X Chromosome Evolution in Cetartiodactyla
Abstract
:1. Introduction
2. Materials and Methods
2.1. Species
2.2. Chromosome Preparation
2.3. BAC Clones
2.4. Fluorescence In-Situ Hybridization (FISH)
2.5. Bioinformatics Analysis
2.6. Ancestral Chromosome Deduction
3. Result
3.1. BACs Localization
3.2. Intrachromosome Rearrangements
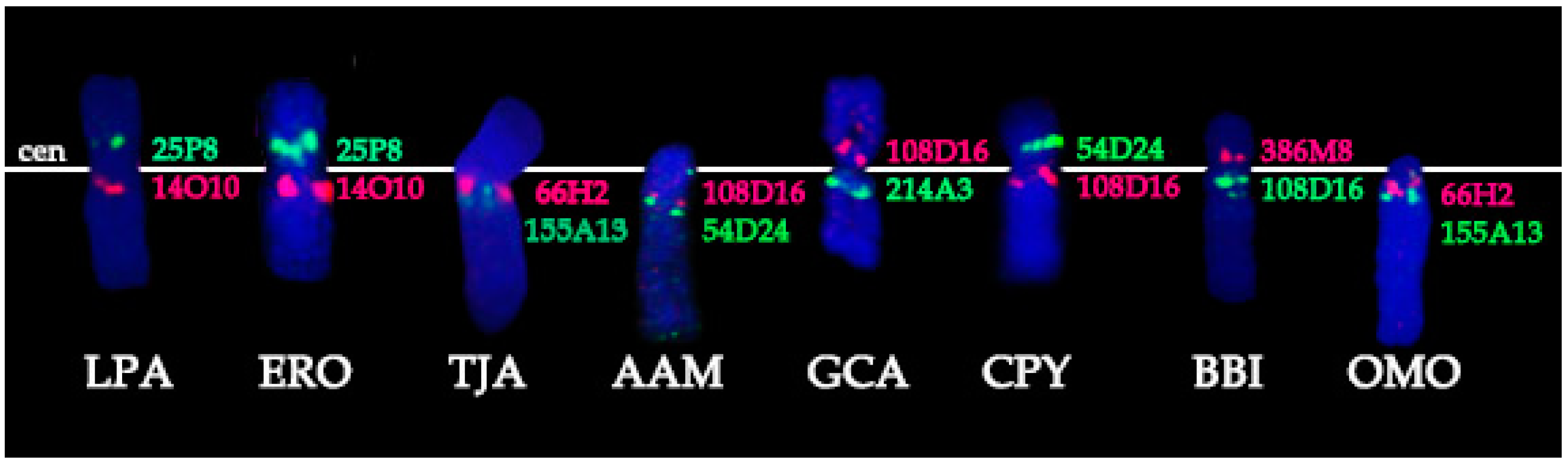
- Conservation: no change in the BAC order and no change of the centromere position. We identified a group of four basal cetartiodactyl species (gray whale (ERO), common hippopotamus (HAM), alpaca (LPA), and pig (SSC)) that have an identical order of the BACs and the same relative position of the centromere (located in XSB1).
- Centromere repositioning: conserved BAC order, changes in the centromere position. Centromere repositions have been shown in roe deer, and mouse-deer, resulting in metacentric (Siberian roe deer (CPY)) and acrocentric (Java mouse-deer (TJA)) X chromosomes, respectively. This event took place prior to a formation of some lineage specific ancestral chromosomes (RAX (Ruminant Ancestral X), AAX (Antilopinae Ancestral X), and CEAX (Cervinae Ancestral X)), indicating that centromere repositioning is one of the key rearrangements of the ruminant X: while maintaining a conserved order of the segments there was a displacement of the centromere (Figure 1).
- Inversion: changes in the BAC order. Three kinds of inversions were identified: (A) syntenic block (SB) flip—this inversion reverses the orientation of the whole syntenic block (TJA, AAM, AAX, CEAX); (B) an inversion inside the syntenic block (goat (CHI), muskox (OMO)); (C) the exchange inversion—inversion that involves several BAC clones from two syntenic blocks (TJA, fallow deer (DDA)) (Figure 2).
