cristescu, S.; Michieli, P.; Wolters-Arts, M.; Bouwmeester, H.; Mariani, C.; Vriezen, W.H.; et al. ABA-deficiency results in reduced plant and fruit size in tomato. J. Plant Physiol. 2012, 169, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Ueguchi-Tanaka, M.; Umemura, K.; Uozu, S.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. Rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 2003, 15, 2900–2910. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, T.; Chuck, G.S.; Fujioka, S.; Klempien, A.; Weizbauer, R.; Potluri, D.P.V.; Choe, S.; Johal, G.S.; Schulz, B. Brassinosteroid control of sex determination in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 19814–19819. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Hagedorn, P.H.; de Torres-Zabala, M.; Grant, M.R.; Rung, J.H.; Collinge, D.B.; Lyngkiaer, M.F. Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defense towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008, 56, 867–880. [Google Scholar] [PubMed]
- Zhang, K.; Gan, S.S. An abscisic acid–AtNAP transcription factor–SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol. 2012, 158, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Y.; Lv, B.; Li, J.; Luo, L.; Lu, S.; Zhang, X.; Ma, H.; Ming, F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T.; et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Tarkowska, D.; Sakuraba, Y.; Balazadeh, S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signaling. Nat. Plants 2016, 2, 16013. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Allu, A.D.; Mehterov, N.; Thirumalaikumar, V.P.; Alseekh, S.; Fernie, A.R.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 exerts conserved control over gibberellin and brassinosteroid metabolism and signaling genes in tomato. Front. Plant Sci. 2017, 8, 214. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought–responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [PubMed]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, D.; Gong, X.; Li, M.; Li, C.; Sun, T.; Ma, F. Overexpression of a Novel Apple NAC Transcription Factor Gene, MdNAC1, Confers the Dwarf Phenotype in Transgenic Apple (Malus domestica). Genes 2018, 9, 229. https://doi.org/10.3390/genes9050229
Jia D, Gong X, Li M, Li C, Sun T, Ma F. Overexpression of a Novel Apple NAC Transcription Factor Gene, MdNAC1, Confers the Dwarf Phenotype in Transgenic Apple (Malus domestica). Genes. 2018; 9(5):229. https://doi.org/10.3390/genes9050229
Chicago/Turabian StyleJia, Dongfeng, Xiaoqing Gong, Mingjun Li, Chao Li, Tingting Sun, and Fengwang Ma. 2018. "Overexpression of a Novel Apple NAC Transcription Factor Gene, MdNAC1, Confers the Dwarf Phenotype in Transgenic Apple (Malus domestica)" Genes 9, no. 5: 229. https://doi.org/10.3390/genes9050229




