Phenotypic and Genotypic Analysis of Antimicrobial Resistance among Listeria monocytogenes Isolated from Australian Food Production Chains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Serotyping of PCR Serogrouping of Isolates
2.3. Pulsed-Field Gel Electrophoresis Subtyping
2.4. Antibiotic Susceptibility Testing
2.5. Whole Genome Sequencing and Multilocus Sequence Typing
2.6. Phylogenetic Analysis of Clonal Complex 1 Isolates
3. Results
3.1. Serotyping
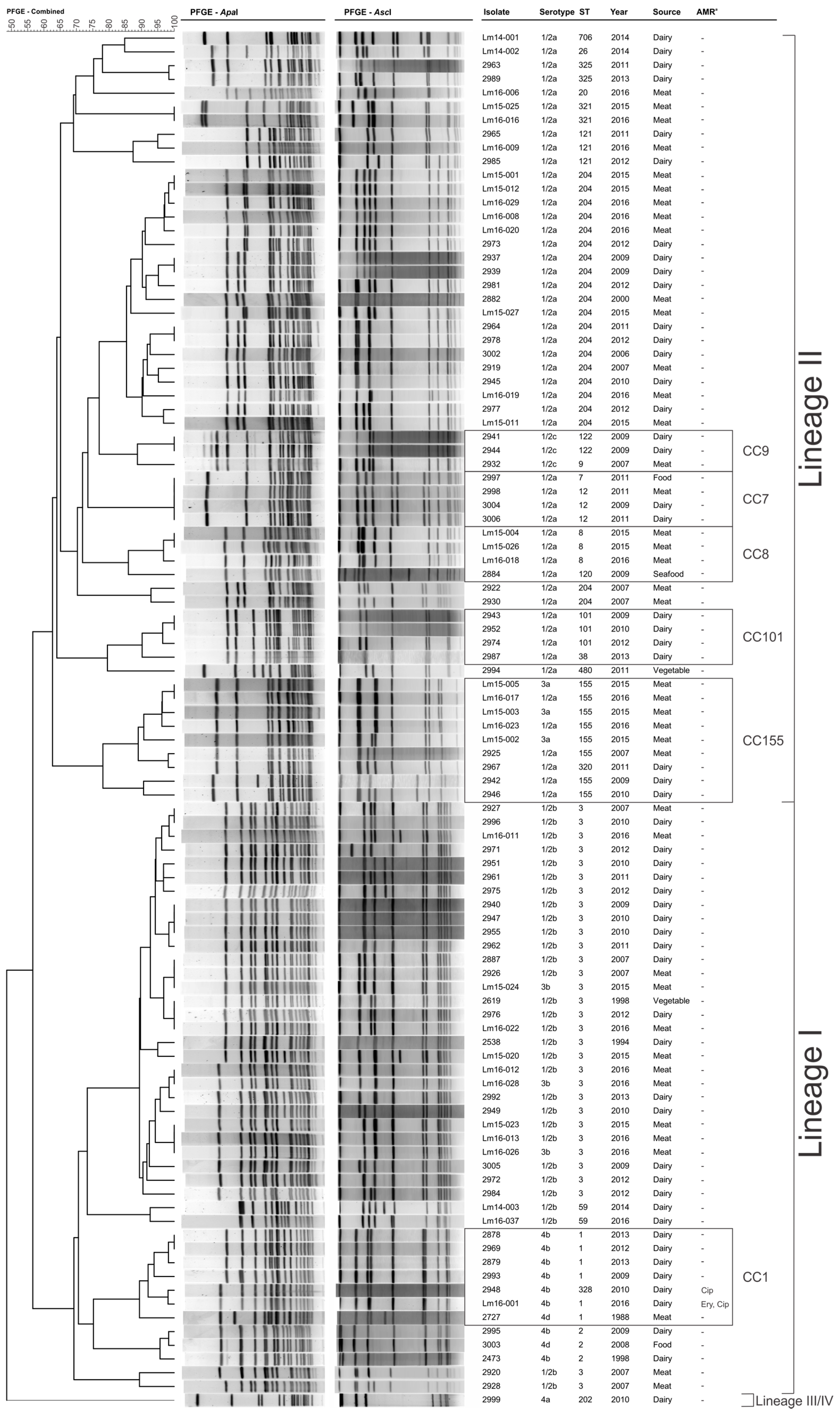
3.2. Pulsed-Field Gel Electrophoresis Subtyping
3.3. Antimicrobial Resistance Characteristics
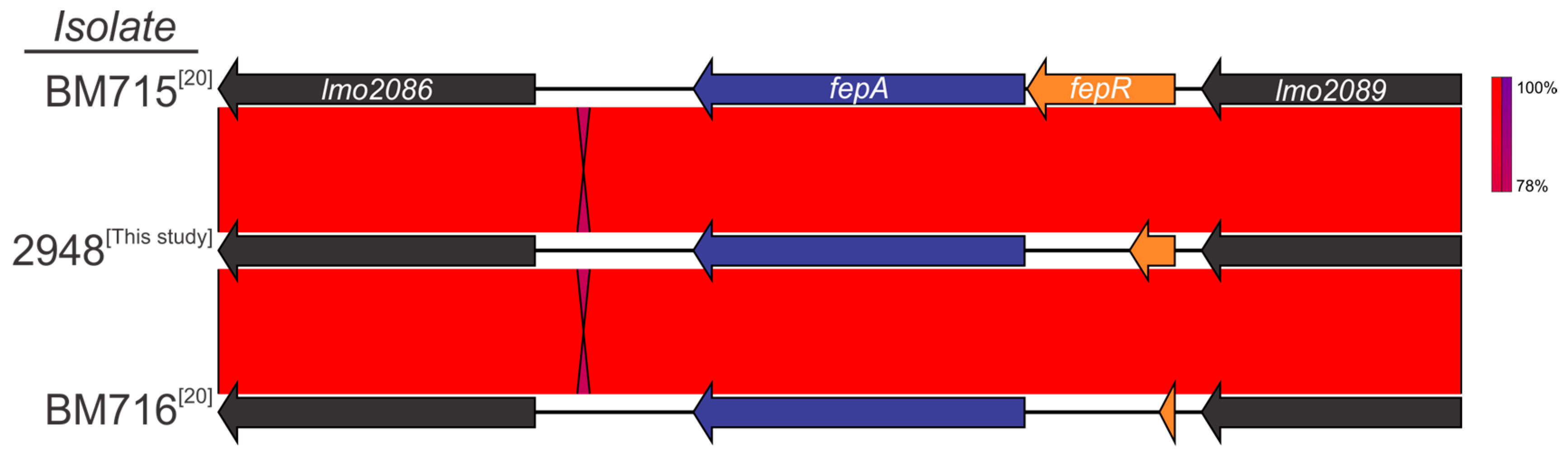
3.4. Detection of Genetic Markers Related to Antimicrobial Resistance
3.5. Clonal Complex 1 Population Structure Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fox, E.M.; Leonard, N.; Jordan, K. Molecular Diversity of Listeria Monocytogenes Isolated from Irish Dairy Farms. Foodborne Pathog. Dis. 2011, 8, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Lappi, V.R.; Thimothe, J.; Nightingale, K.K.; Gall, K.; Scott, V.N.; Wiedmann, M. Longitudinal studies on Listeria in Smoked Fish Plants: Impact of intervention strategies on contamination patterns. J. Food Prot. 2004, 67, 2500–2514. [Google Scholar] [CrossRef] [PubMed]
- Aureli, P.; Ferrini, A.M.; Mannoni, V.; Hodzic, S.; Wedell-Weergaard, C.; Oliva, B. Susceptibility of Listeria Monocytogenes Isolated from Food in Italy to Antibiotics. Int. J. Food Microbiol. 2003, 83, 325–330. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Domı́nguez-Bernal, G.; Goebel, W.; González-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef] [PubMed]
- Drevets, D.A.; Bronze, M.S. Listeria Monocytogenes: Epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol. Med. Microbiol. 2008, 53, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Schuchat, A.; Swaminathan, B.; Broome, C.V. Epidemiology of Human Listeriosis. Clin. Microbiol. Rev. 1991, 4, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Charlier, C.; Perrodeau, E.; Leclercq, A.; Cazenave, B.; Pilmis, B.; Henry, B.; Lopes, A.; Maury, M.; Moura, A.; Net, F.G.; et al. Clinical Features and Prognostic Factors of Listeriosis: The Monalisa national prospective cohort study. Lancet Infect. Dis. 2017, 17, 510–519. [Google Scholar] [CrossRef]
