BSG and MCT1 Genetic Variants Influence Survival in Multiple Myeloma Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Controls
2.2. Genotyping
2.3. Statistical Analysis
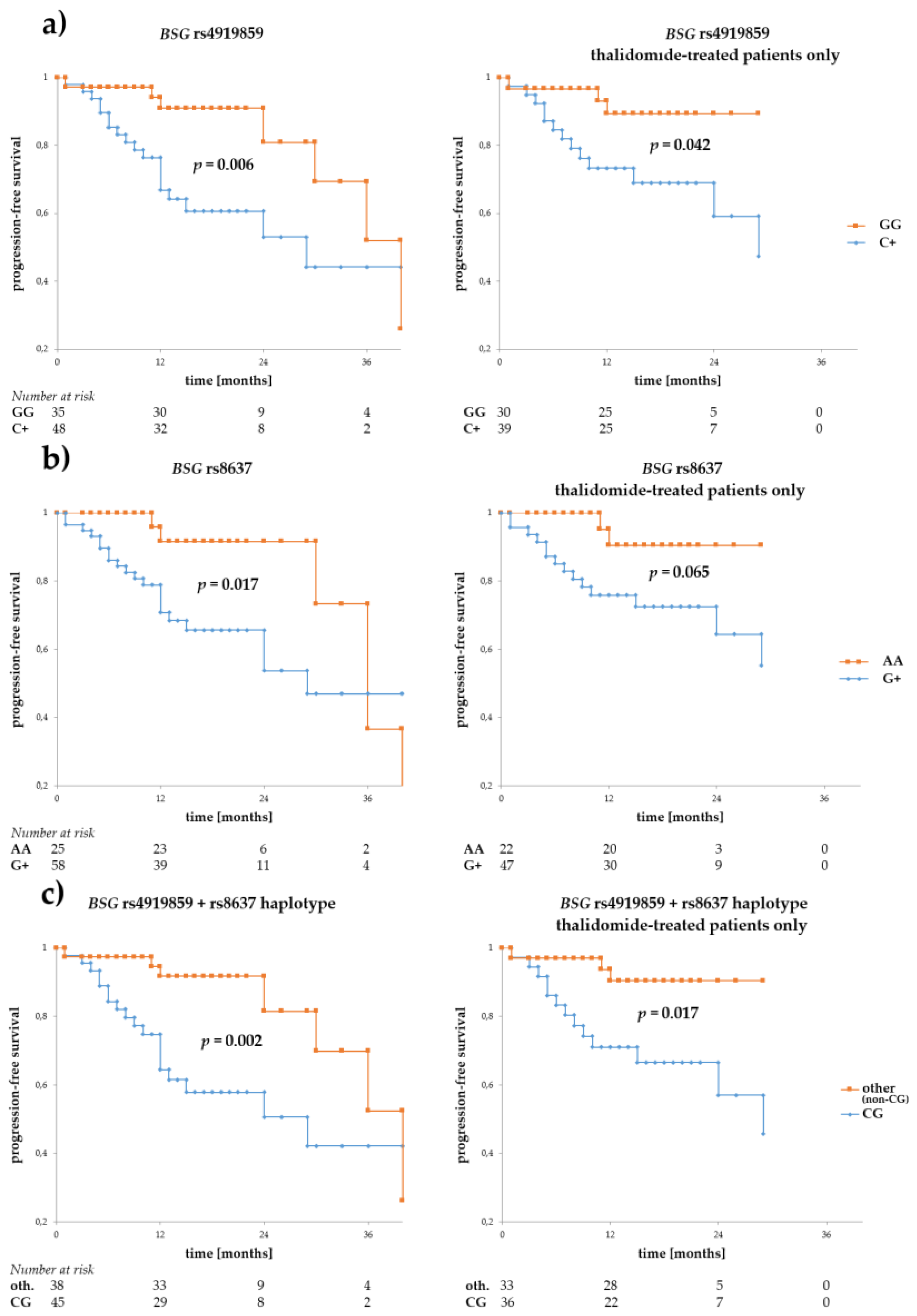
3. Results
3.1. BSG and SLC16A1 Allele and Genotype Frequencies
3.2. Associations of BSG and SLC16A1 Polymorphism with Survival
3.3. Influence of BSG and SLC16A1 Polymorphism on Response to Treatment
3.4. BSG and SLC16A1 Polymorphism and Other Clinical Parameters
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Raab, M.S.; Podar, K.; Breitkreutz, I.; Richardson, P.G.; Anderson, K.C. Multiple myeloma. Lancet 2009, 374, 324–339. [Google Scholar] [CrossRef]
- Rosenberg, P.S.; Barker, K.A.; Anderson, W.F. Future distribution of multiple myeloma in the United States by sex, age, and race/ethnicity. Blood 2015, 125, 410–412. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, T. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J. Biochem. 2016, 159, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Spring, F.A.; Holmes, C.H.; Simpson, K.L.; Mawby, W.J.; Mattes, M.J.; Okubo, Y.; Parsons, S.F. The Oka blood group antigen is a marker for the M6 leukocyte activation antigen, the human homolog of OX-47 antigen, basigin and neurothelin, an immunoglobulin superfamily molecule that is widely expressed in human cells and tissues. Eur. J. Immunol. 1997, 27, 891–897. [Google Scholar] [CrossRef]
- Walters, D.K.; Arendt, B.K.; Jelinek, D.F. CD147 regulates the expression of MCT1 and lactate export in multiple myeloma cells. Cell Cycle 2013, 12, 3175–3183. [Google Scholar] [CrossRef] [PubMed]
- Kirk, P.; Wilson, M.C.; Heddle, C.; Brown, M.H.; Barclay, A.N.; Halestrap, A.P. CD147 is tightly associated with lactate transporters MCT1 and MCT4 and facilitates their cell surface expression. EMBO J. 2000, 19, 3896–3904. [Google Scholar] [CrossRef] [PubMed]
- Arendt, B.K.; Walters, D.K.; Wu, X.; Tschumper, R.C.; Huddleston, P.M.; Henderson, K.J.; Dispenzieri, A.; Jelinek, D.F. Increased expression of extracellular matrix metalloproteinase inducer (CD147) in multiple myeloma: Role in regulation of myeloma cell proliferation. Leukemia 2012, 26, 2286–2296. [Google Scholar] [CrossRef] [PubMed]
