ConGEMs: Condensed Gene Co-Expression Module Discovery Through Rule-Based Clustering and Its Application to Carcinogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Literature Review
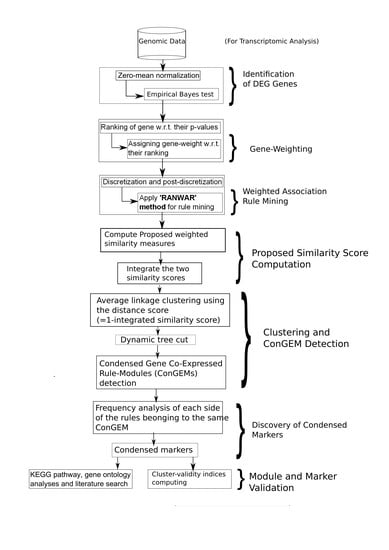
2.2. Proposed Method of Identifying Condensed Rule-Modules
| Algorithm 1 |
| Inputs: (, , , ), where refers to the user-specified cutoff for the corrected p-value, denotes the user-defined cutoff for (weighted) support value (i.e., minimum support), stands for the user-defined cutoff for (weighted) confidence value (i.e., minimum confidence), and be the user-notified number of top (experimental) rules on which the similarity matrix will be computed. Outputs: (Co-expressed rule-modules, Cluster-validity indices, Condensed markers).
|
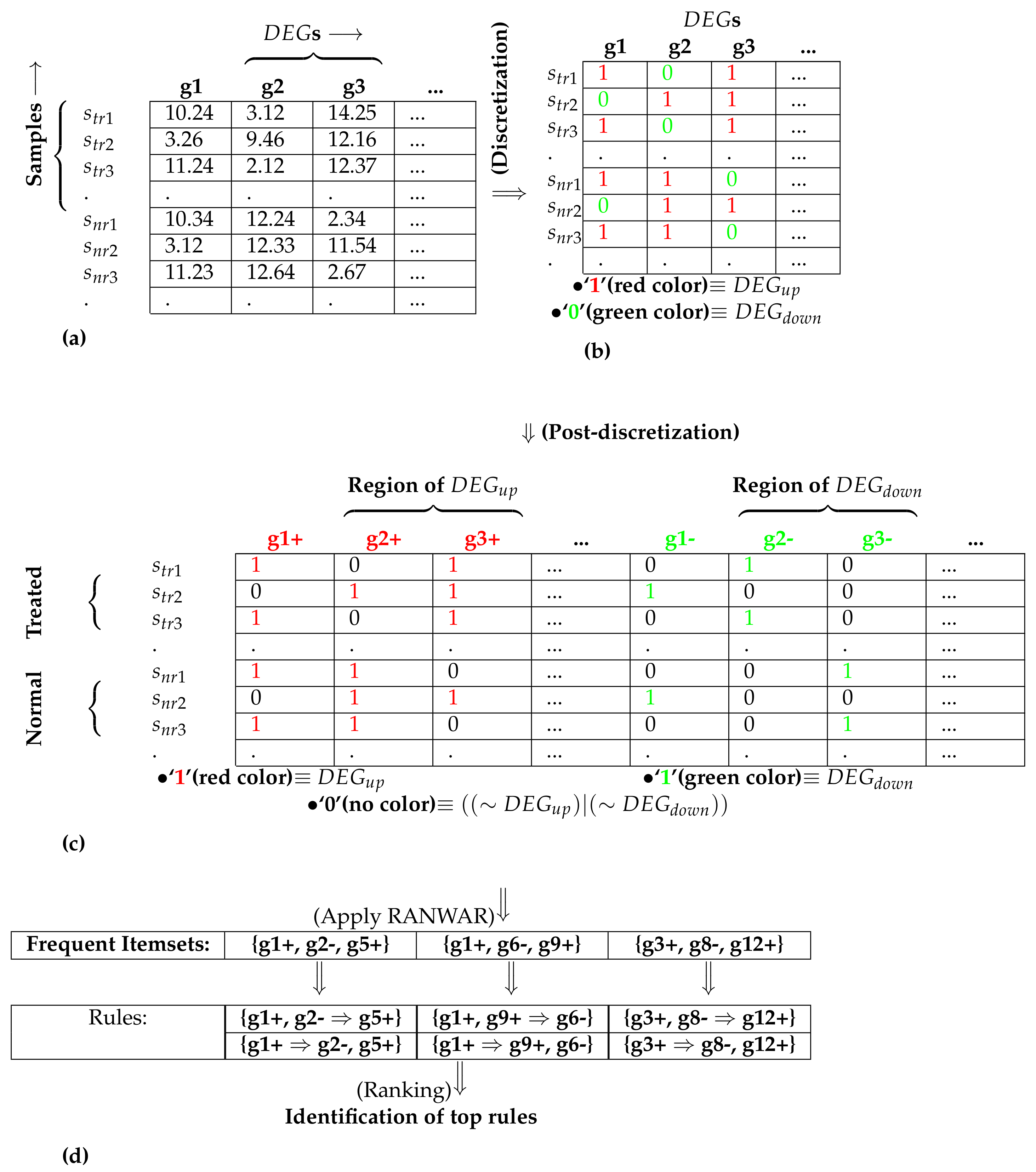
2.2.1. Identification of Differentially Expressed Genes
2.2.2. Assigning Gene-Based Weight
2.2.3. Discretization
2.2.4. Post-Discretization
2.2.5. Identification of Weighted Association Rules
2.2.6. Proposed Weighted Rule-Based Similarity Measures
2.2.7. Clustering Rules Using Proposed Weighted Rule-Based Similarity Measures
2.2.8. Discovery of Condensed Gene Expression Markers
- (i)
- Geneset Equivalent Pruning regulation 1: If there are some genesets whose genes overlapped each other (e.g., “a” and “a, b”), and if each of the genesets had the same frequency, then we considered only the geneset (e.g., “a, b”) that covered all the participating genes belonging to all these genesets, and subsequently the remaining genesets (e.g., “a”) were eliminated from the list.
- (ii)
- Geneset Equivalent Pruning regulation 2: If there are some genesets whose genes overlapped each other (e.g., “a” and “a, b”) and if each of the genesets contained a different frequency, then we considered only the geneset among them which had highest frequency, and subsequently the remaining genesets were omitted.
3. Results and Discussion
3.1. Dataset Information
3.2. Experimental Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Mukhopadhyay, A.; Mandal, M. Identifying Non-redundant Gene Markers from Microarray Data: A Multiobjective Variable Length PSO-based Approach. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shi, I.; Sadraei, N.H.; Duan, Z.H.; Shi, T. Aberrant Signaling Pathways in Squamous Cell Lung Carcinoma. Cancer Inform. 2011, 10, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Shinmura, K.; Igarashi, H.; Kato, H.; Kawanishi, Y.; Inoue, Y.; Nakamura, S.; Ogawa, H.; Yamashita, Y.; Kawase, A.; Funai, K.; et al. CLCA2 as a Novel Immunohistochemical Marker for Differential Diagnosis of Squamous Cell Carcinoma from Adenocarcinoma of the Lung. Dis. Markers 2014, 2014, 619273. [Google Scholar] [CrossRef] [PubMed]