- Emmanuelle, C.; Courvalin, P. Antibiotic Resistance in Listeria Spp. Antimicrob. Agents Chemother. 1999, 43, 2103–2108. [Google Scholar]
- Spitzer, P.G.; Hammer, S.M.; Karchmer, A.W. Treatment of Listeria Monocytogenes Infection with Trimethoprim-Sulfamethoxazole: Case Report and Review of the Literature. Rev. Infect. Dis. 1986, 8, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, L.V.; White, R.L.; Reboli, A.C. Pharmacodynamics of Trimethoprim-Sulfamethoxazole in Listeria Meningitis: A case report. Pharmacother. J. Hum. Pharmacol. Drug Ther. 1990, 10, 301–304. [Google Scholar]
- Troxler, R.; von Graevenitz, A.; Funke, G.; Wiedemann, B.; Stock, I. Natural Antibiotic Susceptibility of Listeria Species: L. Grayi, L. Innocua, L. Ivanovii, L. Monocytogenes, L. Seeligeri and L. Welshimeri Strains. Clin. Microbiol. Infect. 2000, 6, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk-Balska, A.; Markiewicz, Z. The intrinsic Cephalosporin resistome of Listeria Monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics. J. Appl. Microbiol. 2016, 120, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Hof, H. Listeria Infections of the Eye. Eur. J. Ophthalmol. 2017, 27, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Qian, W.; Zhang, X.; Wang, H.; Ye, K.; Bai, Y.; Zhou, G. Prevalence, genetic diversity and antimicrobial resistance of Listeria Monocytogenes isolated from Ready-to-Eat meat products in Nanjing, China. Food Control 2015, 50, 202–208. [Google Scholar] [CrossRef]
- Yan, H.; Neogi, S.B.; Mo, Z.; Guan, W.; Shen, Z.; Zhang, S.; Li, L.; Yamasaki, S.; Shi, L.; Zhong, N. Prevalence and characterization of antimicrobial resistance of foodborne Listeria Monocytogenes isolates in Hebei Province of Northern China, 2005–2007. Int. J. Food Microbiol. 2010, 144, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Fallah, A.A.; Saei-Dehkordi, S.S.; Mahzounieh, M. Occurrence and antibiotic resistance profiles of Listeria Monocytogenes isolated from seafood products and market and processing environments in Iran. Food Control 2013, 34, 630–636. [Google Scholar] [CrossRef]
