Ancient DNA from Giant Panda (Ailuropoda melanoleuca) of South-Western China Reveals Genetic Diversity Loss during the Holocene
Abstract
:1. Introduction
2. Materials and Methods
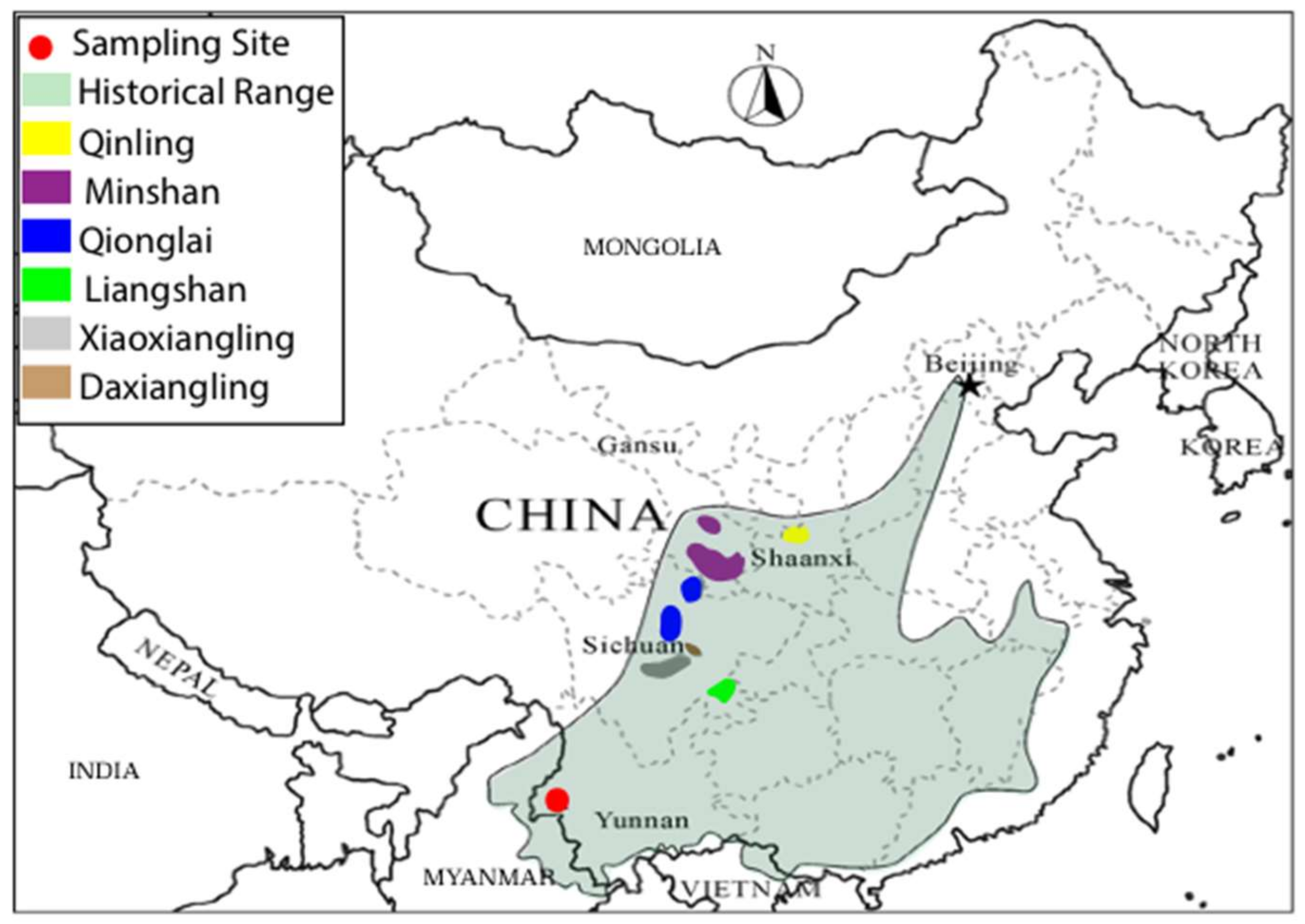
2.1. Samples
2.2. DNA Extraction and Amplification
2.3. Bioinformatic Analyses
2.4. Diversity Comparisons
2.5. Phylogenetic and Network Analysis
2.6. Demographic Inference
- A uniform prior on the basal divergence of the Ursidae with an upper limit of 12 million years, representing the age of a well-documented fossil representative of the Ailuropodinae, and a lower limit of 20 million years based on a previous molecular dating study.