- Man, Y.; Cao, J.; Jin, S.; Xu, G.; Pan, B.; Shang, L.; Che, D.; Yu, Q.; Yu, Y. Newly Identified Biomarkers for Detecting Circulating Tumor Cells in Lung Adenocarcinoma. Tohoku J. Exp. Med. 2014, 234, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Mallik, S.; Bhadra, T.; Maulik, U. Identifying Epigenetic Biomarkers using Maximal Relevance and Minimal Redundancy Based Feature Selection for Multi-Omics Data. IEEE Trans. Nanobiosci. 2017, 16, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Mallik, S.; Sen, S.; Maulik, U. IDPT: Insights into Potential Intrinsically Disordered Proteins Through Transcriptomic Analysis of Genes for Prostate Carcinoma Epigenetic Data. Gene 2016, 586, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Mallik, S. Towards integrated oncogenic marker recognition through mutual information-based statistically significant feature extraction: An association rule mining based study on cancer expression and methylation profiles. Quant. Biol. 2017, 5, 302–327. [Google Scholar] [CrossRef]
- Ruiza, R.; Riquelmea, J.C.; Aguilar-Ruizb, J.S. Incremental wrapper-based gene selection from microarray data for cancer classification. Pattern Recognit. 2006, 39, 2383–2392. [Google Scholar] [CrossRef]
- Agrawal, R.; Imielinski, T.; Swami, A. Mining Association Rules between Sets of Items in large Databases. In Proceedings of the 2017 ACM International Conference on Management of Data; ACM: New York, NY, USA, 1993; pp. 207–216. [Google Scholar]
- Mallik, S.; Mukhopadhyay, A.; Maulik, U. RANWAR: Rank-Based Weighted Association Rule Mining from Gene Expression and Methylation Data. IEEE Trans. NanoBiosci. 2014, 14, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, R.; Srikant, R. Fast algorithms for mining association rules. In Proceedings of the 20th International Conference on Very Large Data Bases, VLDB, San Francisco, CA, USA, 12–15 September 1994; pp. 487–499. [Google Scholar]
