Responsive Proteins in Wheat Cultivars with Contrasting Nitrogen Efficiencies under the Combined Stress of High Temperature and Low Nitrogen
Abstract
:1. Introduction
2. Materials and Methods
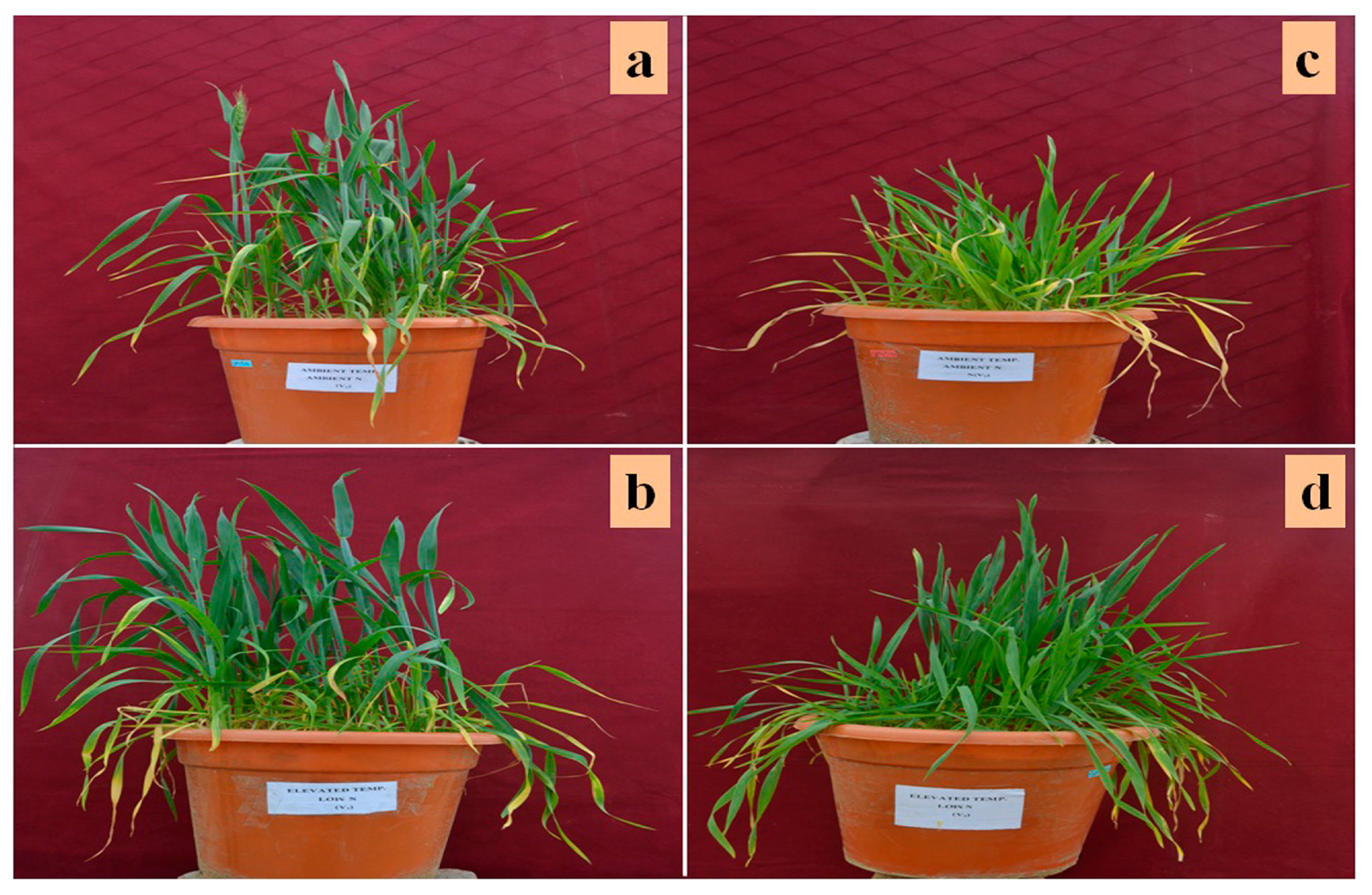
2.1. Plant Culture and Treatments
2.2. Protein Extraction
2.3. Two-Dimensional Gel Electrophoresis
2.4. Gel Analysis
2.5. In-Gel Digestion and Protein Identification
2.6. Statistical Analyses
3. Results
3.1. Protein Profiling and Spatial Categorization of Differential Protein Expression
3.2. Functional Cataloguing and Expression-Profile Analysis
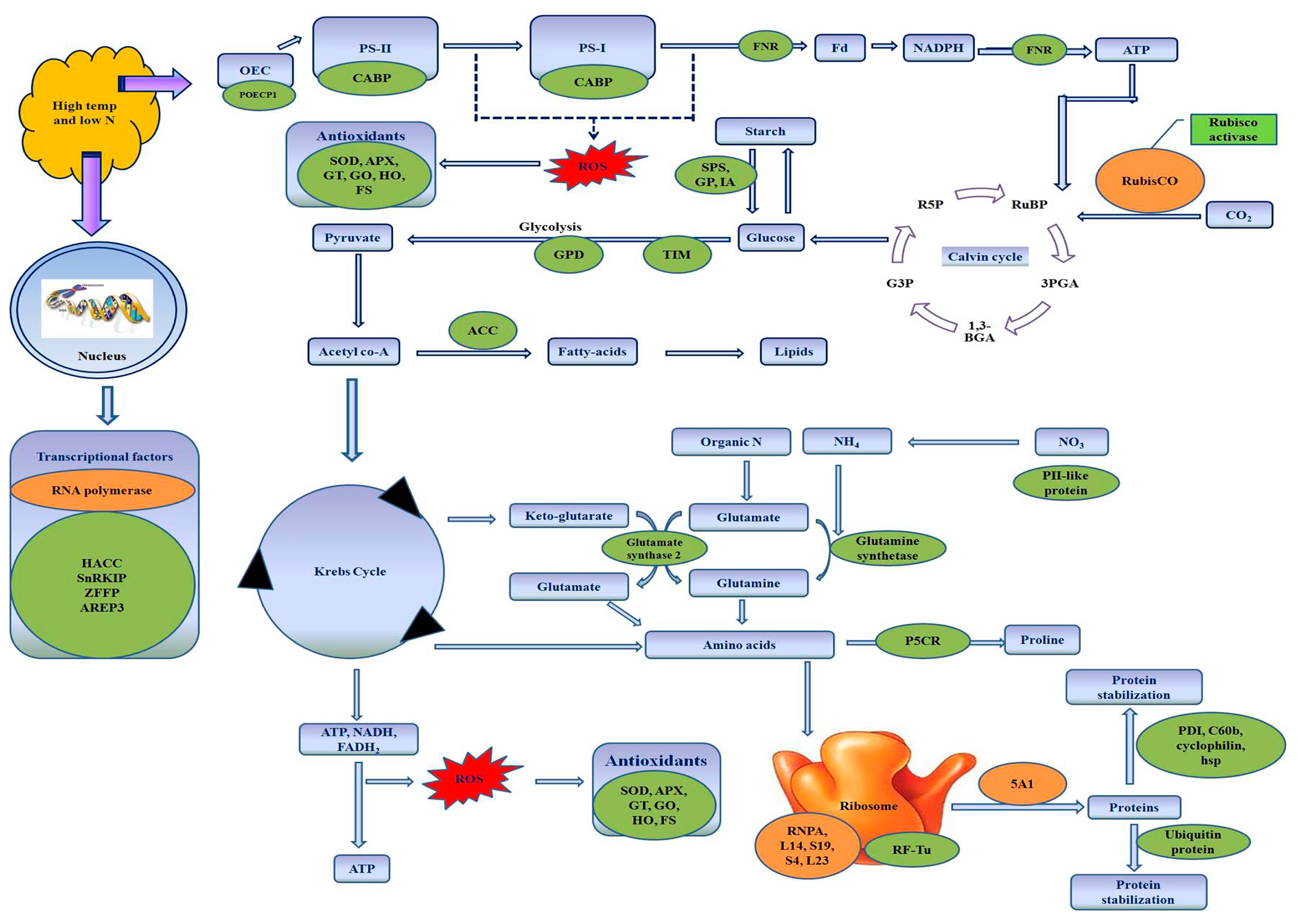
4. Discussion
4.1. Proteins Related to Nitrogen Metabolism
4.2. Proteins Related to Photosynthesis
4.3. Proteins Associated with Osmoregulation
4.4. Proteins Related to Antioxidant Defense System
4.5. Proteins Related to Carbohydrate Metabolism
4.6. Proteins Related to Energy Metabolism and Fatty Acid Metabolism
4.7. Proteins Related to Transcription and Protein Synthesis
4.8. Changes in Proteins Involved in Protein Stabilization and Degradation
4.9. Unclassified Proteins
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fischer, R.; Edmeades, G.O. Breeding and cereal yield progress. Crop Sci. 2010, 50, 85–98. [Google Scholar] [CrossRef]
- Zou, J.; Liu, C.; Chen, X. Proteomics of rice in response to heat stress and advances in genetic engineering for heat tolerance in rice. Plant Cell Rep. 2011, 30, 2155–2165. [Google Scholar] [CrossRef] [PubMed]