3.3. Bioinformatic Analysis of Mammalian X Chromosomes
4. Discussion
4.1. Ancestral X Chromosome
4.2. Ancestral Form of Ruminantia-Pecora X-Chromosome
4.3. Cervidae
4.4. Bovidae
4.5. X Chromosome Rearrangements
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ohno, S.; Beçak, W.; Beçak, M.L. X-autosome ratio and the behavior pattern of individual X-chromosomes in placental mammals. Chromosoma 1964, 15, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Pathak, S.; Stock, A.D. The X chromosomes of mammals: karyological homology as revealed by banding techniques. Genetics 1974, 78, 703–714. [Google Scholar] [PubMed]
- Murphy, W.J.; Larkin, D.M.; Everts-Van Der Wind, A.; Bourque, G.; Tesler, G.; Auvil, L.; Beever, J.E.; Chowdhary, B.P.; Galibert, F.; Gatzke, L.; et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 2005, 309, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Graphodatsky, A.S. Conserved and variable elements of mammalian chromosomes. Cytogenet. Anim. 1989, 95–124. [Google Scholar]
- Ferguson-Smith, M.A. History and evolution of cytogenetics. Mol. Cytogenet. 2015, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Griffin, D.K.; O’Brien, P.C.M.; Yang, F.; Lin, C.C.; Ferguson-Smith, M.A. Defining the anatomy of the Rangifer tarandus sex chromosomes. Chromosoma 1998, 107, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chi, J.; Nie, W.; Wang, J.; Yang, F. Phylogenomics of several deer species revealed by comparative chromosome painting with Chinese muntjac paints. Genetica 2006, 127, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Kulemzina, A.I.; Trifonov, V.A.; Perelman, P.L.; Rubtsova, N.V.; Volobuev, V.; Ferguson-Smith, M.A.; Stanyon, R.; Yang, F.; Graphodatsky, A.S. Cross-species chromosome painting in Cetartiodactyla: Reconstructing the karyotype evolution in key phylogenetic lineages. Chromosome Res. 2009, 17, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Kulemzina, A.I.; Yang, F.; Trifonov, V.A.; Ryder, O.A.; Ferguson-Smith, M.A.; Graphodatsky, A.S. Chromosome painting in Tragulidae facilitates the reconstruction of Ruminantia ancestral karyotype. Chromosome Res. 2011, 19, 531. [Google Scholar] [CrossRef] [PubMed]
- Kulemzina, A.I.; Perelman, P.L.; Grafodatskaya, D.A.; Nguyen, T.T.; Thompson, M.; Roelke-Parker, M.E.; Graphodatsky, A.S. Comparative chromosome painting of pronghorn (Antilocapra americana) and saola (Pseudoryx nghetinhensis) karyotypes with human and dromedary camel probes. BMC Genet. 2014, 15, 68. [Google Scholar] [CrossRef] [PubMed]
- Kulemzina, A.I.; Proskuryakova, A.A.; Beklemisheva, V.R.; Lemskaya, N.A.; Perelman, P.L.; Graphodatsky, A.S. Comparative Chromosome Map and Heterochromatin Features of the Gray Whale Karyotype (Cetacea). Cytogenet. Genome Res. 2016, 148, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.J.; Harrison, W.R.; Ponce de Leon, F.A.; Davis, S.K.; Elder, F.F.B. A molecular cytogenetic analysis of X chromosome repatterning in the Bovidae: transpositions, inversions, and phylogenetic inference. Cytogenet. Genome Res. 1998, 80, 179–184. [Google Scholar] [CrossRef]
- Rubes, J.; Musilova, P.; Kopecna, O.; Kubickova, S.; Cernohorska, H.; Kulemsina, A.I. Comparative molecular cytogenetics in Cetartiodactyla. Cytogenet. Genome Res. 2012, 137, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Stanyon, R.; Archidiacono, N.; Rocchi, M. Comparative Primate Molecular Cytogenetics: Revealing Ancestral Genomes, Marker Order, and Evolutionary New Centromeres. In Post-Genome Biology of Primates; Hirai, H., Imai, H., Go, Y., Eds.; Springer: Tokyo, Japan, 2012; pp. 193–216. ISBN 978-4-431-54010-6. [Google Scholar]