- Sugiri, Y.D.; Golz, G.; Meeyam, T.; Baumann, M.P.; Kleer, J.; Chaisowwong, W.; Alter, T. Prevalence and antimicrobial susceptibility of Listeria Monocytogenes on Chicken Carcasses in Bandung, Indonesia. J. Food Prot. 2014, 77, 1407–1410. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Who Global Strategy for Containment of Antimicrobial Resistance; WHO: Geneva, Switzerland, 2001; pp. 1–100. [Google Scholar]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Guerin, F.; Galimand, M.; Tuambilangana, F.; Courvalin, P.; Cattoir, V. Overexpression of the Novel MATE Fluoroquinolone Efflux Pump FepA in Listeria Monocytogenes is driven by inactivation of its local repressor FepR. PLoS ONE 2014, 9, e106340. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. Mechanisms of resistance to Quinolones. Clin. Infect. Dis. 2005, 41, S120–S126. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.Z.; Paixão, R.; Gobbi, D.D.S.; Raimundo, D.C.; Ferreira, T.P.; Moreno, A.M.; Hofer, E.; Reis, C.M.F.; Matté, G.R.; Matté, M.H. Characterization of Antibiotic Resistance in Listeria Spp. Isolated from Slaughterhouse Environments, Pork and Human Infections. J. Infect. Dev. Ctries. 2014, 8, 416–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lampidis, R.; Kostrewa, D.; Hof, H. Molecular Characterization of the Genes Encoding DNA Gyrase and Topoisomerase iv of Listeria Monocytogenes. J. Antimicrob. Chemother. 2002, 49, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Facinelli, B.; Giovanetti, E.; Varaldo, P.E. Transferable Erythromycin Resistance in Listeria Spp. Isolated from Food. Appl. Environ. Microbiol. 1996, 62, 269–270. [Google Scholar] [PubMed]
- Flamm, R.K.; Hinrichs, D.J.; Thomashow, M.F. Introduction of Pam Beta 1 into Listeria Monocytogenes by Conjugation and Homology between Native L. Monocytogenes Plasmids. Infect. Immun. 1984, 44, 157–161. [Google Scholar] [PubMed]