- A uniform prior on the divergence of the Tremarctinae and Ursinae clades of 7 to 14 million years, based on the fossil Tremarctine bear Plionarctos.
- A uniform prior on the basal divergence of the Ursinae of 4.3 to 6 million years, based on reported ages of Ursus minimus.
- A uniform prior on the divergence of brown and polar bears of 0.48 to 1.1 million years, based on previous studies. This prior was applied to the common ancestor of a polar bear and a Finnish brown bear, likely representing the initial divergence of these respective species, rather than more recent gene flow events.
3. Results
3.1. Sequence Data Recovery
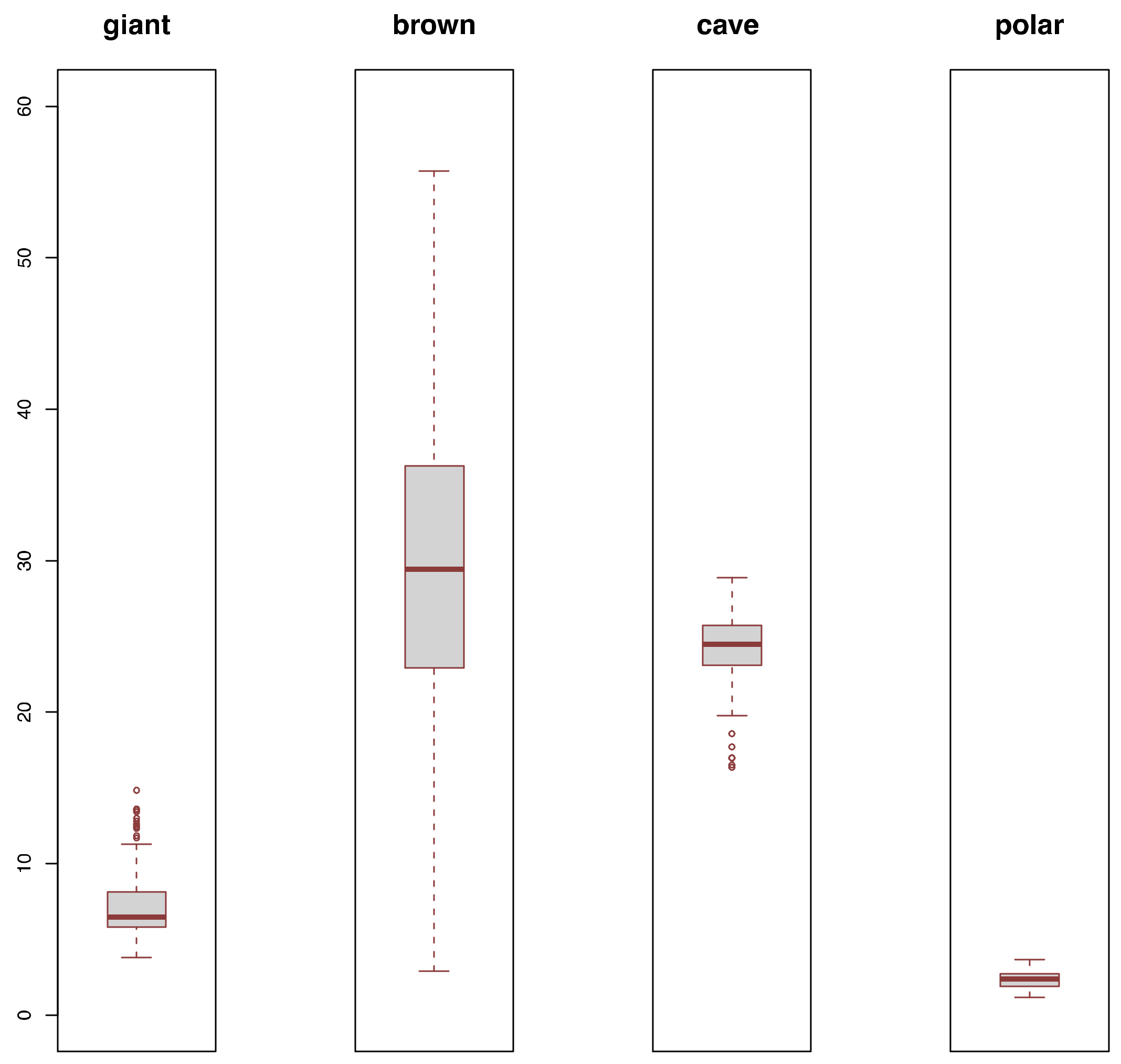
3.2. Diversity Comparisons
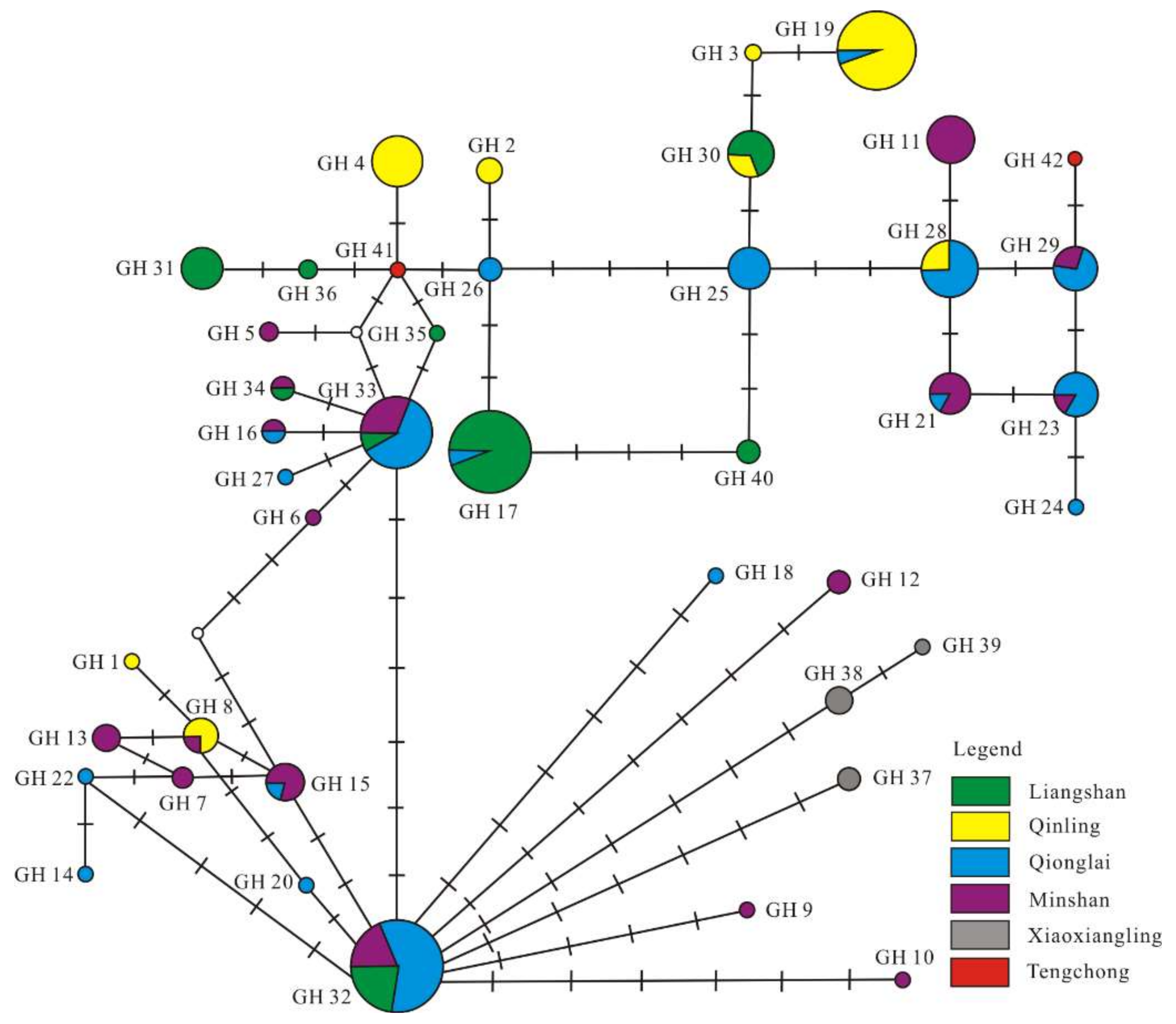
3.3. Phylogenetic and Network Analysis
3.4. Demographic Analysis
4. Discussion
4.1. Genetic Diversity and Demographic History of Giant Pandas
4.2. Impacts of Human Activities and Climate Change
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, J. On the taxonomic status of species, geological distribution and evolutionary history of Ailuropoda. Acta Zool. Sin. 1974, 20, 191–201. [Google Scholar]
- Qiu, Z.; Qi, G. Ailuropod found from the late miocene deposits in Lufeng, Yunnan. Vertebr. Palasiat. 1989, 27, 153–169. [Google Scholar]
- Pei, W. A brief history of the development of giant panda. Acta Zool. Sin. 1974, 20, 188–190. [Google Scholar]
- Jin, C.; Ciochon, R.L.; Dong, W.; Hunt, R.M., Jr.; Liu, J.; Jaeger, M.; Zhu, Q. The first skull of the earliest giant panda. Proc. Natl. Acad. Sci. USA 2007, 104, 10932–10937. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Long, Z. The vicissitudes of the giant panda. Acta Zool. Sin. 1983, 29, 93–104. [Google Scholar]
- Wen, H.; He, Y. The distribution of giant panda in Henan, Hubei, Hunan and Sichuan in last five thousands years. J. Southwest Teach. Coll. 1981, 1, 87–93. [Google Scholar]
- Li, T.; Lai, X.; Wang, W.; Zhou, X. Taxonomy and evolution of giant panda. Geol. Sci. Technol. Inf. 2004, 23, 40–46. [Google Scholar]
- Hu, J.C. Research on the Giant Panda; Shanghai Science and Technology Publishing House: Shanghai, China, 2001. [Google Scholar]
- Guo, J. Wildlife conservation. Giant panda numbers are surging—Or are they? Science 2007, 316, 974–975. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Johnson, W.E.; Menotti-Raymond, M.; Yuhki, N.; Martenson, J.S.; Mainka, S.; Shi-Qiang, H.; Zhihe, Z.; Li, G.; Pan, W.; et al. Patterns of genetic diversity in remaining giant panda populations. Conserv. Biol. 2001, 15, 1596–1607. [Google Scholar] [CrossRef]