- Madan, P.; Jagadish, S.V.; Craufurd, P.Q.; Fitzgerald, M.; Lafarge, T.; Wheeler, T.R. Effect of elevated CO2 and high temperature on seed-set and grain quality of rice. J. Exp. Bot. 2012, 63, 3843–3852. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, B.; Rasmussen, A.; Porter, J.R. Temperatures and the growth and development of maize and rice, a review. Glob. Chang. Biol. 2014, 20, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Intergovernmental Panel on Climate Change. Climate Change 2007: The Physical Science Basis; Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2007; p. 996. [Google Scholar]
- Intergovernmental Panel on Climate Change. Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part A, Global and Sectoral Aspects; Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2014; p. 1132. [Google Scholar]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment, scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glass, A.D.M. Nitrogen use efficiency of crop plants, physiological constraints upon nitrogen absorption. Crit. Rev. Plant Sci. 2003, 22, 453–470. [Google Scholar] [CrossRef]
- Good, A.G.; Shrawat, A.K.; Muench, D.G. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 2004, 9, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.; Bonnett, D.; Chapman, S.C.; Furbank, R.T.; Manes, Y.; Mather, D.E.; Parry, M.A.J. Raising yield potential of wheat. I. Overview of a consortium approach and breeding strategies. J. Exp. Bot. 2011, 62, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Parry, M.A.J.; Hawkesford, M.J. An integrated approach to crop genetic improvement. J. Integr. Plant Biol. 2012, 54, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era, past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Chew, Y.H.; Halliday, K.J. A stress-free walk from Arabidopsis to crops. Curr. Opin. Biotechnol. 2011, 22, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; de Vos, R.C.H.