- Chiatante, G.; Capozzi, O.; Svartman, M.; Perelman, P.; Centrone, L.; Romanenko, S.S.; Ishida, T.; Valeri, M.; Roelke-Parker, M.E.; Stanyon, R. Centromere repositioning explains fundamental number variability in the New World monkey genus Saimiri. Chromosoma 2016, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Trifonov, V.A.; Kosyakova, N.; Romanenko, S.A.; Stanyon, R.; Graphodatsky, A.S.; Liehr, T. New insights into the karyotypic evolution in muroid rodents revealed by multicolor banding applying murine probes. Chromosome Res. 2010, 18, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Trifonov, V.A.; Musilova, P.; Kulemsina, A.I. Chromosome evolution in Perissodactyla. Cytogenet. Genome Res. 2012, 137, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Hassanane, M.S.; Chaudhary, R.; Chowdhary, B.P. Microdissected bovine X chromosome segment delineates homoeologous chromosomal regions in sheep, goat and buffalo. Chromosome Res. 1998, 6, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Piumi, F.; Schibler, L.; Vaiman, D.; Oustry, A.; Cribiu, E.P. Comparative cytogenetic mapping reveals chromosome rearrangements between the X chromosomes of two closely related mammalian species (cattle and goats). Cytogenet. Genome Res. 1998, 81, 36–41. [Google Scholar] [CrossRef]
- Iannuzzi, L.; Di Meo, G.P.; Perucatti, A.; Incarnato, D.; Schibler, L.; Cribiu, E.P. Comparative FISH mapping of bovid X chromosomes reveals homologies and divergences between the subfamilies Bovinae and Caprinae. Cytogenet. Genome Res. 2000, 89, 171–176. [Google Scholar] [CrossRef]
- Iannuzzi, L.; King, W.A.; Di Berardino, D. Chromosome evolution in domestic bovids as revealed by chromosome banding and FISH-mapping techniques. Cytogenet. Genome Res. 2009, 126, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Perucatti, A.; Genualdo, V.; Iannuzzi, A.; Rebl, A.; Di Berardino, D.; Goldammer, T.; Iannuzzi, L. Advanced comparative cytogenetic analysis of X chromosomes in river buffalo, cattle, sheep, and human. Chromosome Res. 2012, 20, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, D.S.; Davis, S.K.; De Donato, M.; Burzlaff, J.D.; Womack, J.E.; Taylor, J.F.; Kumamoto, A.T. A Molecular Cytogenetic Analysis of the Tribe Bovini (Artiodactyla: Bovidae: Bovinae) with an Emphasis on Sex Сhromosome Morphology and NOR Distribution. Chromosome Res. 1999, 7, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Cernohorska, H.; Kubickova, S.; Kopecna, O.; Kulemzina, A.I.; Perelman, P.L.; Elder, F.F.; Robinson, T.J.; Graphodatsky, A.S.; Rubes, J. Molecular cytogenetic insights to the phylogenetic affinities of the giraffe (Giraffa camelopardalis) and pronghorn (Antilocapra americana). Chromosome Res. 2013, 21, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, J.; Kubickova, S.; Musilova, P.; Cernohorska, H.; Muskova, H.; Rubes, J. A Comparative Study of Pygmy Hippopotamus (Choeropsis liberiensis) Karyotype by Cross-Species Chromosome Painting. J. Mamm. Evol. 2016, 1–10. [Google Scholar] [CrossRef]
- Yang, F.; Graphodatsky, A.S. Animal probes and ZOO-FISH. In Fluorescence in Situ Hybridization (FISH); Liehr, T., Ed.; Springer: Berlin, Germany, 2017; pp. 323–346. [Google Scholar]
- Seabright, M. A rapid banding technique for human chromosomes. Lancet 1971, 2, 971–972. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Siepel, A.; Bejerano, G.; Pedersen, J.S.; Hinrichs, A.S.; Hou, M.; Rosenbloom, K.; Clawson, H.; Spieth, J.; Hillier, L.W.; Richards, S.; et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005, 15, 1034–1050. [Google Scholar] [CrossRef] [PubMed]
- Therneau, T.; Atkinson, B.; Ripley, B. Recursive Partitioning and Regression Trees. R Package Version 4.1–10; Mayo Foundation: Rochester, MN, USA, 2015. [Google Scholar]
- Proskuryakova, A.A.; Kulemzina, A.I.; Graphodatsky, A.S. Localisation of NOR on sex chromosome of Tragulus javanicus. Cytogenet. Genome Res. 2018. in preparation. [Google Scholar]
- Ohno, S.; Wolf, U.; Atkin, N.B. Evolution from fish to mammals by gene duplication. Hereditas 1968, 59, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Raudsepp, T.; Lee, E.-J.; Kata, S.R.; Brinkmeyer, C.; Mickelson, J.R.; Skow, L.C.; Womack, J.E.; Chowdhary, B.P. Exceptional conservation of horse–human gene order on X chromosome revealed by high-resolution radiation hybrid mapping. Proc. Natl. Acad. Sci. USA 2004, 101, 2386–2391. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.J.; Sun, S.; Chen, Z.