- Zhan, X.; Li, M.; Zhang, Z.; Goossens, B.; Chen, Y.; Wang, H.; Bruford, M.W.; Wei, F. Molecular censusing doubles giant panda population estimate in a key nature reserve. Curr. Biol. 2006, 16, R451–R452. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.J.; Zhang, Z.J.; Wu, H.; Goossens, B.; Li, M.; Jiang, S.W.; Bruford, M.W.; Wei, F.W. Molecular analysis of dispersal in giant pandas. Mol. Ecol. 2007, 16, 3792–3800. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, M.; Zhang, Z.; Goossens, B.; Zhu, L.; Zhang, S.; Hu, J.; Bruford, M.W.; Wei, F. Genetic viability and population history of the giant panda, putting an end to the “evolutionary dead end”? Mol. Biol. Evolut. 2007, 24, 1801–1810. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Fan, W.; Tian, G.; Zhu, H.; He, L.; Cai, J.; Huang, Q.; Cai, Q.; Li, B.; Bai, Y.; et al. The sequence and de novo assembly of the giant panda genome. Nature 2010, 463, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wu, Q.; Ma, S.; Ma, T.; Shan, L.; Wang, X.; Nie, Y.; Ning, Z.; Yan, L.; Xiu, Y.; et al. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc. Natl. Acad. Sci. USA 2017, 114, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wei, W.; Nie, Y.; Zhou, W.; Hu, Y.; Wu, Q.; Wei, F. Distinctive diet-tissue isotopic discrimination factors derived from the exclusive bamboo-eating giant panda. Integr. Zool. 2016, 11, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Sgro, C.M. Climate change and evolutionary adaptation. Nature 2011, 470, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Koch, P.L.; Barnosky, A.D. Late quaternary extinctions: State of the debate. Ann. Rev. Ecol. Evolut. Syst. 2006, 37, 215–250. [Google Scholar] [CrossRef]
- Stuart, A.J.; Kosintsev, P.A.; Higham, T.F.; Lister, A.M. Pleistocene to holocene extinction dynamics in giant deer and woolly mammoth. Nature 2004, 431, 684–689. [Google Scholar] [CrossRef] [PubMed]
- Alroy, J. A multispecies overkill simulation of the end-pleistocene megafaunal mass extinction. Science 2001, 292, 1893–1896. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.L.; Turney, C.; Barnett, R.; Martin, F.; Bray, S.C.; Vilstrup, J.T.; Orlando, L.; Salas-Gismondi, R.; Loponte, D.; Medina, M.; et al. Synergistic roles of climate warming and human occupation in patagonian megafaunal extinctions during the last deglaciation. Sci. Adv. 2016, 2, e1501682. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.N.; Alroy, J.; Beeton, N.J.; Bird, M.I.; Brook, B.W.; Cooper, A.; Gillespie, R.; Herrando-Perez, S.; Jacobs, Z.; Miller, G.H.; et al. What caused extinction of the pleistocene megafauna of sahul? Proc. Biol. Sci. Royal. Soc. 2016, 283, 20152399. [Google Scholar] [CrossRef] [PubMed]