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Mitler, R.; Finka, A.; Goloubinoff, P. How do plants feel the heat? Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, P.Y.; Ahmad, A.; Hemant Ganie, A.H.; Iqbal, M. Potassium and calcium application ameliorates growth and oxidative homeostasis in salt-stressed Indian mustard plants. Pak. J. Bot. 2015, 47, 1629–1639. [Google Scholar]
- Yousuf, P.Y.; Ahmad, A.; Aref, I.M.; Ozturk, M.; Hemant Ganie, A.H.; Iqbal, M. Salt-stress-responsive chloroplast proteins in Brassica juncea genotypes with contrasting salt tolerance and their quantitative PCR analysis. Protoplasma 2016, 253, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Larkindale, J.; Vierling, E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008, 146, 748–761. [Google Scholar] [CrossRef] [PubMed]
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M.R. Gene expression profiling of plants under salt stress. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Li, H.; Payne, W.A.; Michels, G.J.; Charles, M.R. Reducing plant abiotic and biotic stress, drought and attacks of greenbugs, corn leaf aphids and virus disease in dryland sorghum. Environ. Exp. Bot. 2008, 63, 305–316. [Google Scholar] [CrossRef]
- Yousuf, P.Y.; Ahmad, A.; Ganie, A.H.; Iqbal, M. Salt stress-induced modulations in the shoot proteome of Brassica juncea genotypes. Environ. Sci. Pollut. Res. 2016, 23, 2391–2401. [Google Scholar] [CrossRef] [PubMed]
- Leff, B.; Ramankutty, N.; Foley, J.A. Geographic distribution of major crops across the world. Glob. Biogeochem. Cycles 2004, 18, GB1009. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. FAOSTAT, Food Balance Sheets. 2012. Available online: faostat.fao.org (accessed on 15 April 2013).