-Q.; Pecon-Slattery, J.; O’Brien, S.J. Extensive Conservation of Sex Chromosome Organization Between Cat and Human Revealed by Parallel Radiation Hybrid Mapping. Genome Res. 1999, 9, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Delgado, C.L.; Waters, P.D.; Gilbert, C.; Robinson, T.J.; Graves, J.A.M. Physical mapping of the elephant X chromosome: conservation of gene order over 105 million years. Chromosome Res. 2009, 17, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Quilter, C.R.; Blott, S.C.; Mileham, A.J.; Affara, N.A.; Sargent, C.A.; Griffin, D.K. A mapping and evolutionary study of porcine sex chromosome gene. Mamm. Genome 2002, 13, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Avila, F.; Baily, M.P.; Perelman, P.; Das, P.J.; Pontius, J.; Chowdhary, R.; Owens, E.; Johnson, W.E.; Merriwether, D.A.; Raudsepp, T. A Comprehensive Whole-Genome Integrated Cytogenetic Map for the Alpaca (Lama pacos). Cytogenet. Genome Res. 2014, 144, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Adega, F.; Chaves, R.; Guedes-Pinto, H. Chromosomal evolution and phylogenetic analyses in Tayassu pecari and Pecari tajacu (Tayassuidae): tales from constitutive heterochromatin. J. Genet. 2007, 86, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, N.O.; Larramendy, M.L.; Bianchi, M.S.; Cortes, L. Karyological conservatism in South American camelids. Cell. Mol. Life Sci. 1986, 42, 622–624. [Google Scholar] [CrossRef]
- Bunch, T.D.; Foote, W.C.; Maciulis, A. Chromosome banding pattern homologies and NORs for the Bactrian camel, guanaco, and llama. J. Hered. 1985, 76, 115–118. [Google Scholar] [CrossRef]
- Balmus, G.; Trifonov, V.A.; Biltueva, L.S.; O’Brien, P.C.; Alkalaeva, E.S.; Fu, B.; Skidmore, J.A.; Allen, T.; Graphodatsky, A.S.; Yang, F.; et al. Cross-species chromosome painting among camel, cattle, pig and human: further insights into the putative Cetartiodactyla ancestral karyotype. Chromosome Res. 2007, 15, 499–514. [Google Scholar] [CrossRef] [PubMed]
- Árnason, Ú. Comparative chromosome studies in Cetacea. Hereditas 1974, 77, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.J.; Pringle, T.H.; Crider, T.A.; Springer, M.S.; Miller, W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007, 17, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Price, S.A.; Bininda-Emonds, O.R.; Gittleman, J.L. A complete phylogeny of the whales, dolphins and even-toed hoofed mammals (Cetartiodactyla). Biol. Rev. 2005, 80, 445–473. [Google Scholar] [CrossRef] [PubMed]
- Hassanin, A.; Delsuc, F.; Ropiquet, A.; Hammer, C.; van Vuuren, B.J.; Matthee, C.; Ruiz-Garcia, M.; Catzeflis, F.; Areskoug, V.; Nguyen, T.T.; et al. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. C. R. Biol. 2012, 335, 32–50. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.J.; Menninger, J.C.; Nash, W.G. Atlas of Mammalian Chromosomes; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Chi, J.; Fu, B.; Nie, W.; Wang, J.; Graphodatsky, A.S.; Yang, F. New insights into the karyotypic relationships of Chinese muntjac (Muntiacus reevesi), forest musk deer (Moschus berezovskii) and gayal (Bos frontalis). Cytogenet. Genome Res. 2005, 108, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Müller, S.; Just, R.; Ferguson-Smith, M.A.; Wienberg, J. Comparative chromosome painting in mammals: human and the Indian muntjac (Muntiacus muntjak vaginalis). Genomics 1997, 39, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; O’Brien, P.C. M.; Wienberg, J.; Ferguson-Smith, M.A. Evolution of the black muntjac (Muntiacus crinifrons) karyotype revealed by comparative chromosome painting. Cytogenet. Genome Res. 1997, 76, 159–163. [Google Scholar] [CrossRef]
- Yang, F.; O’Brien, P.C. M.; Wienberg, J.; Neitzel, H.; Lin, C.C.; Ferguson-Smith, M.A. Chromosomal evolution of the Chinese muntjac (Muntiacus reevesi). Chromosoma 1997, 106, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Kingdon, J. The Kingdon Field Guide to African Mammals; Bloomsbury Publishing: London, UK, 2015. [Google Scholar]
- Buckland, R.A.; Evans, H.J. Cytogenetic aspects of phylogeny in the Bovidae. Cytogenet. Genome Res. 1978, 21, 42–63. [Google Scholar] [CrossRef]
