Genetic Variation and Hybridisation among Eight Species of kōwhai (Sophora: Fabaceae) from New Zealand Revealed by Microsatellite Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Microsatellite Markers
2.3. Genetic Analyses
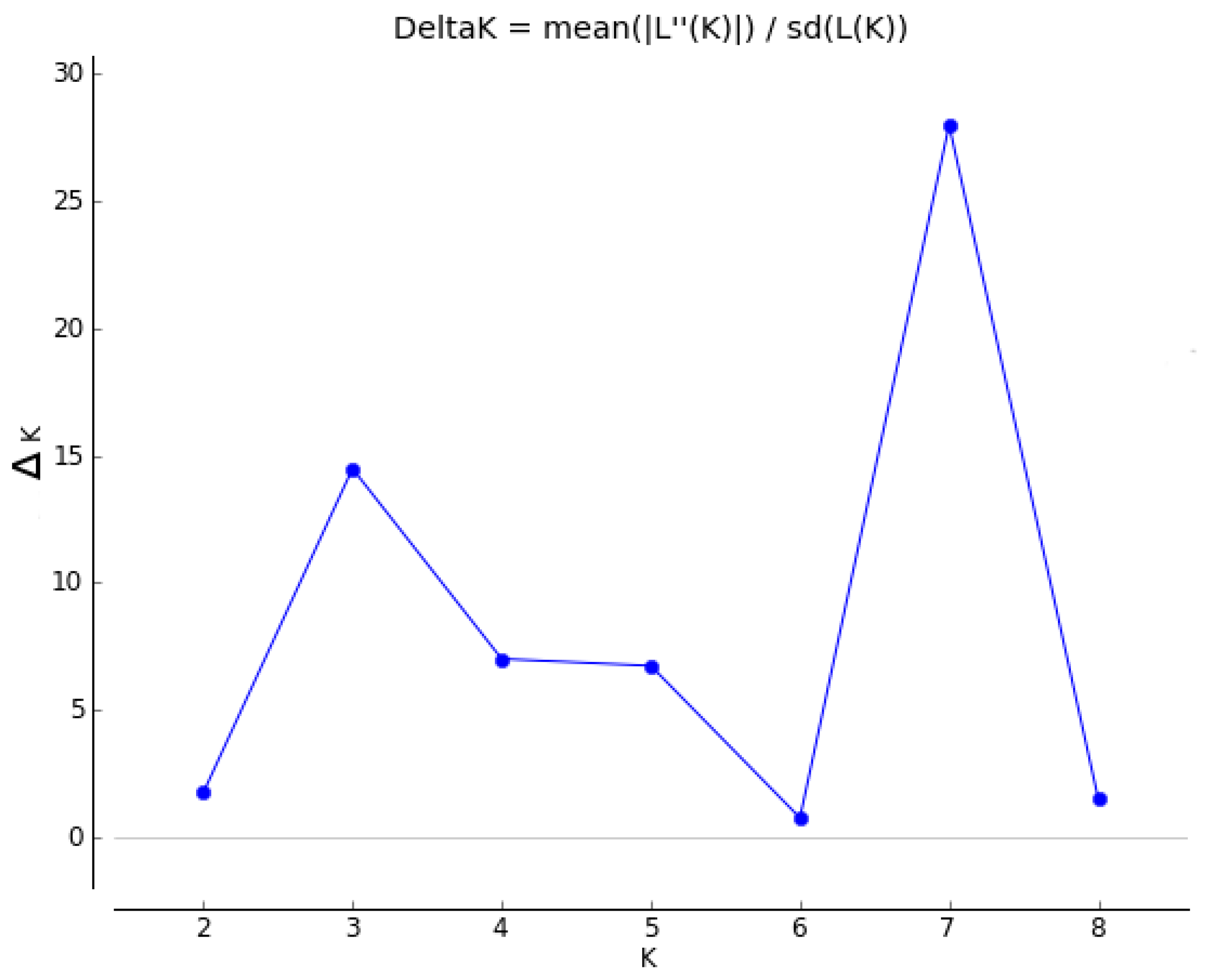
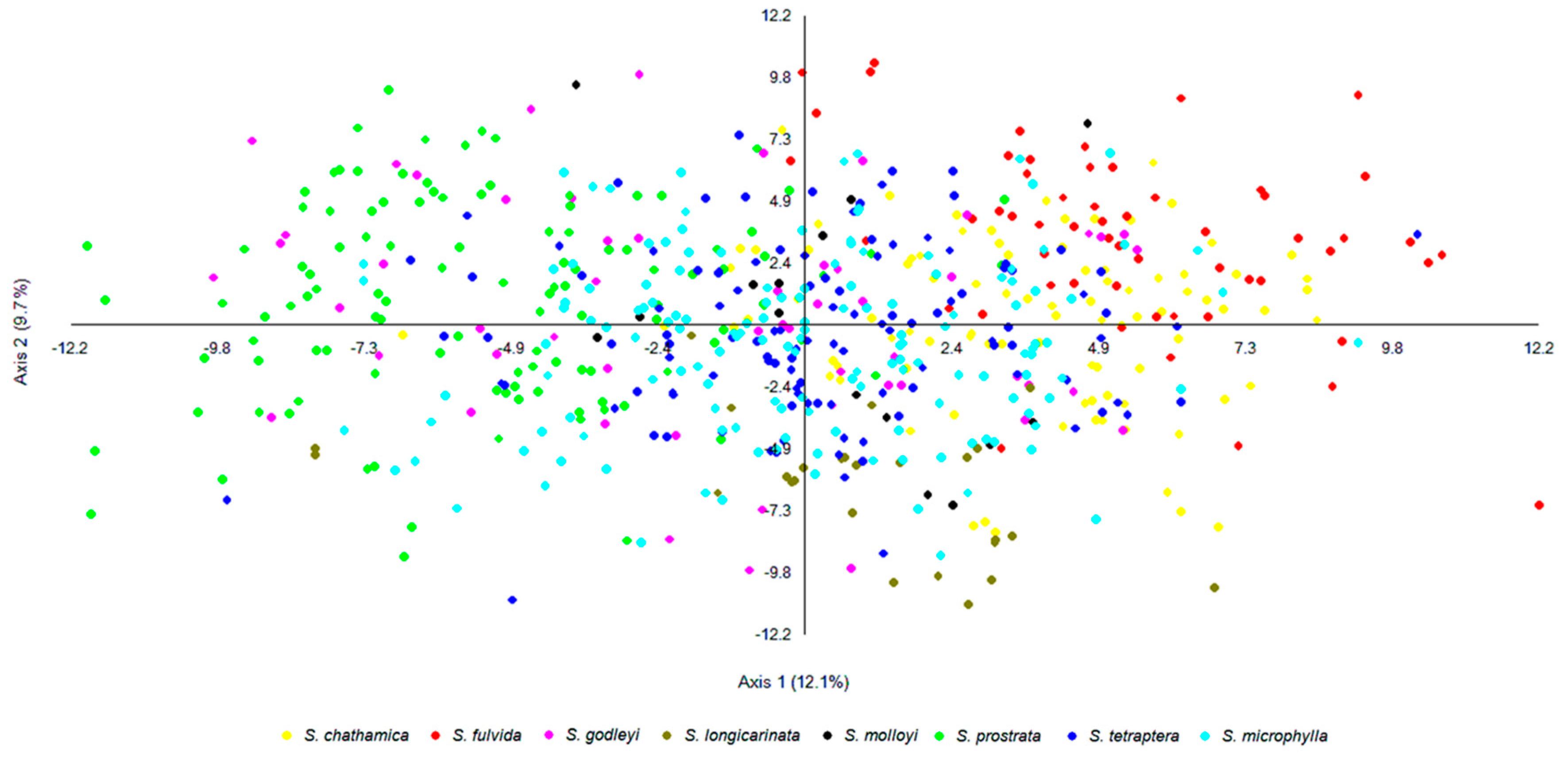
3. Results
4. Discussion
4.1. Structuring of Genetic Diversity and Taxonomic Concepts
4.2. Hybridisation
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mitchell, A.D.; Heenan, P.B. Sophora sect. Edwardsia (Fabaceae): further evidence from nrDNA sequence data of a recent and rapid radiation around the Southern Oceans. Bot. J. Linn. Soc. 2002, 140, 435–441. [Google Scholar] [CrossRef]
- Shepherd, L.D.; Heenan, P.B. Evidence for both long-distance dispersal and isolation in the Southern Oceans: molecular phylogeny of Sophora sect Edwardsia (Fabaceae). N. Z. J. Bot. 2017, 55, 334–346. [Google Scholar] [CrossRef]
- Heenan, P.B.; Dawson, M.I.; Wagstaff, S.J. The relationship of Sophora sect. Edwardsia (Fabaceae) to Sophora tomentosa, the type species of the genus Sophora, observed from DNA sequence data and morphological characters. Bot. J. Linn. Soc. 2004, 146, 439–446. [Google Scholar] [CrossRef]
- Heenan, P.B. Reinstatement of Sophora longicarinata (Fabaceae) from northern South Island, and typification of S. microphylla. N. Z. J. Bot. 1998, 36, 369–379. [Google Scholar] [CrossRef]