- Gourdji, S.M.; Sibley, A.M.; Lobell, D.B. Global crop exposure to critical high temperatures in the reproductive period, historical trends and future projections. Environ. Res. Lett. 2013, 8, 24–41. [Google Scholar] [CrossRef]
- Koehler, A.K.; Challinor, A.J.; Hawkins, E.; Asseng, S. Influences of increasing temperature on Indian wheat, quantifying limits to predictability. Environ. Res. Lett. 2013, 8, 034016. [Google Scholar] [CrossRef]
- Teixeira, E.I.; Fischer, G.; van Velthuizen, H.; Walter, C.; Ewert, F. Global hot-spots of heat stress on agricultural crops due to climate change. Agric. For. Meteorol. 2013, 170, 206–215. [Google Scholar] [CrossRef]
- Kharel, T.P.; Clay, D.E.; Clay, S.A.; Beck, D.; Reese, C.; Carlson, G.; Park, H. Nitrogen and water stress affect winter wheat yield and dough quality. Agron. J. 2011, 103, 1389–1396. [Google Scholar] [CrossRef]
- Asseng, S.; Ewert, F.; Martre, P.; Rötter, R.P.; Lobell, D.B.; Cammarano, D.; Kimball, B.A.; Ottman, M.J.; Wall, G.W.; White, J.W.; et al. Rising temperatures reduce global wheat production. Nat. Clim. Chang. 2015, 5, 143–147. [Google Scholar] [CrossRef]
- Tjoelker, M.G.; Reich, P.B.; Oleksyn, J. Changes in leaf nitrogen and carbohydrates underlie temperature and CO2 acclimation of dark respiration in five boreal tree species. Plant Cell Environ. 1999, 22, 767–778. [Google Scholar] [CrossRef]
- Semenov, M.A.; Jamieson, P.D.; Martre, P. Deconvoluting nitrogen use efficiency in wheat: A simulation study. Eur. J. Agron. 2007, 26, 283–294. [Google Scholar] [CrossRef]
- Chandna, R.; Kaur, G.; Iqbal, M.; Khan, I.; Ahmad, A. Differential response of wheat genotypes to applied nitrogen, biochemical and molecular analysis. Arch. Agron. Soil Sci. 2012, 58, 915–929. [Google Scholar] [CrossRef]
- Isaacson, T.; Damasceno, C.M.; Saravanan, R.S.; He, Y.; Catala, C.; Saladie, M.; Rose, J.K. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 2006, 1, 769–774. [Google Scholar] [CrossRef] [PubMed]
- O’Farrel, P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar]
- National Center for Biotechnology Information (NCBI). Available online: https://www.ncbi.nlm.nih.gov (accessed on 14 November 2016).