- Mallik, S.; Mukhopadhyay, A.; Maulik, U.; Bandyopadhyay, S. Integrated Analysis of Gene Expression and Genome-wide DNA Methylation for Tumor Prediction: An Association Rule Mining-based Approach. In Proceedings of the IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), IEEE Symposium Series on Computational Intelligence (SSCI), Singapore, 16–19 April 2013; pp. 120–127. [Google Scholar]
- Zhao, Z.; Xu, J.; Chen, J.; Kim, S.; Reimers, M.; Bacanu, S.A.; Yu, H.; Liu, C.; Sun, J.; Wang, Q.; et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol. Psychiatry 2015, 20, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, S.; Vosa, U.; van der Graaf, A.; Franke, L.; de Magalhaes, J.P. Gene co-expression analysis for functional classification and gene-disease predictions. Brief. Bioinform. 2017. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, T.; Mallik, S.; Bandyopadhyay, S. Identification of Multi-View Gene Modules using Mutual Information Based Hypograph Mining. IEEE Trans. Syst. Man Cybern. Syst. 2017. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, H.; Quan, X.; Liu, Z.; Yin, Y. Disease-related gene module detection based on a multi-label propagation clustering algorithm. PLoS ONE 2017, 12, e0178006. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, T.; Konagaya, A. KnowledgeEditor: A new tool for interactive modeling and analyzing biological pathways based on microarray data. Bioinformatics 2003, 19, 433–434. [Google Scholar] [CrossRef] [PubMed]
- Segal, E.; Shapira, M.; Regev, A.; Pe’er, D.; Botstein, D.; Koller, D.; Friedman, N. Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003, 34, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Tornow, S.; Mewes, H.W. Functional modules by relating protein interaction networks and gene expression. Nucleic Acids Res. 2003, 31, 6283–6289. [Google Scholar] [CrossRef] [PubMed]
- Prinz, S.; Avila-Campillo, I.; Aldridge, C.; Srinivasan, A.; Dimitrov, K.; Siegel, A.F.; Galitski, T. Control of Yeast Filamentous-Form Growth by Modules in an Integrated Molecular Network. Genome Res. 2004, 14, 380–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Current Opin. HIV AIDS 2014, 511, 543–550. [Google Scholar] [CrossRef]
- Smyth, G. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3. [Google Scholar] [CrossRef]
- Creighton, C.; Hanash, S. Mining Gene Expression Databases for Association Rules. Bioinformatics 2003, 19, 79–86. [Google Scholar] [CrossRef]
- WGCNA: Weighted gene co-expression network analysis. Available online: http://hms-dbmi.github.io/scw/WGCNA.html (accessed on 12 July 2017).
