Genome Wide Identification, Evolutionary, and Expression Analysis of VQ Genes from Two Pyrus Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Classification of P. bretschneideri and P. communis VQ Motif-Containing Genes
2.2. Chromosome Locations and Microcollinearity Analysis of Pyrus VQ Motif-Containing Genes
2.3. Intron–Exon Structure and Domain Search of P. bretschneideri and P. communis VQ Motif-Containing Genes
2.4. Expression Data Analysis
2.5. Expression Correlation of Homologous VQ Motif-Containing Genes between P. bretschneideri and P. communis
2.6. Plant Material, RNA Extraction, and Real-Time Polymerase Chain Reaction Analysis
2.7 Availability of Data and Materials
3. Results
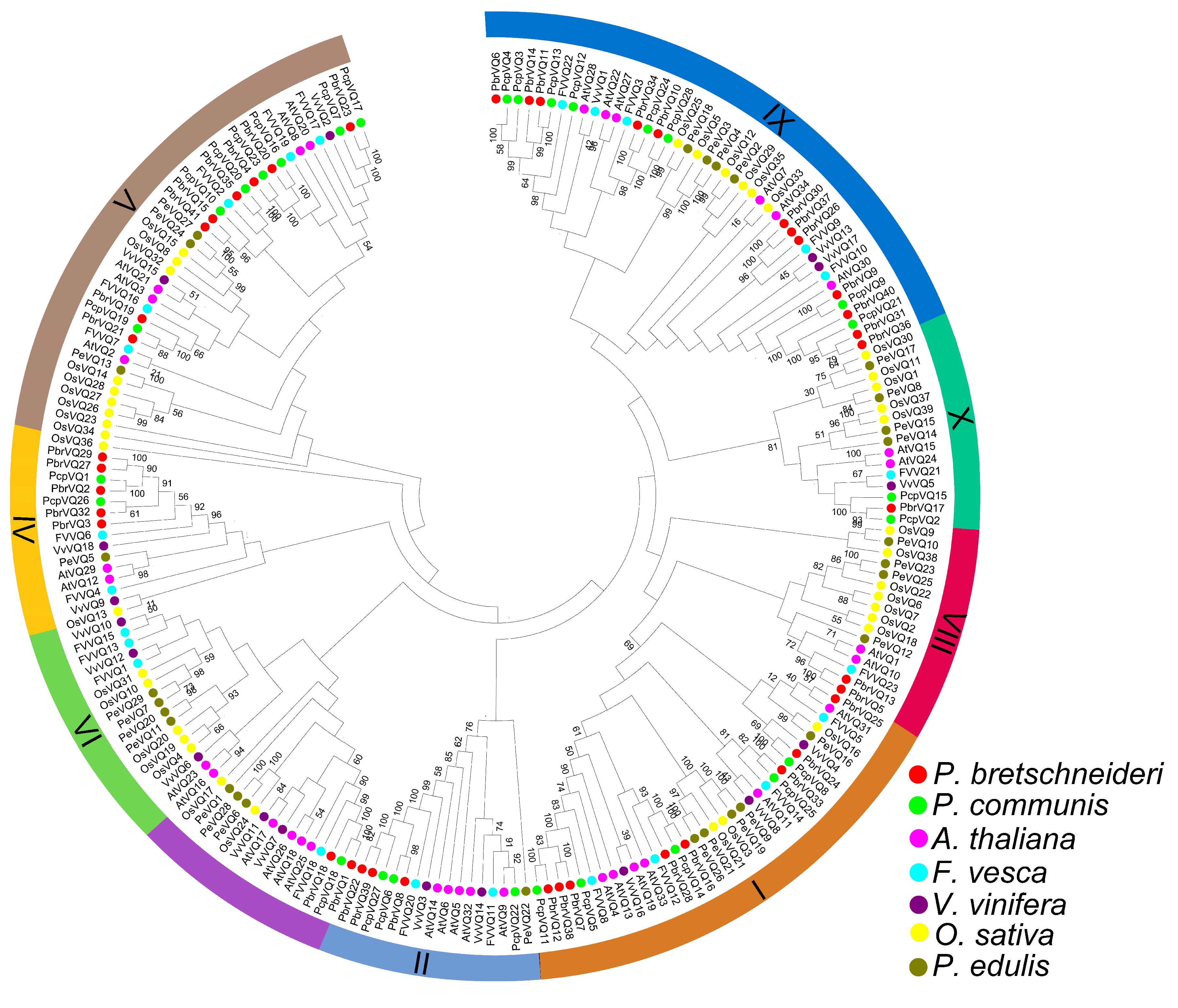
3.1. Identification and Classification of P. bretschneideri and P. communis VQ Motif-Containing Genes
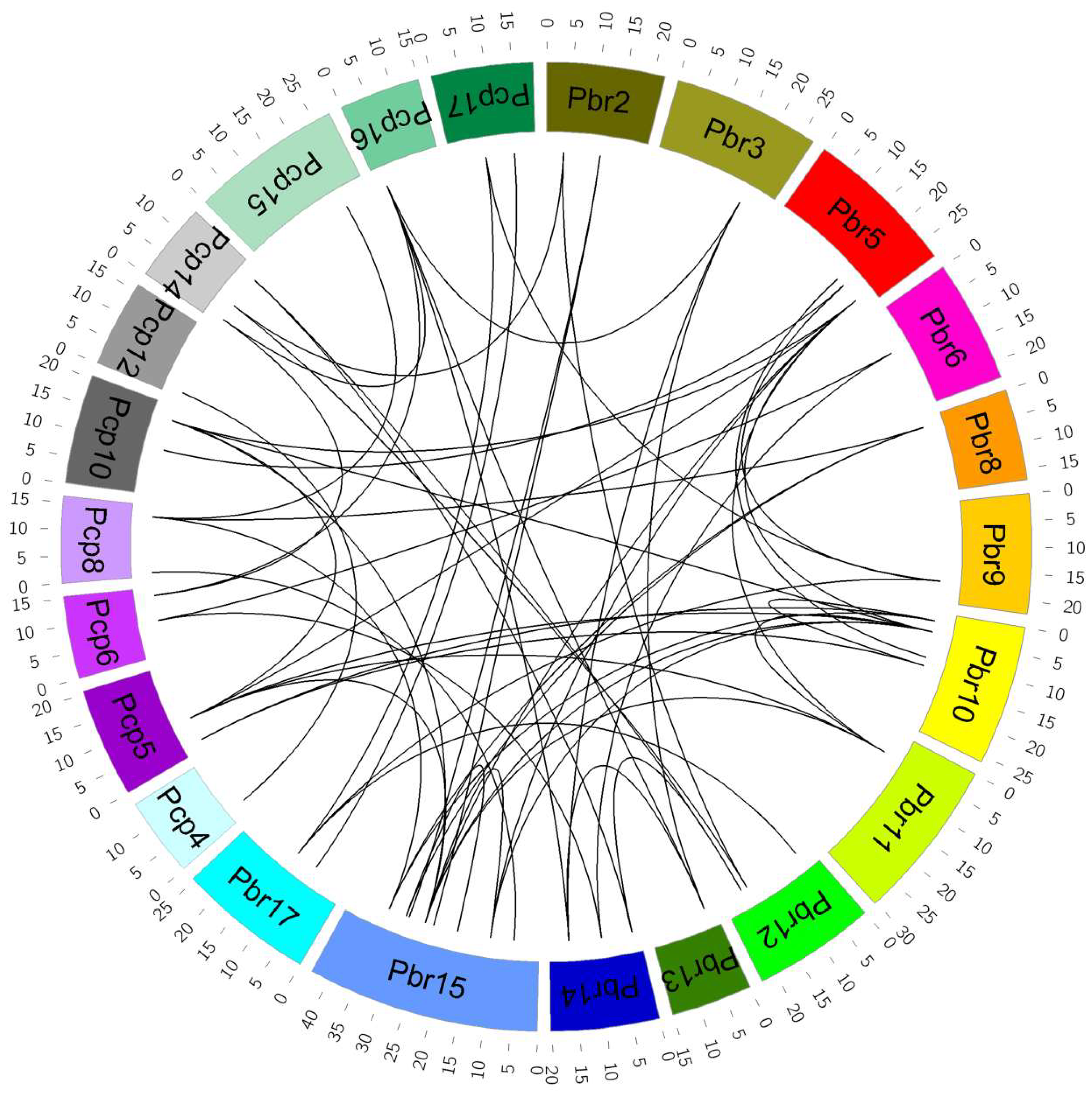
3.2. Chromosome Locations and Duplication Events of P. bretschneideri and P. communis VQ Motif-Containing Genes
3.3. Syntenic Blocks Analysis of P. bretschneideri and P. communis VQ Motif-Containing Genes
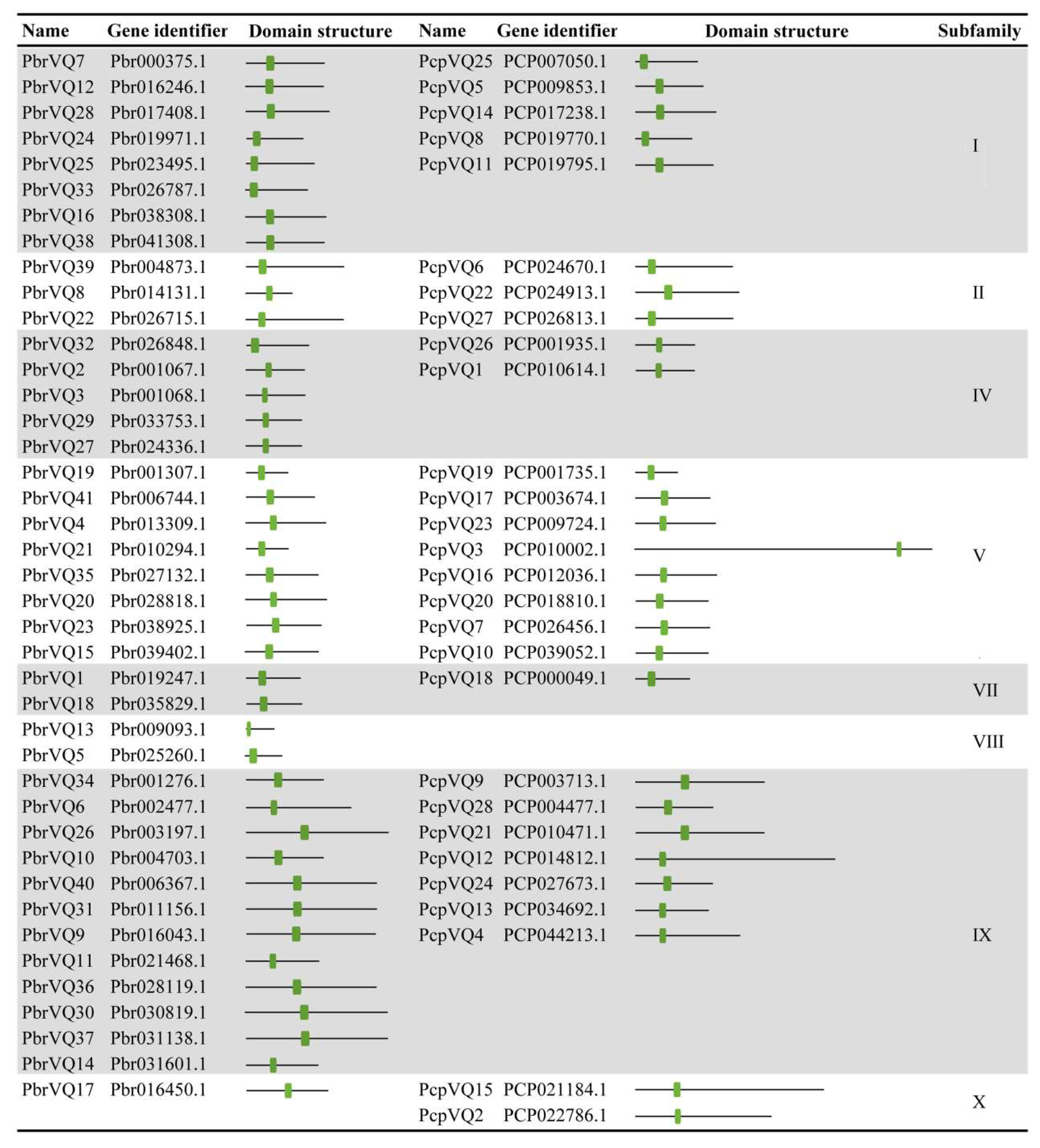
3.4. Gene Structural and Conserved Motifs Analysis of P. bretschneideri and P. communis VQ Motif-Containing Genes
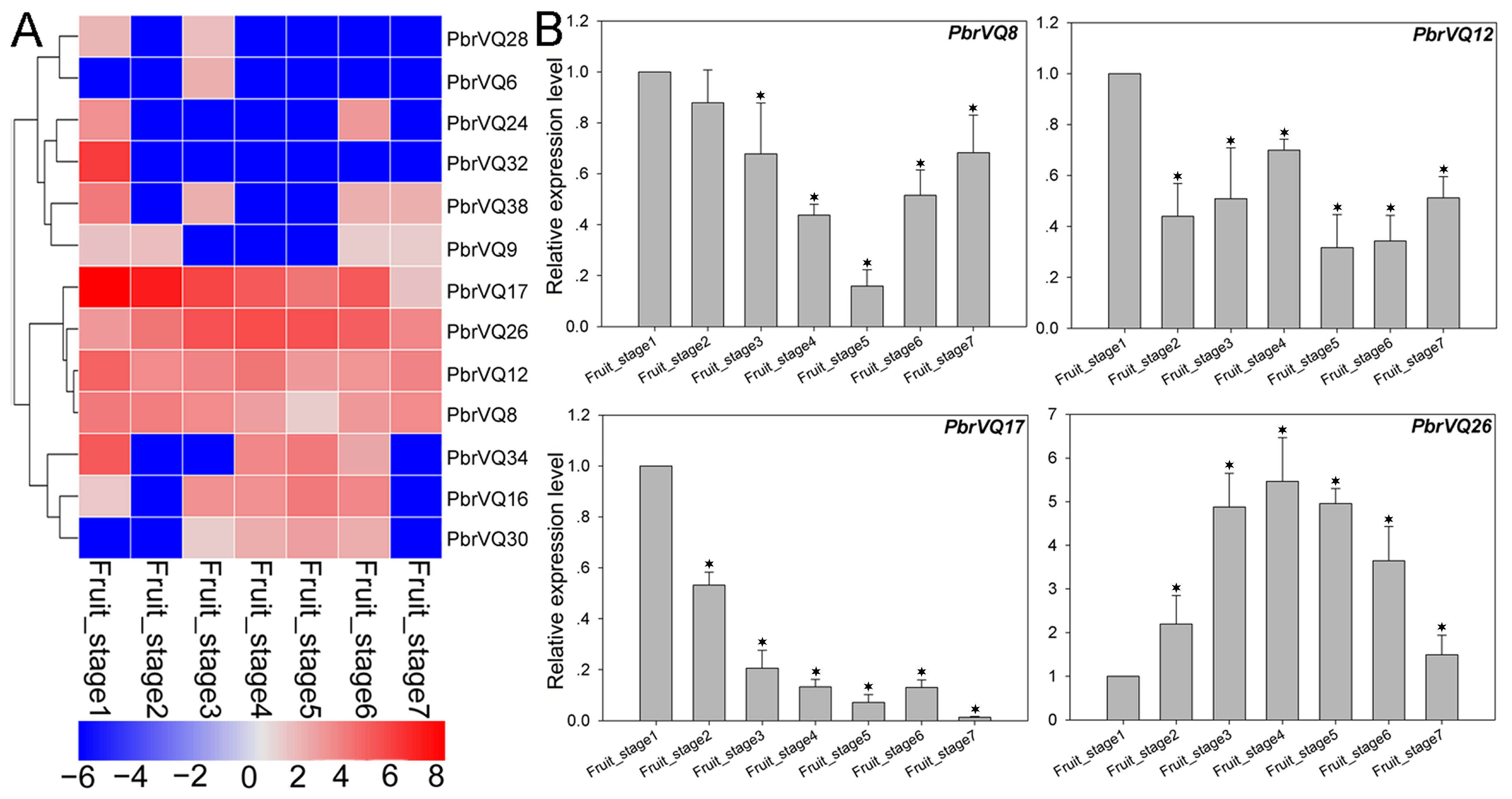
3.5. Expression Pattern Analysis of P. bretschneideri VQ Motif-Containing Genes by Transcriptome Data
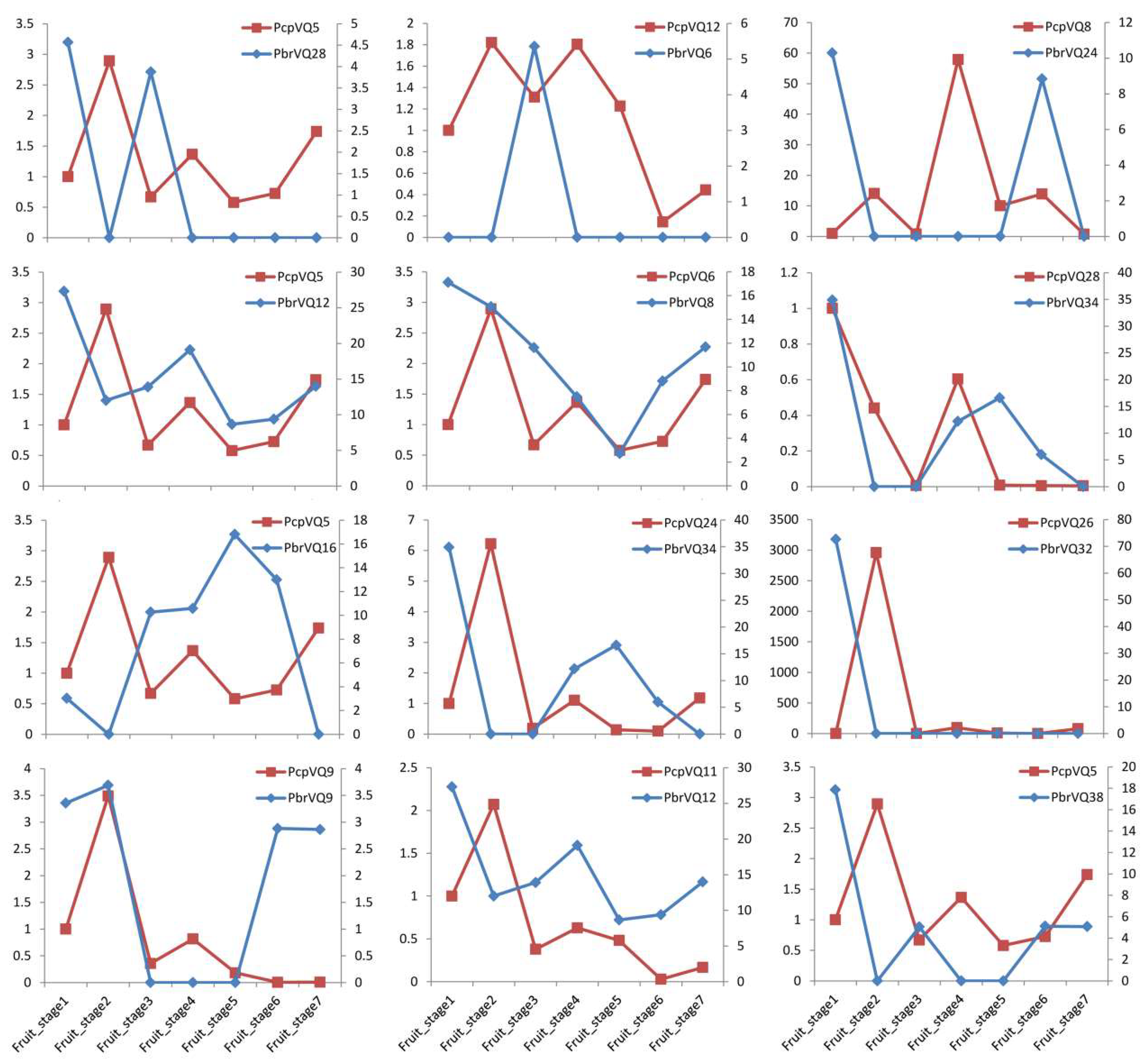