- Vozdova, M.; Ruiz-Herrera, A.; Fernandez, J.; Cernohorska, H.; Frohlich, J.; Sebestova, H.; Kubickova, S.; Rubes, J. Meiotic behaviour of evolutionary sex-autosome translocations in Bovidae. Chromosome Res. 2016, 24, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Iannuzzi, L. Standard karyotype of the river buffalo (Bubalus bubalis L., 2n= 50). Report of the committee for the standardization of banded karyotypes of the river buffalo. Cytogenet. Cell Genet. 1994, 67, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.C.; Benirschke, K. An atlas of Mammalian Chromosomes; Springer Science & Business Media: Berlin, Germany, 2013; Volume 10. [Google Scholar]
- Kingswood, S.C.; Kumamoto, A.T. Madoqua kirkii. Mamm. Species 1997, 1–10. [Google Scholar] [CrossRef]
- Pevzner, P.; Tesler, G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 7672–7677. [Google Scholar] [CrossRef] [PubMed]
- Larkin, D.M.; Pape, G.; Donthu, R.; Auvil, L.; Welge, M.; Lewin, H.A. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 2009, 19, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Rubtsov, N.B.; Rubtsova, N.V.; Anopriyenko, O.V.; Karamysheva, T.V.; Shevchenko, A.I.; Mazurok, N.A.; Nesterova, T.B.; Zakian, S.M. Reorganization of the X chromosome in voles of the genus Microtus. Cytogenet. Genome Res. 2002, 99, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Yamada, F.; Hashimoto, T.; Abe, S.; Matsuda, Y.; Kuroiwa, A. Centromere repositioning in the X chromosome of XO/XO mammals, Ryukyu spiny rat. Chromosome Res. 2008, 16, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Gladkikh, O.L.; Romanenko, S.A.; Lemskaya, N.A.; Serdyukova, N.A.; O’Brien, P.C.; Kovalskaya, J.M.; Smorkatcheva, A.V.; Golenishchev, F.N.; Perelman, P.L.; Trifonov, V.A.; et al. Rapid Karyotype Evolution in Lasiopodomys Involved at Least Two Autosome–Sex Chromosome Translocations. PLoS ONE 2016, 11, e0167653. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, T.; Sung, S.; Lee, C.; Kim, H. Reanalysis of Ohno’s hypothesis on conservation of the size of the X chromosome in mammals. Anim. Cells Syst. 2012, 16, 438–446. [Google Scholar] [CrossRef]
- Murphy, W.J.; Stanyon, R.; O’Brien, S.J. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol. 2001, 2, reviews0005.1–reviews0005.8. [Google Scholar] [CrossRef]


| Scientific Name, Abbreviation | Code | Common Name | Family | Diploid Number | Source of Cell Line |
|---|---|---|---|---|---|
| Sus scrofa | SSC | Pig | Suidae | 38, XX | IMCB SB RAS, Novosibirsk-1* |
| Lama pacos | LPA | Alpaca | Camelidae | 74, XY | 2* |
| Eschrihtius robustus | ERO | Gray whale | Eschrichtiidae (Cetacea) | 44, XY | [11] |
| Hippopotamus amphibius | HAM | Common hippopotamus | Hippopotamidae | 36, XY | [8] |
| Tragulus javanicus | TJA | Java mouse-deer | Tragulidae | 32, XY | Frozen Zoo (San Diego Zoo’s Conservation Research, San Diego, CA, USA) |
| Antilocapra americana | AAM | Pronghorn | Antilocapridae | 58, XY | [10] |
| Giraffa camelopardalis | GCA | Giraffe | Giraffidae | 30, XY | [8] |
| Moschus moschiferus | MMO | Siberian musk deer | Moschidae | 58, XY | [8] |
| Dama dama | DDA | Fallow deer | Cervidae, Cervinae | 68, XX | Catoctin Wildlife Preserve and Zoo, Maryland, USA |
| Elaphurus davidianus | EDA | Pere David’s deer | 68, XX | 3* | |
| Alces alces | AAL | Eurasian elk | Cervidae, Capreolinae | 68, XX | IMCB SB RAS, Novosibirsk |
| Capreolus pygargus | CPY | Siberian roe deer | 70, XX | IMCB SB RAS, Novosibirsk | |
| Ovibos moschatus | OMO | Muskox | Bovidae, Antilopinae | 48, XX | IMCB SB RAS, Novosibirsk |
| Capra hircus | CHI | Goat | 60, XX | Catoctin Wildlife Preserve and Zoo, Maryland, USA | |
| Ovis aries musimon | OAR | Sheep | 54, XX | Catoctin Wildlife Preserve and Zoo, Maryland, USA | |
| Hippotragus niger | HNI | Sable antelope | 60, XX | 3* | |
| Bison bison | BBI | American bison | Bovidae, Bovinae | 60, XX | 4* |
| Bos taurus | BTA | Cattle | 60, XX | IMCB SB RAS, Novosibirsk |
| No. | BAC’s Order and Localization on Cattle X Chromosome | CHORI (CH-240) BACs Localization on Cetartiodactyl X Chromosomes | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Domestic Pig, SSC | Alpaca, LPA | Gray Whale, ERO | Common Hippopota-mus, HAM | Java Mouse-Deer, TJA | Pronghorn, AAM | Giraffe, GCA | Siberian Roe Deer, CPY | Eurasian Elk, AAL | Fallow Deer, DDA | Pere David’s Deer, EDA | Muskox, OMO | Goat, CHI | Sheep, OAR | Sable Antelope, HNI | ||||