- Heenan, P.B. The correct name for Chilean pelu (Fabaceae): the identity of Edwardsia macnabiana and the reinstatement of Sophora cassioides. N. Z. J. Bot. 2001, 39, 167–170. [Google Scholar] [CrossRef]
- Heenan, P.B.; de Lange, P.J.; Wilton, A.D. Sophora (Fabaceae) in New Zealand: taxonomy, distribution, and biogeography. N. Z. J. Bot. 2001, 39, 17–53. [Google Scholar] [CrossRef]
- Hurr, K.A.; Lockhart, P.J.; Heenan, P.B.; Penny, D. Dispersal of the Edwardsia section of Sophora (Leguminosae) around the Southern Oceans: Molecular evidence. J. Biogeogr. 1999, 26, 565–577. [Google Scholar] [CrossRef]
- Maich, B. A biochemical genetic evaluation of taxonomy of Sophora microphylla (Fabaceae Subfamily Papilionoideae). Master’s Thesis, Victoria University, Wellington, New Zealand, 2002. [Google Scholar]
- Song, J. Genetic diversity and flowering in Clianthus and New Zealand Sophora (Fabaceae). Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 2005. [Google Scholar]
- Shepherd, L.D.; de Lange, P.J.; Perrie, L.R.; Heenan, P.B. Chloroplast phylogeography of New Zealand Sophora trees (Fabaceae): extensive hybridization and widespread Last Glacial Maximum survival. J. Biogeogr. 2017, 44, 1640–1651. [Google Scholar] [CrossRef]
- Grierson, E.R.P. The development and genetic variation of Sophora prostrata-A New Zealand divaricating shrub. Master’s Thesis, The University of Waikato, Hamilton, New Zealand, 2014. [Google Scholar]