- Langfelder, P.; Zhang, B.; Horvath, S. Defining Clusters from a Hierarchical Cluster Tree: The Dynamic Tree Cut package for R. Bioinformatics 2007, 24, 719–720. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Mallik, S.; Mukhopadhyay, A. A Survey and Comparative Study of Statistical Tests for Identifying Differential Expression from Microarray Data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 11, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Mallik, S. Integrating Multiple Data Sources for Combinatorial Marker Discovery: A Study in Tumorigenesis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016. [Google Scholar] [CrossRef]
- Yu, H.; Mitra, R.; Yang, J.; Li, Y.Y.; Zhao, Z. Algorithms for network-based identification of differential regulators from transcriptome data: A systematic evaluation. Sci. China 2014, 57, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Ravasz, E.; Somera, A.L.; Mongru, D.A.; Oltvai, Z.N.; Barabási, A.L. Hierarchical Organization of Modularity in Metabolic Networks. Science 2002, 297, 1551–1555. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Yip, A.; Horvath, S. Gene Network Interconnectedness and the Generalized Topological Overlap Measure. BMC Bioinform. 2007, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Sethi, P.; Alagiriswamy, S. Association Rule Based Similarity Measures for the Clustering of Gene Expression Data. Open Med. Inform. J. 2010, 4, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Alaimo, S.; Marceca, G.P.; Ferro, A.; Pulvirenti, A. Detecting Disease Specific Pathway Substructures through an Integrated Systems Biology Approach. Non-Coding RNA 2017, 3, 20. [Google Scholar] [CrossRef]
- Alaimo, S.; Giugno, R.; Acunzo, M.; Veneziano, D.; Ferro, A.; Pulvirenti, A. Post-transcriptional knowledge in pathway analysis increases the accuracy of phenotypes classification. Oncotarget 2016, 7, 54572–54582. [Google Scholar] [CrossRef] [PubMed]
- Kuner, R.; Muley, T.; Meister, M.; Ruschhaupt, M.; Buness, A.; Xu, E.C.; Schnabel, P.; Warth, A.; Poustka, A.; Sültmann, H.; et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer 2009, 63, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Jones, A.; Lee, S.H.; Ng, E.; Fiegl, H.; Zikan, M.; Cibula, D.; Sargent, A.; Salvesen, H.B.; Jacobs, I.J.; et al. The dynamics and prognostic potential of DNA methylation changes at stem cell gene loci in women’s cancer. PLoS Genet. 2012, 8, e1002517. [Google Scholar] [CrossRef]
- Chen, S.C.; Tsai, T.H.; Chung, C.H.; Li, W.H. Dynamic association rules for gene expression data analysis. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ochs, M.F.; Ahn, S.M.; Hennessey, P.; Tan, M.; Soudry, E.; Gaykalova, DA.; Uemura, M.; Brait, M.; Shao, C.; et al. Expression Microarray Analysis Reveals Alternative Splicing of LAMA3 and DST Genes in Head and Neck Squamous Cell Carcinoma. PLoS ONE 2014, 9, e91263. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Pan, L. Identification of Logic Relationships between Genes and Subtypes of Non-Small Cell Lung Cancer. PLoS ONE 2014, 9, e94644. [Google Scholar] [CrossRef] [PubMed]
- Ben-Hamo, R.; Boue, S.; Martin, F.; Talikka, M.; Efroni, S. Classification of lung adenocarcinoma and squamous cell carcinoma samples based on their gene expression profile in the sbv IMPROVER Diagnostic Signature Challenge. Syst. Biomed. 2013, 1, 268–277. [Google Scholar] [CrossRef]
- Ge, L.; Liu, S.; Xie, L.; Sang, L.; Ma, C.; Li, H. Differential mRNA expression profiling of oral squamous cell carcinoma by high-throughput RNA sequencing. J. Biomed. Res. 2015, 29, 397–404. [Google Scholar]
- Cai, B.; Jiang, X. Revealing Biological Pathways Implicated in Lung Cancer from TCGA Gene Expression Data Using Gene Set Enrichment Analysis. Cancer Inform. 2014, 13 (Suppl. S1), 113–121. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.C.; Secrist, H.; Bangur, C.S.; Wang, T.; Zhang, X.; Harlan, D.; Goodman, G.E.; Houghton, R.L.; Persing, D.H.; Zehentner, B.K. Multigene Real-time PCR Detection of Circulating Tumor Cells in Peripheral Blood of Lung Cancer Patients. Anticancer Res. 2006, 26, 1567–1575. [Google Scholar] [PubMed]
- Raponi, M.; Yu, J. Lung Cancer Prognostics. U.S. Patents US20060252057 A1, 2006. Available online: http://www.google.co.in/patents/US20060252057 (accessed on 14 July 2017).