- Charpentier, E.; Courvalin, P. Emergence of the Trimethoprim resistance Gene dfrD in Listeria Monocytogenes Bm4293. Antimicrob. Agents Chemother. 1997, 41, 1134–1136. [Google Scholar] [PubMed]
- Granier, S.A.; Moubareck, C.; Colaneri, C.; Lemire, A.; Roussel, S.; Dao, T.; Courvalin, P.; Brisabois, A. Antimicrobial Resistance of Listeria Monocytogenes Isolates from Food and the Environment in France over a 10-Year Period. Appl. Environ. Microbiol. 2011, 77, 2788–2790. [Google Scholar] [CrossRef] [PubMed]
- Iain, B.M.; Sørensen, S.J.; Hansen, L.H.; Licht, T.R. Effect of Tetracycline on Transfer and Establishment of the Tetracycline-Inducible Conjugative Transposon Tn916 in the Guts of Gnotobiotic Rats. Appl. Environ. Microbiol. 2004, 70, 758–764. [Google Scholar]
- Bertsch, D.; Muelli, M.; Weller, M.; Uruty, A.; Lacroix, C.; Meile, L. Antimicrobial susceptibility and Antibiotic Resistance Gene Transfer Analysis of Foodborne, Clinical, and Environmental Listeria Spp. isolates including Listeria Monocytogenes. MicrobiologyOpen 2014, 3, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Poyart-Salmeron, C.; Trieu-Cuot, P.; Carlier, C.; MacGowan, A.; McLauchlin, J.; Courvalin, P. Genetic Basis of Tetracycline Resistance in Clinical Isolates of Listeria Monocytogenes. Antimicrob. Agents Chemother. 1992, 36, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.; Duffy, G.; Sheridan, J.J.; Blair, I.S.; McDowell, D.A. Antibiotic Resistance among Listeria, Including Listeria Monocytogenes, in Retail Foods. J. Appl. Microbiol. 2011, 90, 517–522. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; le Monnier, A.; Brisse, S. A new perspective on Listeria Monocytogenes Evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Buchrieser, C.; Glasser, P.; Jacquet, C.; Martin, P. Differentiation of the Major Listeria Monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [PubMed]
- USA PulseNet. Standard Operating Procedure for Pulsenet PFGE of Listeria Monocytogenes. Available online: http://www.pulsenetinternational.org/assets/PulseNet/uploads/pfge/PNL04_ListeriaPFGEProtocol.pdf (accessed on 7 February 2018).