3.6. Comparison of the Expression Patterns of P. bretschneideri VQ Motif-Containing Genes and Their Orthologous in P. communis during Fruit Development
4. Discussion
4.1. Phylogeny of P. bretschneideri and P. communis VQ Motif-Containing Gene Family
4.2. Expansion and Duplication of P. bretschneideri and P. communis VQ Motif-Containing Gene Family
4.3. Expression Analysis of P. bretschneideri VQ Motif-Containing Genes
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Morikawa, K.; Shiina, T.; Murakami, S.; Toyoshima, Y. Novel nuclear-encoded proteins interacting with a plastid sigma factor, Sig1, in Arabidopsis Thaliana. FEBS Lett. 2002, 514, 300–304. [Google Scholar] [CrossRef]
- Wang, A.; Garcia, D.; Zhang, H.; Feng, K.; Chaudhury, A.; Berger, F.; Peacock, W.J.; Dennis, E.S.; Luo, M. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 2010, 63, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhou, Y.; Yang, Y.; Chi, Y.; Zhou, J.; Chen, J.; Wang, F.; Fan, B.; Shi, K.; Zhou, Y. Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 2012, 159, 810–825. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Kwon, S.I.; Choi, C.Y.; Lee, H.; Ahn, I.; Park, S.R.; Bae, S.; Lee, S.; Hwang, D.J. Expression analysis of rice VQ genes in response to biotic and abiotic stresses. Gene 2013, 529, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Sun, G.; Jin, Y.; Qiu, L. Identification of active VQ motif-containing genes and the expression patterns under low nitrogen