- Swaisgood, R.; Wang, D.; Wei, F. Ailuropoda melanoleuca, giant panda. In IUCN Red List of Threatened Species; International Union for Conservation of Nature and Natural Resources: Gland, Switzerland, 2016. [Google Scholar]
- Xu, W.; Vina, A.; Kong, L.; Pimm, S.L.; Zhang, J.; Yang, W.; Xiao, Y.; Zhang, L.; Chen, X.; Liu, J.; et al. Reassessing the conservation status of the giant panda using remote sensing. Nat. Ecol. Evol. 2017, 1, 1635. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; McShea, W.J.; Wang, D.; Li, S.; Zhao, Q.; Wang, H.; Lu, Z. Evaluating landscape options for corridor restoration between giant panda reserves. PLoS ONE 2014, 9, e105086. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zheng, P.; Dong, S.; Zhan, X.; Wu, Q.; Guo, X.; Hu, Y.; He, W.; Zhang, S.; Fan, W.; et al. Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat. Genet. 2013, 45, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Shi, L.; He, G.; Zhang, A.; Song, Y.; Zhong, S.; Fei, L. Genetic diversity in the giant panda: Evidence from protein electrophoresis. Chin. Sci. Bull. 1994, 39, 742–745. [Google Scholar]
- Zhang, Y.; Ryder, O.A.; Fan, Z.; Zhang, H.; He, T.; He, G.; Zhang, A.; Fei, L.; Zhong, S.; Chen, H.; et al. Sequence variation and genetic diversity in the giant panda. Sci. China Ser. C Life Sci. 1997, 40, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Feng, W.; Zhang, A.; Li, S.; Yu, J.; Huang, X.; He, G.; Fei, L. The comparative analysis on the genetic diversity of giant pandas between Liangshan and Xiaoxiangling Mountains. Acta Theriol. Sin. 1997, 17, 248–252. [Google Scholar]
- Hu, Y.; Qi, D.; Wang, H.; Wei, F. Genetic evidence of recent population contraction in the southernmost population of giant pandas. Genetica 2010, 138, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Hu, Y.; Yan, L.; Nie, Y.; Wu, Q.; Zhang, Z. Giant pandas are not an evolutionary cul-de-sac: Evidence from multidisciplinary research. Mol. Biol. Evolut. 2015, 32, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Basler, N.; Xenikoudakis, G.; Westbury, M.V.; Song, L.; Sheng, G.; Barlow, A. Reduction of the contaminant fraction of DNA obtained from an ancient giant panda bone. BMC Res. Notes 2017, 10, 754. [Google Scholar] [CrossRef] [PubMed]