- Almudena, H.M.; Payeras-Cifre, A. What Is New in Listeriosis? BioMed. Res. Int. 2014, 2014, 358051. [Google Scholar] [CrossRef]
- Grumbach, N.M.; Mylonakis, E.; Wing, E.J. Development of Listerial Meningitis during Ciprofloxacin Treatment. Clin. Infect. Dis. 1999, 29, 1340–1341. [Google Scholar] [CrossRef] [PubMed]
- Clinical & Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI Document M100-S24; CLSI: Wayne, PA, USA, 2014. [Google Scholar]
- European Committee of Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of Mics and Zone Diameters Version 5.0.; EUCAST: Växjö, Sweden, 2015. [Google Scholar]
- Fox, E.M.; Allnutt, T.; Bradbury, M.I.; Fanning, S.; Chandry, P.S. Comparative Genomics of the Listeria Monocytogenes St204 Subgroup. Front. Microbiol. 2016, 7, 2057. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Jennison, A.V.; Masson, J.J.; Fang, N.; Graham, R.M.; Bradbury, M.I.; Fegan, N.; Gobius, K.S.; Graham, T.M.; Guglielmino, C.J.; Brown, J.L.; et al. Analysis of the Listeria Monocytogenes Population Structure among Isolates from 1931 to 2015 in Australia. Front. Microbiol. 2017, 8, 603. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.N.; Slezak, T.; Hall, B.G. Ksnp3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 2015, 31, 2877–2878. [Google Scholar] [CrossRef] [PubMed]
- Wiedmann, M.; Bruce, J.L.; Keating, C.; Johnson, A.E.; McDonough, P.L.; Batt, C.A. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 1997, 65, 2707–2716. [Google Scholar] [PubMed]
- Thea, K.; Cole, M.; Farber, J.M.; Eisenbrand, G.; Zabaras, D.; Fox, E.M.; Hill, J.P. Food safety for food security: Relationship between global megatrends and developments in food safety. Trends Food Sci. Technol. 2017, 68, 160–175. [Google Scholar]
- Fox, E.M.; Leonard, N.; Jordan, K. Physiological and transcriptional characterization of persistent and nonpersistent Listeria Monocytogenes Isolates. Appl. Environ. Microbiol. 2011, 77, 6559–6569. [Google Scholar] [CrossRef] [PubMed]
- Annette, F.; Langsrud, S.; Schirmer, B.C.T.; Møretrø, T.; Heir, E. Genome Analysis of Listeria Monocytogenes Sequence Type 8 Strains Persisting in Salmon and Poultry Processing Environments and Comparison with Related Strains. PLoS ONE 2016, 11, e0151117. [Google Scholar] [CrossRef] [Green Version]
- Leong, D.; NicAogáin, K.; Luque-Sastre, L.; McManamon, O.; Hunt, K.; Alvarez-Ordóñez, A.; Scollard, J.; Schmalenberger, A.; Fanning, S.; O’Byrne, C.; et al. A 3-Year Multi-Food Study of the Presence and Persistence of Listeria Monocytogenes in 54 Small Food Businesses in Ireland. Int. J. Food Microbiol. 2017, 249, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Morvan, A.; Moubareck, C.; Leclercq, A.; Hervé-Bazin, M.; Bremont, S.; Lecuit, M.; Courvalin, P.; le Monnier, A. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob. Agents Chemother. 2010, 54, 2728–2731. [Google Scholar] [CrossRef] [PubMed]
- Alicia, A.-H.; Prieto, M.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Increase over time in the prevalence of multiple antibiotic resistance among isolates of Listeria monocytogenes from poultry in Spain. Food Control 2012, 23, 37–41. [Google Scholar]