- Shepherd, L.D.; Heenan, P.B. Origins of beach-cast Sophora seeds from the Kermadec and Chatham Islands. N. Z. J. Bot. 2017, 55, 241–248. [Google Scholar] [CrossRef]
- Van Etten, M.L.; Houliston, G.J.; Mitchell, C.M.; Heenan, P.B.; Robertson, A.W.; Tate, J.A. Sophora microphylla (Fabaceae) microsatellite markers and their utility across the genus. Appl. Plant Sci. 2014, 2, 1300081. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, M.L.; Tate, J.A.; Anderson, S.H.; Kelly, D.; Ladley, J.J.; Merrett, M.F.; Peterson, P.G.; Robertson, A.W. The compounding effects of high pollen limitation, selfing rates and inbreeding depression leave a New Zealand tree with few viable offspring. Ann. Bot. 2015, 116, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6: Genetic Analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.F.; Willis, D.P.M.; Shipley, P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Resour. 2004, 535–538. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, K.; Naama, M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across. Mol. Ecol. Resour. 2015, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Kovach, W.L. MVSP—A multivariate statistical package for Windows; Kovach Computing Services: Pentraeth, UK, 2007. [Google Scholar]
- Environmental Systems Research Institute. Esri: GIS Mapping Software, Spatial Data Analytics & Location Platform. Available online: http://www.esri.com (accessed on 13 February 2018).