| 1 | X syntenic block 2 (XSB2) | CH240-514O22 | Start 1949353, End 2129088 | 66H2 | 66H2 | 66H2 | 66H2 | 66H2 | 108D16 | 386M8 | 386M8 | 386M8 | 93K24 | 514O22 | 66H2 | 66H2 | 66H2 | 66H2 |
| 2 | CH240-287O21 | Start 7324034, End 7488466 | 155A13 | 155A13 | 155A13 | 155A13 | 155A13 | 54D24 | 103E10 | 103E10 | 103E10 | 122N13 | 287O21 | 155A13 | 155A13 | 155A13 | 155A13 | |
| 3 | CH240-128C9 | Start 8233624, End 8391009 | 90L14 | 90L14 | 90L14 | 90L14 | 90L14 | 93K24 | 229I15 | 229I15 | 229I15 | 195J23 | 128C9 | 90L14 | 90L14 | 90L14 | 90L14 | |
| 4 | CH240-106A3 | Start 13345128, End 13540519 | 373L23 | 373L23 | 373L23 | 373L23 | 373L23 | 122N13 | 106A3 | 106A3 | 106A3 | 316D2 | 106A3 | 373L23 | 373L23 | 373L23 | 373L23 | |
| 5 | CH240-229I15 | Start 13805346, End 13950311 | 62M10 | 62M10 | 62M10 | 62M10 | 62M10 | 195J23 | 128C9 | 128C9 | 128C9 | 386M8 | 229I15 | 62M10 | 62M10 | 62M10 | 62M10 | |
| 6 | CH240-103E10 | Start 20150516, End 20286173 | 122P17 | 122P17 | 122P17 | 122P17 | 122P17 | 316D2 | 287O21 | 287O21 | 287O21 | 103E10 | 103E10 | 122P17 | 122P17 | 122P17 | 122P17 | |
| 7 | CH240-386M8 | Start 33395588, End 33587168 | 252G15 | 252G15 | 252G15 | 252G15 | 252G15 | 514O22 | 514O22 | 514O22 | 514O22 | 229I15 | 386M8 | 252G15 | 252G15 | 252G15 | 252G15 | |
| 8 | X syntenic block 3 (XSB3) | CH240-108D16 | Start 48672324, End 48917704 | 375C5 | 375C5 | 375C5 | 375C5 | 375C5 | 287O21 | 316D2 | 316D2 | 316D2 | 106A3 | 108D16 | 375C5 | 375C5 | 375C5 | 375C5 |
| 9 | CH240-54D24 | Start 53219586, End 53351583 | 130I15 | 130I15 | 130I15 | 130I15 | 130I15 | 128C9 | 195J23 | 195J23 | 195J23 | 229I15 | 54D24 | 130I15 | 130I15 | 130I15 | 130I15 | |
| 10 | CH240-93K24 | Start 57734547, End 57947720 | 118P13 | 118P13 | 118P13 | 118P13 | 118P13 | 106A3 | 122N13 | 122N13 | 122N13 | 287O21 | 93K24 | 118P13 | 118P13 | 118P13 | 118P13 | |
| 11 | CH240-122N13 | Start 62228039, End 62371946 | 25P8 | 25P8 | 25P8 | 25P8 | 25P8 | 229I15 | 93K24 | 93K24 | 93K24 | 514O22 | 122N13 | 25P8 | 25P8 | 25P8 | 25P8 | |
| 12 | CH240-195J23 | Start 62982639, End 63183460 | 14O10 | 14O10 | 14O10 | 14O10 | 14O10 | 103E10 | 54D24 | 54D24 | 54D24 | 54D24 | 195J23 | 14O10 | 14O10 | 14O10 | 14O10 | |
| 13 | CH240-316D2 | Start 68490278, End 68678635 | 214A3 | 214A3 | 214A3 | 214A3 | 214A3 | 386M8 | 108D16 | 108D16 | 108D16 | 108D16 | 316D2 | 214A3 | 214A3 | 214A3 | 214A3 | |
| 14 | X syntenic block 1 (XSB1) | CH240-214A3 | Start 84397606, End 84521707 | 108D16 | 108D16 | 108D16 | 108D16 | 316D2 | 214A2 | 214A3 | 214A3 | 214A3 | 214A3 | 214A3 | 386M8 | 386M8 | 386M8 | 386M8 |
| 15 | CH240-14O10 | Start 85224265, End 85389684 | 54D24 | 54D24? | 54D24 | 54D24 | 195J23 | 14O9 | 14O10 | 14O10 | 14O10 | 14O10 | 14O10 | 103E10 | 103E10 | 103E10 | 103E10 | |
| 16 | CH240-25P8 | Start 90681870, End 90861947 | 93K24 | 93K24 | 93K24 | 93K24 | 122N13 | 25P7 | 25P8 | 25P8 | 25P8 | 25P8 | 25P8 | 128C9 | 128C9 | 128C9 | 229I15 | |
| 17 | CH240-118P13 | Start 92264186, End 92429310 | 122N13 | 122N13 | 122N13 | 122N13 | 93K24 | 118P12 | 118P13 | 118P13 | 118P13 | 118P13 | 118P13 | 106A3 | 106A3 | 106A3 | 106A3 | |
| 18 | CH240-130I15 | Start 95938488, End 96135558 | 195J23 | 195J23 | 195J23 | 195J23 | 54D24 | 130I14 | 130I15 | 130I15 | 130I15 | 130I15 | 130I15 | 229I15 | 229I15 | 229I15 | 128C9 | |
| 19 | CH240-375C5 | Start 103959199, End 104119579 | 316D2 | 316D2 | 316D2 | 316D2 | 514O22 | 375C4 | 375C5 | 375C5 | 375C5 | 375C5 | 375C5 | 287O21 | 287O21 | 287O21 | 287O21 | |
| 20 | CH240-252G15 | Start 108195394, End 108349350 | 514O22 | 514O22 | 514O22 | 514O22 | 287O21 | 252G14 | 252G15 | 252G15 | 252G15 | 252G15 | 252G15 | 514O22 | 514O22 | 514O22 | 514O22 | |
| 21 | CH240-122P17 | Start 110284444, End 110450903 | 287O21 | 287O21 | 287O21 | 287O21 | 128C9 | 122P16 | 122P17 | 122P17 | 122P17 | 122P17 | 122P17 | 316D2 | 316D2 | 316D2 | 316D2 | |
| 22 | CH240-62M10 | Start 111125731, End 111275450 | 128C9? | 128C9 | 128C9 | 128C9? | 106A3 | 62M9 | 62M10 | 62M10 | 62M10 | 62M10 | 62M10 | 195J23 | 195J23 | 195J23 | 195J23 | |
| 23 | CH240-373L23 | Start 117191008, End 117371368 | 106A3 | 106A3 | 106A3 | 106A3 | 229I15 | 373L22 | 373L23 | 373L23 | 373L23 | 373L23 | 373L23 | 122N13 | 122N13 | 122N13 | 122N13 | |