- Hofreiter, M. Long DNA sequences and large data sets: Investigating the quaternary via ancient DNA. Quat. Sci. Rev. 2008, 27, 2586–2592. [Google Scholar] [CrossRef]
- Ji, X.; Jablonski, N.G.; Liu, H.; Lawrence, J.F.; Li, Z.; Li, Z.; Liu, J. The discovery of the holocene giant panda skeletons from a sinkhole of Jiangdongshan, Tengchong, Yunnan province and its significances. In Proceedings of the Eleventh Annual Meeting of the Chinese Society of Vertebrate Paleontology; Dong, W., Ed.; China Ocean Press: Beijing, China, 2008; pp. 119–125. [Google Scholar]
- Jin, J.J.H.; Jablonski, N.G.; Flynn, L.J.; Chaplin, G.; Xueping, J.; Zhicai, L.; Xiaoxue, S.; Guihua, L. Micromammals from an early holocene archaeological site in Southwest China: Paleoenvironmental and taphonomic perspectives. Quat. Int. 2012, 281, 58–65. [Google Scholar] [CrossRef]
- Jablonski, N.G.; Ji, X.; Chaplin, G.; Wang, L.; Yang, S.; Li, G.; Li, Z. A preliminary report on new and previously known vertebrate paleontological sites in baoshan prefecture, Yunnan province, China. Proc. Calif. Acad. Sci. 2003, 54, 209–224. [Google Scholar]
- Jablonski, N.G.; Ji, X.; Liu, H.; Li, Z.; Flynn, L.J.; Li, Z. Remains of holocene giant pandas from jiangdong mountain (Yunnan, China) and their relevance to the evolution of quaternary environments in south-Western China. Hist. Biol. 2012, 24, 527–536. [Google Scholar] [CrossRef]
- Rohland, N.; Hofreiter, M. Ancient DNA extraction from bones and teeth. Nat. Protoc. 2007, 2, 1756–1762. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Zeng, B.; Meng, X.; Yue, B.; Zhang, Z.; Zou, F. The complete mitochondrial genome and phylogenetic analysis of the giant panda (Ailuropoda melanoleuca). Gene 2007, 397, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Römpler, H.; Dear, P.H.; Krause, J.; Meyer, M.; Rohland, N.; Schoneberg, T.; Spriggs, H.; Stiller, M.; Hofreiter, M. Multiplex amplification of ancient DNA. Nat. Protoc. 2006, 1, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Hofreiter, M.; Jaenicke, V.; Serre, D.; von Haeseler, A.; Paabo, S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001, 29, 4793–4799. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. Flash: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing Subgroup. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, J.; Yu, J.; Gibbs, R.A.; Yu, F. An integrative variant analysis pipeline for accurate genotype/haplotype inference in population NGS data. Genome Res. 2013, 23, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Korneliussen, T.S.; Albrechtsen, A.; Nielsen, R. Angsd: Analysis of next generation sequencing data. BMC Bioinform. 2014, 15, 356. [Google Scholar] [CrossRef] [PubMed]