- Elaine, S.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar]
- Haubert, L.; Mendonça, M.; Lopes, G.V.; Cardoso, M.R.d.; da Silva, W.P. Listeria monocytogenes Isolates from Food and Food Environment Harbouring Tetm and Ermb Resistance Genes. Lett. Appl. Microbiol. 2016, 62, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Lyon, S.A.; Berrang, M.E.; Fedorka-Cray, P.J.; Fletcher, D.L.; Meinersmann, R.J. Antimicrobial resistance of Listeria monocytogenes isolated from a poultry further processing plant. Foodborne Pathog. Dis. 2008, 5, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Khen, B.K.; Lynch, O.A.; Carroll, J.; McDowell, D.A.; Duffy, G. Occurrence, Antibiotic resistance and molecular characterization of Listeria monocytogenes in the beef chain in the Republic of Ireland. Zoonoses Public Health 2015, 62, 11–17. [Google Scholar] [PubMed]
- Mauro, C.; Paludi, D.; Zanardi, E.; Ghidini, S.; Vergara, A.; Ianieri, A. Characterization of Antimicrobial Resistance of Foodborne Listeria Monocytogenes. Int. J. Food Microbiol. 2009, 128, 497–500. [Google Scholar]
- Davis, J.A.; Jackson, C.R. Comparative Antimicrobial Susceptibility of Listeria Monocytogenes, L. Innocua, and L. Welshimeri. Microb. Drug Resist. 2009, 15, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Hossein, J.; Paydar, M.; Ismail, S.; Looi, C.Y.; Wong, W.F.; Radmehr, B.; Abedini, A. Prevalence, Antimicrobial Susceptibility and Virulotyping of Listeria Species and Listeria Monocytogenes Isolated from Open-Air Fish Markets. BMC Microbiol. 2015, 15, 144. [Google Scholar] [CrossRef]
- Legesse, G.; Taddese, A.; Biru, T.; Nigatu, S.; Kebede, E.; Ejo, M.; Fikru, A.; Birhanu, T. Prevalence and antimicrobial susceptibility profile of Listeria Species from Ready-to-Eat foods of animal origin in Gondar Town, Ethiopia. BMC Microbiol. 2015, 15, 100. [Google Scholar] [CrossRef]
- Li, L.; Olsen, R.H.; Ye, L.; Wang, W.; Shi, L.; Yan, H.; Meng, H. Characterization of antimicrobial resistance of Listeria Monocytogenes strains isolated from a pork processing plant and its respective meat markets in Southern China. Foodborne Pathog. Dis. 2016, 13, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Gamboa-Marín, A.; Mejía-Wagner, D.C.; Moreno-Ocampo, P.A.; Buitrago, S.M.; Pérez-Pérez, K.I.; Ruiz-Bolivar, Z.; Poutou-Piñales, R.A.; Carrascal-Camacho, A.K.; Velasco-Briceño, A.; Ocampo-Guerrero, M.L. Antimicrobial susceptibility of Listeria Monocytogenes, Listeria Ivanovii, and Listeria Species isolated from swine processing facilities in Colombia. J. Swine Health Prod. 2013, 21, 10–21. [Google Scholar]
- Zeki, A.; Ardıç, M. Occurrence and antibiotic susceptibility of Listeria Species in turkey meats. Korean J. Food Sci. Anim. Resour. 2015, 35, 669–673. [Google Scholar]
- Sanjita, S.; Sharma, V.; Dahiya, D.K.; Khan, A.; Mathur, M.; Sharma, A. Prevalence, Virulence Potential, and Antibiotic Susceptibility Profile of Listeria Monocytogenes isolated from Bovine Raw milk samples obtained from Rajasthan, India. Foodborne Pathog. Dis. 2017, 14, 132–140. [Google Scholar]