| 24 | CH240-90L14 | Start 126821940, End 127050706 | 229I15 | 229I15 | 229I15 | 229I15 | 108D16 | 90L13 | 90L14 | 90L14 | 90L14 | 90L14 | 90L14 | 93K24 | 93K24 | 93K24 | 93K24 | |
| 25 | CH240-155A13 | Start 128339848, End 128504608 | 103E10 | 103E10 | 103E10 | 103E9 | 103E10 | 155A12 | 155A13 | 155A13 | 155A13 | 155A13 | 155A13 | 54D24 | 54D24 | 54D24 | 54D24 | |
| 26 | CH240-66H2 | Start 141101222, End 141358968 | 386M8 | 386M8 | 386M8 | 386M7 | 386M8 | 66H1 | 66H2 | 66H2 | 66H2 | 66H2 | 66H2 | 108D16 | 108D16 | 108D16 | 108D16 | |
| No. | Laurasiatheria | Euarchontoglires | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BAC Clones in Cattle Genome | BAC Clones in Sheep Genome | BAC Clones in Horse Genome) | BAC Clones in Human Genome | BAC Clones in Mouse Genome | BAC Clones in Rat Genome | |||||||
| 1 | 514O22 | Start 1949353 | 66H2 | Start 10045822 | 66H2 | Start 8367618 | 66H2 | Start 12497685 | 118P13 | Start 7554450 | 25P8 | Start 1711095 |
| End 2129088 | End 10306770 | End 8624882 | End 12794877 | End 7697987 | End 1907049 | |||||||
| 2 | 287O21 | Start 7324034 | 155A13 | Start 19299853 | 155A13 | Start 16543677 | 155A13 | Start 22069138 | 62M10 | Start 9209615 | 375C5 | Start 4672236 |
| End 7488466 | End 19464920 | End 16698622 | End 22228453 | End 9317028 | End 4863802 | |||||||
| 3 | 128C9 | Start 8233624 | 373L23 | Start 28630482 | 373L23 | Start 24698243 | 373L23 | Start 31328065 | 122P17 | Start 10195810 | 252G15 | Start 10936630 |
| End 8391009 | End 28810179 | End 24857300 | End 31509266 | End 10370080 | End 11107682 | |||||||
| 4 | 106A3 | Start 13345128 | 62M10 | Start 34891275 | 62M10 | Start 30220096 | 62M10 | Start 31328065 | 252G15 | Start 12644301 | 122P17 | Start 13483272 |
| End 13540519 | End 35037294 | End 30342071 | End 31509266 | End 12803364 | End 14335671 | |||||||
| 5 | 229I15 | Start 13805346 | 122P17 | Start 35738657 | 122P17 | Start 30907267 | 122P17 | Start 38298814 | 375C5 | Start 18235010 | 62M10 | Start 14415064 |
| End 13950311 | End 35910824 | End 31039631 | End 38458494 | End 18480200 | End 14541523 | |||||||
| 6 | 103E10 | Start 20150516 | 252G15 | Start 37830134 | 252G15 | Start 32879937 | 252G15 | Start 40611820 | 25P8 | Start 20507324 | 118P13 | Start 15650399 |
| End 20286173 | End 37981845 | End 33007527 | End 40767797 | End 20696050 | End 15784402 | |||||||
| 7 | 386M8 | Start 33395588 | 375C5 | Start 41973255 | 375C5 | Start 36512266 | 375C5 | Start 45036869 | 514O22 | Start 23213727 | 130I15 | Start 22235385 |
| End 33587168 | End 42128838 | End 36698919 | End 45234319 | End 23316229 | End 22435973 | |||||||
| 8 | 108D16 | Start 48672324 | 130I15 | Start 49649383 | 25P8 | Start 38190847 | 25P8 | Start 47047149 | 287O21 | Start 41535889 | 66H2 | Start 27957571 |
| End 48917704 | End 49847996 | End 38327897 | End 47226311 | End 41677049 | End 28439737 | |||||||
| 9 | 54D24 | Start 53219586 | 118P13 | Start 52564228 | 118P13 | Start 39580949 | 118P13 | Srart 49122932 | 128C9 | Start 42491010 | 155A13 | Start 40510641 |
| End 53351583 | End 52727917 | End 39734268 | End 49608099 | End 42653374 | End 40710667 | |||||||
| 10 | 93K24 | Start 57734547 | 25P8 | Start 54170178 | 130I15 | Start 44962739 | 130I15 | Start 53053920 | 106A3 | Start 47802786 | 373L23 | Start 53052665 |
| End 57947720 | End 54331345 | End 45135718 | End 53291737 | End 48012093 | End 53277814 | |||||||
| 11 | 122N13 | Start 62228039 | 14O10 | Start 59810734 | 14O10 | Start 52316685 | 14O10 | Start 70333575 | 229I15 | Start 48279488 | 14O10 | Start 70503930 |
| End 62371946 | End 59977176 | End 52471298 | End 70530493 | End 48451406 | End 70671925 | |||||||
| 12 | 195J23 | Start 62982639 | 214A3 | Start 60702841 | 214A3 | Start 53269385 | 214A3 | Start 71438703 | 103E10 | Start 57106307 | 214A3 | Start 71468323 |
| End 63183460 | End 60821565 | End 53389760 | End 71567090 | End 57244888 | End 71575467 | |||||||
| 13 | 316D2 | Start 68490278 | 386M8 | Start 80094458 | 108D16 | Start 76549898 | 108D16 | Start 97540872 | 386M8 | Start 71145260 | 108D16 | Start 100451494 |