- Molina-Pinelo, S.; Gutierrez, G.; Pastor, M.D.; Hergueta, M.; Moreno-Bueno, G.; García-Carbonero, R.; Nogal, A.; Suárez, R.; Salinas, A.; Pozo-Rodríguez, F.; et al. MicroRNA-Dependent Regulation of Transcription in Non-Small Cell Lung Cancer. PLoS ONE 2014, 9, e90524. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.; Langfelder, P. Tutorial for the WGCNA package for R: III. Using Simulated Data to Evaluate Di Erent Module Detection Methods and Gene Screening Approaches. Available online: https://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/Simulated-05-NetworkConstruction.pdf (accessed on 12 July 2017).
- Scutari, M.; Nagarajan, R. Identifying significant edges in graphical models of molecular networks. Artif. Intell. Med. 2013, 57, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, R.; Scutari, M. Impact of noise on molecular network inference. PLoS ONE 2013, 8, e80735. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Yan, X.; Huang, Y.; Han, J.; Zhou, X.J. Mining coherent dense subgraphs across massive biological networks for functional discovery. Bioinformatics 2005, 21 (Suppl. S1), i213–i221. [Google Scholar] [CrossRef] [PubMed]

| Rank | Condensed Marker () | Module Label | Availability of Biological Evidence | Status of Condensed Marker |
|---|---|---|---|---|
| 1 | DST- | purple (consequent) | Available | Existing |
| 2 | TP63- | blue, brown (consequent) | Available | Existing |
| 3 | BNC1- | pink (consequent) | Available | Existing |
| 4 | CLCA2- | yellow (consequent) | Available | Existing |
| 5 | GJB5- | dark red (consequent) | Available | Existing |
| 6 | {DSC3-, KRT5-} | dark turquoise (antecedent) | Available for both | Existing |
| 7 | {CGN+, DSC3-} | salmon (antecedent) | Available for both | Existing |
| 8 | {KRT5-, NTRK2-} | blue (antecedent) | Available for both | Existing |
| 9 | {CGN+, KRT5-} | light green (antecedent) | Available for both | Existing |
| 10 | {DSC3-, TMEM40-, NTRK2-} | yellow (antecedent) | Available for DSC3 and NTRK2, not found for TMEM40 | Novel |
| Individual Gene | p-Value | Literature Evidence | KEGG Pathway and GO-Terms (p-Value) |
|---|---|---|---|
| DST | 9.26 | [39,40,41] | GO:BPs: response to wounding (GO:0009611) (p-value = 0.005237) [42], extracellular matrix organization (GO:0030198) (p-value = 0.006151), extracellular structure organization (GO:0043062) (p-value = 0.006327) [42]; GO:CCs: extracellular vesicular exosome (GO:0070062) (p-value = 1.02 ), extracellular matrix part (GO:0044420) (p-value = 0.001283) [42]; GO:MF: calcium ion binding (GO:0005509) (8.51 ). |