- Obaidat, M.M.; Salman, A.E.B.; Lafi, S.Q.; Al-Abboodi, A.R. Characterization of Listeria Monocytogenes from three countries and antibiotic resistance differences among countries and Listeria Monocytogenes Serogroups. Lett. Appl. Microbiol. 2015, 60, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.; Seamus, F. Antimicrobial Resistance in Foodborne Pathogens—A Cause for Concern? Curr. Drug Targets 2008, 9, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Barbuddhe, S.B.; Doijad, S.P.; Goesmann, A.; Hilker, R.; Poharkar, K.V.; Rawool, D.B.; Kurkure, N.V.; Kalorey, D.R.; Malik, S.S.; Shakuntala, I.; et al. Presence of a Widely Disseminated Listeria Monocytogenes Serotype 4b Clone in India. Emerg. Microbes Infect. 2016, 5, e55. [Google Scholar] [CrossRef] [PubMed]
- Harakeh, S.; Saleh, I.; Zouhairi, O.; Baydoun, E.; Barbour, E.; Alwan, N. Antimicrobial resistance of Listeria Monocytogenes isolated from dairy-based food products. Sci. Total. Environ. 2009, 407, 4022–4027. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Tackling Antibiotic Resistance from a Food Safety Perspective in Europe; WHO: Copenhagen, Denmark, 2011; p. 88. [Google Scholar]


| Location | Collection Start Date | Sample Size | Ciprofloxacin (% Resistant) | Erythromycin (% Resistant) | Penicillin (% Resistant) | Tetracycline (% Resistant) | Study |
|---|---|---|---|---|---|---|---|
| Brazil | 1978 | 26 | 7.1 | 0 | 0 | 0 | [22] |
| France | 1989 | 4668 | 0.43 | 0.02 | NT a | 0.7 | [48] |
| Brazil | 2003 | 50 | 0 | 6 | 0 | 2 | [51] |
| China | 2005 | 90 | 17.8 | 2.22 | 1.11 | 15.6 | [15] |
| USA | 2005 | 157 | 3 | 0 | 0 | 3 | [52] |
| Switzerland | 2006 | 383 | 7 | 0 | 0 | 1.3 | [29] |
| Ireland | 2007 | 191 | 0 | 0 | 4 | 1 | [53] |
| Italy | 2008 | 120 | 1.7 | 0 | 0 | 0.8 | [54] |
| USA | 2009 | 90 | 2 | 0 | 0 | 1 | [55] |
| Ireland | 2011 | 51 | 0 | 0 | 0 | 0 | [1] |
| Iran | 2012 | 43 | NT | 14 | 16.3 | 27.9 | [56] |
| Ethiopia | 2012 | 24 | NT | NT | 66.7 | 37.5 | [57] |
| Indonesia | 2012 | 29 | 0 | 6.9 | 17.2 | 0 | [17] |
| China | 2013 | 78 | 0 | 1.3 | 1.3 | 20.5 | [58] |
| Colombia | 2013 | 259 | 7.4 | 15.8 | 0 | 6.6 | [59] |
| Turkey | 2014 | 12 | 0 | NT | 66.7 | 0 | [60] |
| India | 2014 | 5 | 0 | NT | 100 | 20 | [61] |
| Yemen | 2015 | 51 | NT | 58.8 | 100 | 56.8 | [62] |
| India | 2015 | 21 | NT | 85.7 | 100 | 90.5 | [62] |
| Egypt | 2015 | 32 | NT | 62.5 | 93.8 | 59.4 | [62] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilson, A.; Gray, J.; Chandry, P.S.; Fox, E.M. Phenotypic and Genotypic Analysis of Antimicrobial Resistance among Listeria monocytogenes Isolated from Australian Food Production Chains. Genes 2018, 9, 80. https://doi.org/10.3390/genes9020080
Wilson A, Gray J, Chandry PS, Fox EM. Phenotypic and Genotypic Analysis of Antimicrobial Resistance among Listeria monocytogenes Isolated from Australian Food Production Chains. Genes. 2018; 9(2):80. https://doi.org/10.3390/genes9020080
Chicago/Turabian StyleWilson, Annaleise, Jessica Gray, P. Scott Chandry, and Edward M. Fox. 2018. "Phenotypic and Genotypic Analysis of Antimicrobial Resistance among Listeria monocytogenes Isolated from Australian Food Production Chains" Genes 9, no. 2: 80. https://doi.org/10.3390/genes9020080