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar]
- Guda, C. pTARGET: A web server for predicting protein subcellular localization. Nucleic Acids Res. 2006, 34, W210–W213. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Meng, Q.; Zhao, R.; Li, F.; Chen, X.; Zhang, F.; Cui, Z. Critical nitrogen dilution curve for optimizing nitrogen management of winter wheat production in the north China plain. Agron. J. 2012, 104, 523–529. [Google Scholar] [CrossRef]
- Christophe, S.; Jean-Christophe, A.; Annabelle, L.; Alain, O.; Marion, P.; Anne-Sophie, V. Plant N fluxes and modulation by nitrogen, heat and water stresses: A review based on comparison of legumes and non-legume plants. In Abiotic Stress in Plants—Mechanisms and Adaptations; Shanker, A., Venkateswarlu, B., Eds.; InTech: Rijeka, Croatia, 2011. [Google Scholar]
- Huergo, L.F.; Chandra, G.; Merrick, M. PII signal transduction proteins, nitrogen regulation and beyond. FEMS Microbiol. Rev. 2013, 37, 251–283. [Google Scholar] [CrossRef] [PubMed]
- Kichey, T.; Le Gouis, J.; Sangwan, B.; Hirel, B.; Dubois, F. Changes in the cellular and subcellular localization of glutamine synthetase and glutamate dehydrogenase during flag leaf senescence in wheat (Triticum aestivum L.). Plant Cell Physiol. 2005, 46, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, T.; Gaufichon, L.; Boutet-Mercey, S.; Christ, A.; Masclaux-Daubresse, C. Enzymatic and metabolic diagnostic of nitrogen deficiency in Arabidopsis thaliana Wassileskija accession. Plant Cell Physiol. 2008, 49, 1056–1065. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Bi, Y.; Rothstein, S.J. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 2011, 62, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Aranjuelo, I.; Molero, G.; Erice, G.; Avice, J.C.; Nogues, S. Plant physiology and proteomics reveals the leaf response to drought in alfalfa (Medicago sativa L.). J. Exp. Bot. 2011, 62, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, H.; Tanaka, Y.; Hibino, T.; Hayashi, Y.; Tanaka, A.; Takabe, T.; Takabe, T. Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol. Biol. 2000, 43, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Pascual, M.B.; Jing, Z.P.; Kirby, E.G.; Canovas, F.M.; Gallardo, F. Response of transgenic poplar overexpressing cytosolic glutamine synthetase to phosphinothricin. Phytochemistry 2008, 69, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Liu, R.; Yan, L.; Liu, Z.Q.; Jiang, S.C.; Shen, Y.Y.; Wang, X.F.; Zhang, D.P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2012, 63, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, P.Y.; Ahmad, A.; Ganie, A.H.; Sareer, O.; Krishnapriya, V.; Aref, I.M.; Iqbal, M. Antioxidant response and proteomic modulations in Indian mustard grown under salt stress. Plant Growth Regul. 2016, 81, 31–50. [Google Scholar] [CrossRef]
- Yan, S.; Tang, Z.; Su, W.; Sun, W. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 2005, 5, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Rollins, J.A.; Habte, E.; Templer, S.E.; Colby, T.; Schmidt, J.; von Korff, M. Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeum vulgare L.). J. Exp. Bot. 2013, 64, 3201–3212. [Google Scholar] [CrossRef] [PubMed]
- Kosova, K.; Vitamvas, P.; Prasil, I.T. Proteomics of stress responses in wheat and barley—search for potential protein markers of stress tolerance. Front. Plant Sci. 2014, 5, 711. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.C.; Peng, Z.Y.; Li, C.L.; Li, F.; Liu, C.; Xia, G.M. Proteomic analysis on a high salt tolerance introgression strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 2008, 8, 1470–1489. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.P.; Sauve, R.J.; Liu, Z.; Reddy, S.; Bhatti, S.; Hucko, S.D.; Fish, T.; Thannhauser, T.W. Identification of salt-induced changes in leaf and root proteomes of the wild tomato, Solanum chilense. J. Am. Soc. Hortic. Sci. 2011, 136, 288–302. [Google Scholar]