| TP63 | 1.27 | [40] | GO:BPs: regulation of Notch signaling pathway (GO:0008593) (p-value = 0.024357) [43], positive regulation of Notch signaling pathway (GO:0045747) (p-value = 0.040422) [43]; GO:MF: RNA polymerase II transcription regulatory region sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription (GO:0001228) (p-value = 0.036160). |
| BNC1 | 2.82 | [40] | GO:BP: response to wounding (GO:0009611) (p-value = 0.005237) [42]. |
| CLCA2 | 1.28 | [2,3,4,40,41,44,45] | KEGG pathways: Pancreatic (p-value = 0.020269), Renin (p-value = 0.037037); GO:CC: extracellular region (GO:0005576) (p-value = 0.000225) [42]. |
| GJB5 | 1.94 | [40] | |
| CGN | 1.96 | [46] | KEGG pathway: Tight (p-value = 0.04347487); GO:CC: cell-cell junction (GO:0005911) (p-value = 0.000681). |
| DSC3 | 3.08 | [40,41,45] | GO:BPs: cell–cell adhesion via plasma-membrane adhesion molecules (GO:0098742) (p-value = 1.15978 ), cell–cell adhesion (GO:0098609) (p-value = 1.28951 ); GO:CCs: extracellular region (GO:0005576) (p-value = 0.000225) [42], cell–cell junction (GO:0005911) (p-value = 0.000681); GO:MF: calcium ion binding (GO:0005509) (p-value = 8.51 ). |
| KRT5 | 6.50 | [40,41] | GO:BP: regulation of Rac GTPase activity (GO:0032314) (p-value = 0.020821), positive regulation of neuron projection development (GO:0010976) (p-value = 0.031048); GO:CC: extracellular vesicular exosome (GO:0070062) (p-value = 1.02 ). |
| NTRK2 | 1.47 | [40] | GO:BPs: regulation of Rac GTPase activity (GO:0032314) (p-value = 0.020821), positive regulation of neuron projection development (GO:0010976) (p-value = 0.031048); GO:MF: growth factor binding (GO:0019838) (p-value = 0.011462) [42]. |
| TMEM40 | 1.29 | - |
| Validty Index | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| avgDI ⇑ | 3.82 | Inf | Inf | Inf | Inf | - | - | - | - | - | - |
| avgSW ⇑ | 7.82 | 9.97 | 3.94 | 6.55 | 2.89 | - | - | - | - | - | - |
| avgSC ⇓ | 6.80 | 4.24 | 3.41 | 3.39 | 3.02 | 9.41 | 6.3 | 1.66 | 6.53 | 1.65 | 6.53 |
| avgCC ⇑ | 2.53 | 1.52 | 8.74 | 1.30 | 8.64 | 8.86 | 5.18 | 1.47 | 5.38 | 1.47 | 5.38 |
| avgMAR ⇑ | 2.99 | 1.26 | 8.24 | 1.24 | 1.00 | - | - | - | - | - | - |
| Density ⇑ | 2.21 | 9.83 | 3.99 | 6.42 | 2.90 | 7.782 | 3.18 | 2.40 | 3.30 | 2.40 | 3.30 |
| Centralization ⇑ | 1.04 | 1.36 | 7.85 | 1.28 | 6.83 | 7.64 | 4.83 | 1.24 | 4.82 | 1.24 | 4.82 |
| Rand index ⇑ | 2.21 | - | - | - | - | - | - | - | - | - | - |
| Adjusted Rand index ⇑ | - | - | - | - | - | - | - | - | - | - | - |
| Validty Index | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| avgDI ⇑ | 3.47 | Inf | Inf | Inf | Inf | Inf | - | Inf | - | Inf | - |