- Arnason, U.; Gullberg, A.; Janke, A.; Kullberg, M. Mitogenomic analyses of caniform relationships. Mol. Phylogenet. Evolut. 2007, 45, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Krause, J.; Unger, T.; Nocon, A.; Malaspinas, A.S.; Kolokotronis, S.O.; Stiller, M.; Soibelzon, L.; Spriggs, H.; Dear, P.H.; Briggs, A.W.; et al. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the miocene-pliocene boundary. BMC Evolut. Biol. 2008, 8, 220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Gilbson, T.J.; Plewnlak, F.; Jeanmougin, F.; Higgins, D.G. The clustal_x windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evolut. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Fortes, G.G.; Grandal-d’Anglade, A.; Kolbe, B.; Fernandes, D.; Meleg, I.N.; Garcia-Vazquez, A.; Pinto-Llona, A.C.; Constantin, S.; de Torres, T.J.; Ortiz, J.E.; et al. Ancient DNA reveals differences in behaviour and sociality between brown bears and extinct cave bears. Mol. Ecol. 2016, 25, 4907–4918. [Google Scholar] [CrossRef] [PubMed]
- Stiller, M.; Baryshnikov, G.; Bocherens, H.; Grandal d’Anglade, A.; Hilpert, B.; Munzel, S.C.; Pinhasi, R.; Rabeder, G.; Rosendahl, W.; Trinkaus, E.; et al. Withering away—25,000 years of genetic decline preceded cave bear extinction. Mol. Biol. Evolut. 2010, 27, 975–978. [Google Scholar] [CrossRef] [PubMed]
- Cahill, J.A.; Stirling, I.; Kistler, L.; Salamzade, R.; Ersmark, E.; Fulton, T.L.; Stiller, M.; Green, R.E.; Shapiro, B. Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears. Mol. Ecol. 2015, 24, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the raxml web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the cipres science gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evolut. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.-J.; Forster, P.; Rohl, A. Median-joining network for inferring intraspecific phylogenies. Mol. Biol. Evolut. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evolut. 2005, 22, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with beauti and the beast 1.7. Mol. Biol. Evolut. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Lammers, F.; Bidon, T.; Pfenninger, M.; Kolter, L.; Nilsson, M.; Janke, A. The evolutionary history of bears is characterized by gene flow across species. Sci. Rep. 2017, 7, 46487. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer v1.6. Available online: http://tree.bio.ed.ac.uk/software/tracer/2014 (accessed on 11 December 2013).