treatment in soybean. Gene 2014, 543, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.M.; Vannozzi, A.; Wang, G.G.; Zhong, Y.Y.; Corso, M.; Cavallini, E.; Cheng, Z.M. A comprehensive survey of the grapevine VQ gene family and its transcriptional correlation with WRKY proteins. Front. Plant Sci. 2015, 6, 417. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Zhao, H.; Zhang, X.; Lei, L.; Lai, J. Genome-wide identification of VQ motif-containing proteins and their expression profiles under abiotic stresses in maize. Front. Plant Sci. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Guo, C.; Chu, J.; Liu, H.; Cheng, Z.-M. Microevolution of the VQ gene family in six species of Fragaria. Genome 2017, 61, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Lin, R. The VQ motif-containing protein family of plant-specific transcriptional regulators. Plant Physiol. 2015, 169, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jing, Y.; Li, J.; Xu, G.; Lin, R. Arabidopsis VQ MOTIF-CONTAINING PROTEIN29 represses seedling deetiolation by interacting with PHYTOCHROME-INTERACTING FACTOR1. Plant Physiol. 2014, 164, 2068–2080. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, F.; Li, J.; Ding, Q.; Zhang, Y.; Li, H.; Zhang, J.; Gao, J. Genome-wide identification and analysis of the VQ motif-containing protein family in Chinese Cabbage (Brassica rapa L. ssp. Pekinensis). Int. J. Mol. Sci. 2015, 16, 28683–28704. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hu, Y.; Pan, J.; Yu, D. Arabidopsis VQ motif-containing proteins VQ12 and VQ29 negatively modulate basal defense against Botrytis cinerea. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Weyhe, M.; Eschenlippold, L.; Pecher, P.; Scheel, D.; Lee, J. Ménage à trois: The complex relationships between mitogen-activated protein kinases, WRKY transcription factors, and VQ-motif-containing proteins. Plant Signal. Behav. 2014, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Perruc, E.; Charpenteau, M.; Ramirez, B.C.; Jauneau, A.; Galaud, J.; Ranjeva, R.; Ranty, B. A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis Thaliana seedlings. Plant J. 2004, 38, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Andreasson, E.; Jenkins, T.; Brodersen, P.; Thorgrimsen, S.; Petersen, N.H.T.; Zhu, S.; Qiu, J.; Micheelsen, P.O.; Rocher, A.; Petersen, M.; et al. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 2005, 24, 2579–2589. [Google Scholar] [CrossRef] [PubMed]
- Petersen, K.; Qiu, J.; Lutje, J.; Fiil, B.K.; Hansen, S.; Mundy, J.; Petersen, M. Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Brodersen, P.; Naested, H.; Andreasson, E.; Lindhart, U.; Johansen, B.; Nielsen, H.B.; Lacy, M.; Austin, M.J.; Parker, J.E.; et al. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 2000, 103, 1111–1120. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jin, Q.; Lin, Y.; Cai, Y. Genome-wide analysis suggests high level of microsynteny and purifying selection affect the evolution of EIN3/EIL family in Rosaceae. PeerJ 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Pecher, P.; Eschenlippold, L.; Herklotz, S.; Kuhle, K.; Naumann, K.; Bethke, G.; Uhrig, J.; Weyhe, M.; Scheel, D.; Lee, J. The Arabidopsis Thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of ‘VQ-motif’-containing proteins to regulate immune responses. New Phytol. 2014, 203, 592–606. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Chagné, D.; Crowhurst, R.N.; Pindo, M.; Thrimawithana, A.; Deng, C.; Ireland, H.; Fiers, M.; Dzierzon, H.; Cestaro, A.; Fontana, P.; et al. The draft genome sequence of European pear (Pyrus communis L.‘Bartlett’). PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.I.; Yau, C.K.; Looseley, M.; Meyers, B.C. Effects of gene expression on molecular evolution in Arabidopsis Thaliana and Arabidopsis Lyrata. Mol. Biol. Evol. 2004, 21, 1719–1726. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’S conserved domain database. Nucleic Acids Res. 2014, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2011, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with Tophat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Yim, W.C.; Lee, B.-M.; Jang, C.S. Expression diversity and evolutionary dynamics of rice duplicate genes. Mol. Genet. Genom. 