- Hartl, D.L.; Clark, G.C. Principles of Population Genetics, 3rd ed.; Sinauer Associates: Sunderland, MA, USA, 1997. [Google Scholar]
- Allan, H.H. Flora of New Zealand; Government Printer: Wellington, New Zealand, 1961; Volume I.
- Tan, H.W.; Heenan, P.B.; de Meyer, S.E.; Willems, A.; Andrews, M. Diverse novel mesorhizobia nodulate New Zealand native Sophora species. Syst. Appl. Microbiol. 2015, 38, 91–98. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, S.E.; Tan, H.W.; Heenan, P.B.; Andrews, M.; Willems, A. Mesorhizobium waimense sp. nov. isolated from Sophora longicarinata root nodules and Mesorhizobium cantuariense sp. nov. isolated from Sophora microphylla root nodules in New Zealand. Int. J. Syst. Evol. Microbiol. 2015, 65, 3419–3426. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, S.E.; Tan, H.W.; Andrews, M.; Heenan, P.B.; Willems, A. Mesorhizobium calcicolae sp. nov., Mesorhizobium novozelandensis sp. nov., Mesorhizobium sophorae sp. nov., Mesorhizobium prostratae sp. nov. and Mesorhizobium microphyllae sp. nov. isolated from Sophora root nodules in New Zealand. Int. J. Syst. Evol. Microbiol. 2016, 66, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.D.; Heenan, P.B.; de Meyer, S.E.; James, T.K.; Chen, W.-M.; Morton, J.D.; Andrews, M. Genetic diversity and nitrogen fixation of mesorhizobia symbionts of New Zealand endemic Sophora species. N. Z. J. Bot. 2017, 55, 466–478. [Google Scholar] [CrossRef]
- Cockayne, L. Hybridism in the New Zealand flora. New Phytol. 1923, 22, 105–127. [Google Scholar] [CrossRef]
- Cockayne, L.; Allan, H.H. An annotated list of groups of wild hybrids in the New Zealand flora. Ann. Bot. 1934, 48, 1–55. [Google Scholar] [CrossRef]
- De Lange, P.J.; Heenan, P.B.; Rolfe, J. Checklist of vascular plants recorded from Chatham Islands; Department of Conservation, New Zealand Government: Wellington, New Zealand, 2011.
- Anderson, E.C.; Thompson, E.A. A model-based method for identifying species hybrids using multilocus genetic data. Genetics 2002, 160, 1217–1229. [Google Scholar] [PubMed]
- Putman, A.; Carbone, I. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol. Evol. 2014, 4, 4399–4428. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.W.; Kelly, D.; Ladley, J.J. Futile selfing in the tree Fuchsia excorticata (Onagraceae) and Sophora microphylla (Fabaceae): Inbreeding depression over 11 years. Int. J. Plant Sci. 2011, 172, 191–198. [Google Scholar] [CrossRef]
- Godley, E.J. Leonard Cockayne and Evolution. N. Z. J. Bot. 1979, 17, 197–215. [Google Scholar] [CrossRef]
- Anderson, S.H. The relative importance of birds and insects as pollinators of the New Zealand flora. N. Z. J. Ecol. 2003, 27, 83–94. [Google Scholar]
- Wolf, D.E.; Takebayashi, N.; Rieseberg, L.H. Predicting the risk of extinction through hybridisation. Conserv. Biol. 2001, 15, 1039–1053. [Google Scholar] [CrossRef]