| End 68678635 | End 80283568 | End 76701833 | End 97704621 | End 71388925 | End 100747362 | |||||||
| 14 | 214A3 | Start 84397606 | 103E10 | Start 93391997 | 93K24 | Start 81725518 | 54D24 | Start 103662230 | 373L23 | Start 84771898 | 93K24 | Start 107378470 |
| End 84521707 | End 93531761 | End 81893517 | End 103838933 | End 84970050 | End 107552526 | |||||||
| 15 | 14O10 | Start 85224265 | 287O21 | Start 101734836 | 54D24 | Start 83362409 | 93K24 | Start 105651553 | 14O10 | Start 100669857 | 54D24 | Start 109470944 |
| End 85389684 | End 101892691 | End 83480210 | End 105782858 | End 100840304 | End 109865654 | |||||||
| 16 | 25P8 | Start 90681870 | 128C9 | Start 102635193 | 122N13 | Start 86318687 | 122N13 | Start 109356460 | 214A3 | Start 101583273 | 122N13 | Start 113344277 |
| End 90861947 | End 102791210 | End 86434114 | End 109486477 | End 101676469 | End 113475228 | |||||||
| 17 | 118P13 | Start 92264186 | 106A3 | Start 107701336 | 195J23 | Start 86961327 | 195J23 | Start 110108649 | 108D16 | Start 130409135 | 195J23 | Start 114041201 |
| End 92429310 | End 107885819 | End 87148727 | End 110310972 | End 130602938 | End 114226114 | |||||||
| 18 | 130I15 | Start 95938488 | 229I15 | Start 108152302 | 316D2 | Start 89122764 | 316D2 | Start 112670755 | 93K24 | Start 136717423 | 316D2 | Start 116629155 |
| End 96135558 | End 108300381 | End 89285710 | End 112844328 | End 136874331 | End 116812891 | |||||||
| 19 | 375C5 | Start 103959199 | 514O22 | Start 111218064 | 514O22 | Start 93641183 | 514O22 | Start 117884744 | 54D24 | Start 138738349 | 514O22 | Start 121570459 |
| End 104119579 | End 111402064 | End 93800197 | End 118065524 | End 138862889 | End 121690157 | |||||||
| 20 | 252G15 | Start 108195394 | 316D2 | Start 115697759 | 287O21 | Start 97957842 | 287O21 | Start 123321223 | 122N13 | Start 142065201 | 287O21 | Start 127687918 |
| End 108349350 | End 115869258 | End 98099669 | End 123489384 | End 142200659 | End 127828202 | |||||||
| 21 | 122P17 | Start 110284444 | 195J23 | Start 118153613 | 128C9 | Start 98755966 | 128C9 | Start 124334066 | 195J23 | Start 142770604 | 128C9 | Start 128883616 |
| End 110450903 | End 118356580 | End 98902048 | End 124491573 | End 142959710 | End 129042712 | |||||||
| 22 | 62M10 | Start 111125731 | 122N13 | Start 118952818 | 106A3 | Start 102828316 | 106A3 | Start 129437403 | 316D2 | Start 145340226 | 106A3 | Start 134638127 |
| End 111275450 | End 119094106 | End 103004015 | End 129628777 | End 145507925 | End 134841751 | |||||||
| 23 | 373L23 | Start 117191008 | 93K24 | Start 122240643 | 229I15 | Start 103242991 | 229I15 | Start 129926486 | 130I15 | Start 152167108 | 229I15 | Start 135116351 |
| End 117371368 | End 122443675 | End 103384683 | End 130107731 | End 152363913 | End 135281818 | |||||||
| 24 | 155A13 | Start 128339848 | 54D24 | Start 124299513 | 103E10 | Start 108619397 | 103E10 | Start 136556505 | 155A13 | Start 157177353 | 103E10 | Start 159580103 |
| End 128504608 | End 124431515 | End 108750001 | End 136681177 | End 157384607 | End 159734497 | |||||||
| 25 | 66H2 | Start 141101222 | 108D16 | Start 129843594 | 386M8 | Start 119476270 | 386M8 | Start 150502313 | 66H2 | Start 167378561 | ||
| End 141358968 | End 130091258 | End 119683931 | End 150728735 | End 167730488 | ||||||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Proskuryakova, A.A.; Kulemzina, A.I.; Perelman, P.L.; Makunin, A.I.; Larkin, D.M.; Farré, M.; Kukekova, A.V.; Lynn Johnson, J.; Lemskaya, N.A.; Beklemisheva, V.R.; et al. X Chromosome Evolution in Cetartiodactyla. Genes 2017, 8, 216. https://doi.org/10.3390/genes8090216
Proskuryakova AA, Kulemzina AI, Perelman PL, Makunin AI, Larkin DM, Farré M, Kukekova AV, Lynn Johnson J, Lemskaya NA, Beklemisheva VR, et al. X Chromosome Evolution in Cetartiodactyla. Genes. 2017; 8(9):216. https://doi.org/10.3390/genes8090216
Chicago/Turabian StyleProskuryakova, Anastasia A., Anastasia I. Kulemzina, Polina L. Perelman, Alexey I. Makunin, Denis M. Larkin, Marta Farré, Anna V. Kukekova, Jennifer Lynn Johnson, Natalya A. Lemskaya, Violetta R. Beklemisheva, and et al. 2017. "X Chromosome Evolution in Cetartiodactyla" Genes 8, no. 9: 216. https://doi.org/10.3390/genes8090216