- Panchabhai, S.; Schlam, I.; Sebastian, S.; Fonseca, R. PKM2 and other key regulators of Warburg effect positively correlate with CD147 (EMMPRIN) gene expression and predict survival in multiple myeloma. Leukemia 2017, 31, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Eichner, R.; Heider, M.; Fernández-Sáiz, V.; van Bebber, F.; Garz, A.K.; Lemeer, S.; Rudelius, M.; Targosz, B.S.; Jacobs, L.; Knorn, A.M.; et al. Immunomodulatory drugs disrupt the cereblon-CD147-MCT1 axis to exert antitumor activity and teratogenicity. Nat. Med. 2016, 22, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Ando, H.; Suzuki, T.; Ogura, T.; Hotta, K.; Imamura, Y.; Yamaguchi, Y.; Handa, H. Identification of a primary target of thalidomide teratogenicity. Science 2010, 327, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Bolomsky, A.; Hübl, W.; Spada, S.; Müldür, E.; Schlangen, K.; Heintel, D.; Rocci, A.; Weißmann, A.; Fritz, V.; Willheim, M.; et al. IKAROS expression in distinct bone marrow cell populations as a candidate biomarker for outcome with lenalidomide-dexamethasone therapy in multiple myeloma. Am. J. Hematol. 2017, 92, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Butrym, A.; Rybka, J.; Łacina, P.; Gębura, K.; Frontkiewicz, D.; Bogunia-Kubik, K.; Mazur, G. Polymorphisms within β-catenin encoding gene affect multiple myeloma development and treatment. Leuk. Res. 2015, 39, 1462–1466. [Google Scholar] [CrossRef] [PubMed]
- Butrym, A.; Łacina, P.; Rybka, J.; Chaszczewska-Markowska, M.; Mazur, G.; Bogunia-Kubik, K. Cereblon and IRF4 variants affect risk and response to treatment in multiple myeloma. Arch. Immunol. Ther. Exp. 2016, 64, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Taylor, J.A. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009, 37, W600–W605. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- VassarStats: Website for Statistical Computation. Available online: http://vassarstats.net/tab2x2.html (accessed on 23 February 2018).