| Locus | S. chathamica (n = 92) | S. fulvida (n = 57) | S. godleyi (n = 55) | S. longicarinata (n = 27) | S. molloyi (n = 15) | S. prostrata (n = 115) | S. tetraptera (n = 116) | S. microphylla (n = 149) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Ho | He | A | Ho | He | A | Ho | He | A | Ho | He | A | Ho | He | A | Ho | He | A | Ho | He | A | Ho | He | |
| Sop-445 | 7 | 0.29 | 0.37 | 5 | 0.23 | 0.29 | 5 | 0.31 | 0.60 | 4 | 0.19 | 0.21 | 4 | 0.27 | 0.56 | 7 | 0.67 | 0.77 | 7 | 0.53 | 0.53 | 10 | 0.55 | 0.67 |
| Sop-802 | 17 | 0.66 | 0.88 | 12 | 0.45 | 0.80 | 16 | 0.56 | 0.80 | 13 | 0.40 | 0.87 | 9 | 0.13 | 0.82 | 12 | 0.50 | 0.83 | 21 | 0.60 | 0.92 | 20 | 0.71 | 0.91 |
| Sop-816 | 9 | 0.46 | 0.66 | 4 | 0.37 | 0.54 | 11 | 0.72 | 0.78 | 6 | 0.70 | 0.74 | 5 | 0.60 | 0.68 | 8 | 0.60 | 0.66 | 12 | 0.67 | 0.73 | 10 | 0.59 | 0.76 |
| Sop-248 | 21 | 0.78 | 0.90 | 15 | 0.78 | 0.90 | 21 | 0.70 | 0.91 | 16 | 0.81 | 0.90 | 11 | 0.80 | 0.88 | 19 | 0.89 | 0.92 | 20 | 0.79 | 0.90 | 23 | 0.85 | 0.92 |
| Sop-42 | 8 | 0.42 | 0.55 | 5 | 0.21 | 0.26 | 7 | 0.53 | 0.62 | 7 | 0.56 | 0.53 | 4 | 0.40 | 0.50 | 13 | 0.52 | 0.64 | 12 | 0.58 | 0.69 | 15 | 0.54 | 0.64 |
| Sop-806 | 14 | 0.61 | 0.76 | 9 | 0.37 | 0.69 | 16 | 0.49 | 0.84 | 8 | 0.67 | 0.71 | 7 | 0.57 | 0.81 | 30 | 0.69 | 0.92 | 16 | 0.72 | 0.76 | 21 | 0.73 | 0.82 |
| Sop-808 | 14 | 0.45 | 0.56 | 12 | 0.58 | 0.76 | 20 | 0.55 | 0.73 | 10 | 0.81 | 0.73 | 8 | 0.67 | 0.70 | 30 | 0.85 | 0.92 | 22 | 0.67 | 0.77 | 26 | 0.64 | 0.75 |
| Sop-814 | 4 | 0.52 | 0.66 | 3 | 0.44 | 0.50 | 6 | 0.33 | 0.71 | 6 | 0.37 | 0.61 | 4 | 0.27 | 0.70 | 5 | 0.63 | 0.74 | 6 | 0.64 | 0.73 | 7 | 0.55 | 0.74 |
| Sop-825 | 10 | 0.67 | 0.86 | 8 | 0.56 | 0.66 | 11 | 0.71 | 0.86 | 11 | 0.74 | 0.88 | 8 | 0.60 | 0.83 | 11 | 0.83 | 0.83 | 11 | 0.77 | 0.88 | 16 | 0.76 | 0.88 |
| Mean | 11.6 | 0.54 | 0.69 | 8.1 | 0.44 | 0.60 | 12.6 | 0.54 | 0.76 | 9.0 | 0.58 | 0.69 | 6.7 | 0.48 | 0.72 | 15.0 | 0.69 | 0.80 | 14.1 | 0.66 | 0.77 | 16.4 | 0.66 | 0.79 |
| AT | 104 | 73 | 113 | 81 | 60 | 135 | 127 | 148 | ||||||||||||||||
| AP | 3 | 0 | 8 | 1 | 1 | 19 | 8 | 9 | ||||||||||||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heenan, P.; Mitchell, C.; Houliston, G. Genetic Variation and Hybridisation among Eight Species of kōwhai (Sophora: Fabaceae) from New Zealand Revealed by Microsatellite Markers. Genes 2018, 9, 111. https://doi.org/10.3390/genes9020111
Heenan P, Mitchell C, Houliston G. Genetic Variation and Hybridisation among Eight Species of kōwhai (Sophora: Fabaceae) from New Zealand Revealed by Microsatellite Markers. Genes. 2018; 9(2):111. https://doi.org/10.3390/genes9020111
Chicago/Turabian StyleHeenan, Peter, Caroline Mitchell, and Gary Houliston. 2018. "Genetic Variation and Hybridisation among Eight Species of kōwhai (Sophora: Fabaceae) from New Zealand Revealed by Microsatellite Markers" Genes 9, no. 2: 111. https://doi.org/10.3390/genes9020111