2009, 281, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. MYB transcription factors in Chinese Pear (Pyrus bretschneideri Rehd.): Genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jiao, C.; Jin, Q.; Lin, Y.; Cai, Y. B-BOX genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Wines, D.R.; Brady, J.M.; Southard, E.M.; MacDonald, R.J. Evolution of the rat Kallikrein gene family: Gene conversion leads to functional diversity. J. Mol. Evol. 1991, 32, 476–492. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ding, T.; Su, B.; Jiang, H. Genome-wide identification, characterization and expression analysis of the Chalcone Synthase family in maize. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, H.; Zhu, D.; Gao, Y.; Yan, H.; Xiang, Y. Genome-wide analysis of VQ motif-containing proteins in Moso Bamboo (Phyllostachys edulis). Planta 2017, 246, 165–181. [Google Scholar] [CrossRef] [PubMed]
- Louhichi, A.; Fourati, A.; Rebaï, A. IGD: A resource for intronless genes in the human genome. Gene 2011, 488, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Saingery, V.; Chambrier, P.; Mayer, U.; Jürgens, G.; Berger, F. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 2003, 131, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) kinase gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y. Computational approaches to unveiling ancient genome duplications. Nat. Rev. Genet. 2004, 5, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, J.A.; Maere, S.; Van de Peer, Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, L.; Yin, H.; Liu, X.; Wang, D.; Wu, J.; Wu, J.; Zhang, S. Modes of gene duplication and gene family expansion and evolution in Chinese white pear. In Proceedings of the Plant and Animal Genome XXIII Conference, San Diego, CA, USA, 10–14 January 2015. [Google Scholar]
- Gargul, J.M.; Mibus, H.; Serek, M. Manipulation of MKS1 gene expression Affects Kalanchoë blossfeldiana and Petunia hybrida phenotypes. Plant Biotechnol. J. 2015, 13, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Lei, R.; Li, X.; Ma, Z.; Lv, Y.; Hu, Y.; Yu, D. Arabidopsis WRKY2 and WRKY34 transcription factors interact with VQ20 protein to modulate pollen development and function. Plant J. 2017, 91, 962–976. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Meng, D.; Abdullah, M.; Jin, Q.; Lin, Y.; Cai, Y. Genome Wide Identification, Evolutionary, and Expression Analysis of VQ Genes from Two Pyrus Species. Genes 2018, 9, 224. https://doi.org/10.3390/genes9040224
Cao Y, Meng D, Abdullah M, Jin Q, Lin Y, Cai Y. Genome Wide Identification, Evolutionary, and Expression Analysis of VQ Genes from Two Pyrus Species. Genes. 2018; 9(4):224. https://doi.org/10.3390/genes9040224
Chicago/Turabian StyleCao, Yunpeng, Dandan Meng, Muhammad Abdullah, Qing Jin, Yi Lin, and Yongping Cai. 2018. "Genome Wide Identification, Evolutionary, and Expression Analysis of VQ Genes from Two Pyrus Species" Genes 9, no. 4: 224. https://doi.org/10.3390/genes9040224