| avgSW ⇑ | 1.85 | 3.13 | 2.12 | 9.16 | 1.61 | 9.90 | - | 9.90 | - | 9.90 | - |
| avgSC ⇓ | 7.41 | 9.99 | 6.39 | 9.77 | 5.49 | 9.90 | 1.08 | 9.90 | 1.26 | 9.90 | 1.27 |
| avgCC ⇑ | 2.77 | 9.64 | 2.76 | 9.55 | 2.58 | 9.90 | 1.81 | 9.90 | 1.98 | 9.90 | 1.99 |
| avgMAR ⇑ | 3.25 | 9.64 | 2.53 | 9.52 | 2.75 | - | - | - | - | - | - |
| Density ⇑ | 1.87 | 9.64 | 2.11 | 9.49 | 1.59 | 9.80 | 1.01 | 9.80 | 1.12 | 9.80 | 1.26 |
| Centralization ⇑ | 1.26 | 1.04 | 1.21 | 2.31 | 1.33 | 1.01 | 8.54 | 1.01 | 8.80 | 1.01 | 8.84 |
| Rand index ⇑ | 4.29 | - | - | - | - | - | - | - | - | - | - |
| Adjusted Rand index ⇑ | - | - | - | - | - | - | - | - | - | - | - |
| Validty Index | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| avgDI ⇑ | 3.82 | Inf | Inf | Inf | Inf | Inf | Inf | Inf | Inf | Inf | Inf |
| avgSW ⇑ | 5.91 | 2.97 | - | 5.50 | - | 5.54 | 2.80 | 8.66 | 4.51 | 8.96 | 6.11 |
| avgSC ⇓ | 6.74 | 9.01 | 9.10 | 9.10 | 9.10 | 6.92 | 5.01 | 8.83 | 6.83 | 9.06 | 6.81 |
| avgCC ⇑ | 2.51 | 6.55 | 5.29 | 5.29 | 5.29 | 6.70 | 5.14 | 8.71 | 6.16 | 8.96 | 7.20 |
| avgMAR ⇑ | 2.98 | 6.53 | 5.31 | 5.31 | 5.31 | - | - | - | - | - | - |
| Density ⇑ | 2.19 | 6.50 | 5.27 | 5.27 | 5.27 | 5.09 | 1.25 | 7.95 | 2.34 | 8.23 | 3.38 |
| Centralization ⇑ | 1.06 | 7.41 | 5.47 | 5.47 | 5.47 | 1.01 | 9.04 | 1.04 | 1.04 | 8.89 | 8.89 |
| Rand index ⇑ | 7.01 | - | - | - | - | - | - | - | - | - | - |
| Adjusted Rand index ⇑ | 3.53 | - | - | - | - | - | - | - | - | - | - |
| Validty Index | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| avgDI ⇑ | 3.81 | Inf | Inf | Inf | - | - | - | - | - | - | - |
| avgSW ⇑ | 7.11 | 1.24 | 5.51 | 1.00 | - | - | - | - | - | - | - |
| avgSC ⇓ | 6.83 | 5.50 | 4.34 | 4.34 | 4.34 | 1.51 | 9.67 | 2.37 | 1.18 | 2.65 | 1.14 |
| avgCC ⇑ | 2.53 | 1.96 | 1.09 | 1.09 | 1.09 | 1.42 | 7.85 | 1.98 | 9.23 | 2.39 | 9.87 |
| avgMAR ⇑ | 2.99 | 1.71 | 1.05 | 1.05 | 1.05 | - | - | - | - | - | - |
| Density ⇑ | 2.21 | 1.42 | 5.99 | 5.99 | 5.99 | 2.23 | 7.90 | 5.02 | 1.04 | 6.23 | 1.09 |
| Centralization ⇑ | 1.03 | 1.21 | 8.84 | 8.84 | 8.84 | 1.34 | 7.68 | 1.68 | 8.13 | 1.83 | 8.79 |
| Rand index ⇑ | 7.18 | - | - | - | - | - | - | - | - | - | - |
| Adjusted Rand index ⇑ | 1.75 | - | - | - | - | - | - | - | - | - | - |
| Validty Index | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| avgDI ⇑ | 4.23 | Inf | Inf | Inf | Inf | Inf | - | Inf | - | Inf | - |
| avgSW ⇑ | 9.33 | 3.83 | 1.89 | 9.07 | 1.55 | 9.80 | - | 9.80 | - | 9.80 | - |
| avgSC ⇓ | 7.45 | 9.91 | 6.20 | 1.22 | 1.22 | 9.80 | 1.22 | 9.80 | 1.30 | 9.80 | 1.30 |
| avgCC ⇑ | 2.59 | 9.60 | 2.63 | 1.20 | 1.20 | 9.80 | 1.20 | 9.80 | 1.25 | 9.80 | 1.25 |