- Balint, M.; Malnas, K.; Nowak, C.; Geismar, J.; Vancsa, E.; Polyak, L.; Lengyel, S.; Haase, P. Species history masks the effects of human-induced range loss—Unexpected genetic diversity in the endangered giant mayfly Palingenia longicauda. PLoS ONE 2012, 7, e31872. [Google Scholar] [CrossRef] [PubMed]
- Saunders, I.W.; Tavaré, S.; Watterson, G.A. On the genealogy of nested subsamples from a haploid population. Adv. Appl. Probab. 1984, 16, 471–491. [Google Scholar] [CrossRef]
- Barnes, I.; Matheus, P.; Shapiro, B.; Jensen, D.; Cooper, A. Dynamics of Pleistocene population extinction in Beringian brown bears. Science 2002, 295, 2267–2270. [Google Scholar] [CrossRef] [PubMed]
- Hofreiter, M.; Muenzel, S.; Conard, N.J.; Pollack, J.; Slatkin, M.; Weiss, G.; Paabo, S. Sudden replacement of cave bear mitochondrial DNA in the late Pleistocene. Curr. Biol. 2007, 17, 122–123. [Google Scholar] [CrossRef] [PubMed]
- Münzel, S.C.; Stiller, M.; Hofreiter, M.; Mittnik, A.; Conard, N.; Bocherens, H. Pleistocene bears in the Swabian Jura (Germany): Genetic replacement, ecological displacement, extinctions and survival. Quant. Int. 2011, 245, 225–237. [Google Scholar] [CrossRef]
- Leonard, J.; Vila, C.; Fox-Dobbs, K.; Koch, P.L.; Wayne, R.K.; Valkenburgh, B.V. Megafaunal extinctions and the disappearance of a specialized wolf ecomorph. Curr. Biol. 2007, 17, 1146–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilot, M.; Branicki, W.; Jedrzejewski, W.; Goszczynski, J.; Jedrzejewska, B.; Dykyy, I.; Shkvyrya, M.; Tsingarska, E. Phylogeographic history of grey wolves in Europe. BMC Evolut. Biol. 2010, 10, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdiosera, C.E.; Garcia, N.; Anderung, C.; Dalen, L.; Cregut-Bonnoure, E.; Kahlke, R.D.; Stiller, M.; Brandstrom, M.; Thomas, M.G.; Arsuaga, J.L.; et al. Staying out in the cold: Glacial refugia and mitochondrial DNA phylogeography in ancient European brown bears. Mol. Ecol. 2007, 16, 5140–5148. [Google Scholar] [CrossRef] [PubMed]
- Valdiosera, C.E.; Garcia-Garitagoitia, J.L.; Garcia, N.; Doadrio, I.; Thomas, M.G.; Hanni, C.; Arsuaga, J.L.; Barnes, I.; Hofreiter, M.; Orlando, L.; et al. Surprising migration and population size dynamics in ancient Iberian brown bears (Ursus arctos). Proc. Natl. Acad. Sci. USA 2008, 105, 5123–5128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Tang, Z.; Qi, D.; Hu, J. Habitat assessment for giant pandas (Ailuropoda melanoleuca) on the northern slope of the Daxiangling Mountains. Acta Theriol. Sin. 2007, 27, 146–152. [Google Scholar]
- Li, X.; Jiang, G.; Tian, H.; Xu, L.; Yan, C.; Wang, Z.; Wei, F.; Zhang, Z. Human impact and climate cooling caused range contraction of large mammals in China over the past two millennia. Ecography 2015, 38, 74–82. [Google Scholar] [CrossRef]
- Turvey, S.T.; Tong, H.; Stuart, A.J.; Lister, A.M. Holocene survival of Late pleistocene megafauna in China: A critical review of the evidence. Quat. Sci. Rev. 2013, 76, 156–166. [Google Scholar] [CrossRef]