- Lu, G.; Middleton, R.E.; Sun, H.; Naniong, M.; Ott, C.J.; Mitsiades, C.S.; Wong, K.K.; Bradner, J.E.; Kaelin, W.G., Jr. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of IKAROS proteins. Science 2014, 343, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Ni, T.; Chen, M.; Yang, K.; Shao, J.; Fu, Y.; Zhou, W. Association of CD147 genetic polymorphisms with carotid atherosclerotic plaques in a Han Chinese population with cerebral infarction. Thromb. Res. 2017, 156, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.S.; Li, F.F.; Sun, L.D.; Li, D.; Su, J.; Kuang, Y.H.; Chen, G.; Chen, X.P.; Chen, X. A miRNA-492 binding-site polymorphism in BSG (basigin) confers risk to psoriasis in central south Chinese population. Hum. Genet. 2011, 130, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Mao, Y.; Wang, C.; Wang, Z. Association study between an SNP in CD147 and its expression with acute coronary syndrome in a Jiangsu Chinese population. Medicine 2015, 94, e1537. [Google Scholar] [CrossRef] [PubMed]
- Li, M.P.; Hu, X.L.; Yang, Y.L.; Zhang, Y.J.; Zhou, J.P.; Peng, L.M.; Tang, J.; Chen, X.P. Basigin rs8259 polymorphism confers decreased risk of chronic heart failure in a Chinese population. Int. J. Environ. Res. Public Health 2017, 14, 211. [Google Scholar] [CrossRef] [PubMed]
- Cupeiro, R.; Benito, P.J.; Maffulli, N.; Calderón, F.J.; González-Lamuño, D. MCT1 genetic polymorphism influence in high intensity circuit training: A pilot study. J. Sci. Med. Sport 2010, 13, 526–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawczuk, M.; Banting, L.K.; Cięszczyk, P.; Maciejewska-Karłowska, A.; Zarębska, A.; Leońska-Duniec, A.; Jastrzębski, Z.; Bishop, D.J.; Eynon, N. MCT1 A1470T: A novel polymorphism for sprint performance? J. Sci. Med. Sport 2015, 18, 114–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, F.; Guo, X.; Chen, Y.; Liu, X.; Tu, J.; Xing, J.; Chen, Z.; Ji, J.; He, X. Polymorphisms of monocarboxylate transporter genes are associated with clinical outcomes in patients with colorectal cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Chen, C.; Liu, B.; Wu, Y.; Chen, Y.; Zhou, X.; Huang, X.; Li, X.; Yang, H.; Chen, Z.; et al. Genetic variations in monocarboxylate transporter genes as predictors of clinical outcomes in non-small cell lung cancer. Tumour Biol. 2015, 36, 3931–3939. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, S.; Futagi, Y.; Kobayashi, M.; Ogura, J.; Iseki, K. Functional characterization of 5-oxoproline transport via SLC16A1/MCT1. J. Biol. Chem. 2015, 290, 2303–2311. [Google Scholar] [CrossRef]