| avgMAR ⇑ | 3.26 | 9.60 | 2.39 | - | - | - | - | - | - | - | - |
| Density ⇑ | 1.89 | 9.60 | 1.85 | 1.49 | 1.49 | 9.60 | 1.49 | 9.60 | 1.59 | 9.60 | 1.59 |
| Centralization ⇑ | 6.47 | 9.32 | 1.18 | 1.12 | 1.12 | 2.04 | 1.12 | 2.04 | 1.11 | 2.04 | 1.11 |
| Rand index ⇑ | 5.24 | - | - | - | - | - | - | - | - | - | - |
| Adjusted Rand index ⇑ | 4.55 | - | - | - | - | - | - | - | - | - | - |
| Validty Index | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| avgDI ⇑ | 4.27 | Inf | Inf | Inf | Inf | Inf | - | Inf | - | Inf | - |
| avgSW ⇑ | 9.88 | 3.67 | 2.78 | 9.15 | 2.19 | 9.80 | - | 9.80 | - | 9.80 | - |
| avgSC ⇓ | 7.37 | 9.92 | 7.62 | 9.80 | 9.80 | 9.80 | 1.27 | 9.80 | 1.47 | 9.80 | 1.48 |
| avgCC ⇑ | 2.58 | 9.71 | 2.43 | 9.80 | 9.80 | 9.80 | 1.24 | 9.80 | 1.36 | 9.80 | 1.39 |
| avgMAR ⇑ | 3.26 | 9.71 | 3.17 | - | - | - | - | - | - | - | - |
| Density ⇑ | 1.88 | 9.71 | 2.77 | 9.60 | 9.60 | 9.60 | 1.64 | 9.60 | 2.00 | 9.60 | 2.04 |
| Centralization ⇑ | 6.72 | 7.9 | 1.23 | 2.04 | 2.04 | 2.04 | 1.17 | 2.04 | 1.21 | 2.04 | 1.22 |
| Rand index ⇑ | 5.54 | - | - | - | - | - | - | - | - | - | - |
| Adjusted Rand index ⇑ | 3.67 | - | - | - | - | - | - |
| Dataset | Method | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LUSC | Proposed | 5-1-3 | 7-1-1 | 6-1-2 | 7-1-1 | 7-1-1 | 7-1-1 | 6-1-2 | 7-1-1 | 6-1-2 | 7-1-1 |
| Cervical | Proposed | 4-1-4 | 5-1-3 | 4-1-4 | 6-1-2 | 5-1-3 | 7-1-1 | 5-1-3 | 7-1-1 | 5-1-3 | 7-1-1 |
| LUSC sm1 | Proposed | 5-0-4 | 7-0-2 | 6-0-3 | 7-0-2 | 5-0-4 | 5-0-4 | 5-0-4 | 6-0-3 | 6-0-3 | 5-0-4 |
| LUSC sm2 | Proposed | 6-0-3 | 8-0-1 | 7-0-2 | 8-0-1 | 7-0-2 | 8-0-1 | 7-0-2 | 8-0-1 | 7-0-2 | 8-0-1 |
| Cervical sm1 | Proposed | 5-0-4 | 5-0-4 | 6-0-3 | 6-0-3 | 6-0-3 | 8-0-1 | 6-0-3 | 7-0-2 | 6-0-3 | 7-0-2 |
| Cervical sm2 | Proposed | 5-0-4 | 6-0-3 | 6-0-3 | 6-0-3 | 6-0-3 | 7-0-2 | 6-0-3 | 7-0-2 | 6-0-3 | 7-0-2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mallik, S.; Zhao, Z. ConGEMs: Condensed Gene Co-Expression Module Discovery Through Rule-Based Clustering and Its Application to Carcinogenesis. Genes 2018, 9, 7. https://doi.org/10.3390/genes9010007
Mallik S, Zhao Z. ConGEMs: Condensed Gene Co-Expression Module Discovery Through Rule-Based Clustering and Its Application to Carcinogenesis. Genes. 2018; 9(1):7. https://doi.org/10.3390/genes9010007
Chicago/Turabian StyleMallik, Saurav, and Zhongming Zhao. 2018. "ConGEMs: Condensed Gene Co-Expression Module Discovery Through Rule-Based Clustering and Its Application to Carcinogenesis" Genes 9, no. 1: 7. https://doi.org/10.3390/genes9010007