- Sun, Y.; An, Z. Late pliocene-pleistocene changes in mass accumulation rates of eolian deposits on the central Chinese Loess plateau. J. Geophys. Res. 2005, 110, d23. [Google Scholar] [CrossRef]
- Cooper, A.; Turney, c.; Hughen, K.A.; Brook, B.W.; Mcdonald, H.G.; Bradshaw, C.J. paleoecology. abrupt warming events drove late Pleistocene holarctic megafaunal turnover. Science 2015, 349, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Rabanus-Wallace, M.T.; Wooller, M.J.; Zazula, G.D.; Shute, E.; Jahren, A.H.; Kosintsev, P.; Burns, J.A.; Breen, J.; Llamas, B.; Cooper, A. Megafaunal isotopes reveal role of increased moisture on rangeland during late Pleistocene extinctions. Nat. Ecol. Evol. 2017, 1, 125. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Tan, L.; Cheng, H.; An, Z.; Edwards, R.L.; Kelly, M.J.; Kong, X.; Wang, X. The variation of summer monsoon precipitation in Central China since the last deglaciation. Earth Planet. Sci. Lett. 2010, 291, 21–31. [Google Scholar] [CrossRef]
- Ren, G.; Beug, H.-J. Mapping holocene pollen data and vegetation of China. Quat.Sci. Rev. 2002, 21, 1395–1422. [Google Scholar] [CrossRef]
- Hong, Y.T.; Hong, B.; Lin, Q.H.; Zhu, Y.X.; Shibata, Y.; Hirota, M.; Uchida, M.; Leng, X.T.; Jiang, H.B.; Xu, H.; et al. Correlation between Indian Ocean summer monsoon and North Atlantic climate during the Holocene. Earth Planet. Sci. Lett. 2003, 211, 371–380. [Google Scholar] [CrossRef]
- Yao, T.; Thompson, L.G. Trend and features of climatic changes in the past 5000 years recorded by the Dunde icecore. Ann. Glaciol. 1992, 16, 21–24. [Google Scholar]
- Needham, J. Science and Civilisation in China: Civil Engineering and Nautics; Cambridge University Press: Cambridge, UK, 1971; Volume 4. [Google Scholar]
- Ren, G. Decline of the mid- to late holocene forests in china: Climatic change or human impact? J. Quat. Sci. 2000, 15, 273–281. [Google Scholar] [CrossRef]
- Zhu, S. Historic forest changes of the Liangshan Mountains in Sichuan Province. J. Chin. Hist. Geogr. 2007, 22, 43–52. [Google Scholar]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, G.-L.; Barlow, A.; Cooper, A.; Hou, X.-D.; Ji, X.-P.; Jablonski, N.G.; Zhong, B.-J.; Liu, H.; Flynn, L.J.; Yuan, J.-X.; et al. Ancient DNA from Giant Panda (Ailuropoda melanoleuca) of South-Western China Reveals Genetic Diversity Loss during the Holocene. Genes 2018, 9, 198. https://doi.org/10.3390/genes9040198
Sheng G-L, Barlow A, Cooper A, Hou X-D, Ji X-P, Jablonski NG, Zhong B-J, Liu H, Flynn LJ, Yuan J-X, et al. Ancient DNA from Giant Panda (Ailuropoda melanoleuca) of South-Western China Reveals Genetic Diversity Loss during the Holocene. Genes. 2018; 9(4):198. https://doi.org/10.3390/genes9040198
Chicago/Turabian StyleSheng, Gui-Lian, Axel Barlow, Alan Cooper, Xin-Dong Hou, Xue-Ping Ji, Nina G. Jablonski, Bo-Jian Zhong, Hong Liu, Lawrence J. Flynn, Jun-Xia Yuan, and et al. 2018. "Ancient DNA from Giant Panda (Ailuropoda melanoleuca) of South-Western China Reveals Genetic Diversity Loss during the Holocene" Genes 9, no. 4: 198. https://doi.org/10.3390/genes9040198