- Krönke, J.; Kuchenbauer, F.; Kull, M.; Teleanu, V.; Bullinger, L.; Bunjes, D.; Greiner, A.; Kolmus, S.; Köpff, S.; Schreder, M.; et al. IKZF1 expression is a prognostic marker in newly diagnosed standard-risk multiple myeloma treated with lenalidomide and intensive chemotherapy: A study of the German myeloma study group (DSMM). Leukemia 2017, 31, 1363–1367. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, A.K.; Mendy, D.; Waldman, M.; Chen, G.; Rychak, E.; Miller, K.; Gaidarova, S.; Ren, Y.; Wang, M.; Breider, M.; et al. Measuring cereblon as a biomarker of response or resistance to lenalidomide and pomalidomide requires use of standardized reagents and understanding of gene complexity. Br. J. Haematol. 2014, 164, 233–244. [Google Scholar] [CrossRef] [PubMed]


| MM Patients (All) | MM Patients (Thalidomide-Treated) | Controls | MM Patients | MM Patients (Thalidomide-Treated) | Controls | ||
|---|---|---|---|---|---|---|---|
| BSG rs4919859 | SLC16A1 rs9429505 | ||||||
| CC | 15 (11.2%) | 13 (13.1%) | 19 (14.1%) | AA | 77 (57.0%) | 59 (59.0%) | 67 (49.6%) |
| CG | 65 (48.5%) | 47 (47.5%) | 54 (40.0%) | AG | 51 (37.8%) | 34 (34.0%) | 60 (44.4%) |
| GG | 54 (40.3%) | 39 (39.4%) | 62 (45.9%) | GG | 7 (5.2%) | 7 (7.0%) | 8 (5.9%) |
| BSG rs4682 | SLC16A1 rs7169 | ||||||
| CC | 3 (2.2%) | 3 (3.0%) | 4 (3.0%) | CC | 22 (16.3%) | 15 (15.0%) | 23 (17.0%) |
| CT | 45 (33.6%) | 35 (35.4%) | 34 (25.4%) | CT | 63 (46.7%) | 46 (46.0%) | 58 (43.0%) |
| TT | 86 (64.2%) | 61 (61.6%) | 96 (71.6%) | TT | 50 (37.0%) | 39 (39.0%) | 54 (40.0%) |
| BSG rs8637 | SLC16A1 rs1049434 | ||||||
| AA | 44 (32.8%) | 32 (32.3%) | 47 (34.8%) | AA | 51 (37.8%) | 39 (39.0%) | 54 (40.0%) |
| AG | 63 (47.0%) | 45 (45.5%) | 61 (45.2%) | AT | 62 (45.9%) | 46 (46.0%) | 58 (43.0%) |
| GG | 27 (20.2%) | 22 (22.2%) | 27 (20.0%) | TT | 22 (16.3%) | 15 (15.0%) | 23 (17.0%) |
| BSG rs8259 | SLC16A1 rs7556664 | ||||||
| TT | 77 (57.0%) | 57 (57.0%) | 78 (57.8%) | AA | 50 (37.0%) | 39 (39.0%) | 54 (40.0%) |
| TA | 49 (36.3%) | 35 (35.0%) | 49 (36.3%) | AT | 63 (46.7%) | 46 (46.0%) | 58 (43.0%) |
| AA | 9 (6.7%) | 8 (8.0%) | 8 (5.9%) | TT | 22 (16.3%) | 15 (15.0%) | 23 (17.0%) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łacina, P.; Butrym, A.; Mazur, G.; Bogunia-Kubik, K. BSG and MCT1 Genetic Variants Influence Survival in Multiple Myeloma Patients. Genes 2018, 9, 226. https://doi.org/10.3390/genes9050226
Łacina P, Butrym A, Mazur G, Bogunia-Kubik K. BSG and MCT1 Genetic Variants Influence Survival in Multiple Myeloma Patients. Genes. 2018; 9(5):226. https://doi.org/10.3390/genes9050226
Chicago/Turabian StyleŁacina, Piotr, Aleksandra Butrym, Grzegorz Mazur, and Katarzyna Bogunia-Kubik. 2018. "BSG and MCT1 Genetic Variants Influence Survival in Multiple Myeloma Patients" Genes 9, no. 5: 226. https://doi.org/10.3390/genes9050226