- Hanke, G.T.; Okutani, S.; Satomi, Y.; Takao, T.; Suzuki, A.; Hase, T. Multiple iso-proteins of FNR in Arabidopsis, evidence for different contributions to chloroplast function and nitrogen assimilation. Plant Cell Environ. 2005, 28, 1146–1157. [Google Scholar] [CrossRef]
- Chauhan, H.; Khurana, N.; Tyagi, A.K.; Khurana, J.P.; Khurana, P. Identification and characterization of high temperature stress responsive genes in bread wheat (Triticum aestivum L.) and their regulation at various stages of development. Plant Mol. Biol. 2011, 75, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Usha, K. Salicylic acid induced physiological and biochemical changes in wheat seedlings under water stress. Plant Growth Regul. 2003, 39, 137–141. [Google Scholar] [CrossRef]
- Portis, A.R. Rubisco activase—Rubisco’s catalytic chaperone. Photosynth. Res. 2003, 75, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Ahsan, N.; Lee, S.H.; Kang, K.Y.; Bahk, J.D.; Lee, I.J.; Lee, B.H. A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 2007, 7, 3369–3383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, G.; Huang, W.; Bi, T.; Chen, G.; Tang, Z.; Su, W.; Sun, W. Proteomic study of Carissa spinarum in response to combined heat and drought stress. Proteomics 2010, 10, 3117–3129. [Google Scholar] [CrossRef] [PubMed]
- Bahrman, N.; Le Gouis, J.; Negroni, L.; Amilhat, L.; Leroy, P.; Laine, A.L.; Jaminon, O. Differential protein expression assessed by two-dimensional gel electrophoresis for two wheat varieties grown at four nitrogen levels. Proteomics 2004, 4, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.M.; Mestre, T.C.; Mittler, R.; Rubio, F.; Garcia-Sanchez, F.; Martinez, V. The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant Cell Environ. 2014, 37, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Murahama, M.; Yoshida, T.; Hayashi, F.; Ichino, T.; Sanada, Y.; Wada, K. Purification and characterization of δ1-pyrroline-5-carboxylate reductase isoenzymes, indicating differential distribution in spinach (Spinacia oleracea L.) leaves. Plant Cell Physiol. 2001, 42, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Higuchi, M.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Kato, T.; Tabata, S.; Shinozaki, K.; Kakimoto, T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 2001, 409, 1060–1063. [Google Scholar] [CrossRef] [PubMed]
- Anderberg, R.J.; Walker-Simmons, M.K. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc. Natl. Acad. Sci. USA 1992, 89, 10183–10187. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Edwards, R. Glutathione Transferases. Arabidopsis Book 2010, 8, e0131. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Hasanuzzaman, M.; Fujita, M. Coordinate induction of antioxidant defense and glyoxalase system by exogenous proline and glycine betaine is correlated with salt tolerance in mung bean. Front. Agric. China 2011, 5, 1–14. [Google Scholar] [CrossRef]
- Yadav, S.K.; Singla-Pareek, S.L.; Ray, M.; Reddy, M.K.; Sopory, S.K. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 2005, 337, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, G.S.; Verma, K. Heme oxygenase (HO), an overlooked enzyme of plant metabolism and defence. J. Exp. Bot. 2010, 61, 2255–2270. [Google Scholar] [CrossRef] [PubMed]
- Fini, A.; Brunetti, C.; Di Ferdinando, M.; Ferrini, F.; Tattini, M. Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal Behav. 2011, 6, 709–711. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.R.; Kossmann, J.; Ritte, G. Leaf starch degradation comes out of the shadows. Trends Plant Sci. 2005, 10, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Sobhanian, H.; Aghaei, K.; Komatsu, S. Changes in the plant proteome resulting from salt stress, toward the creation of salt-tolerant crops? J. Proteom. 2011, 74, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Balderas, R.E.; Garcia-Ponce, B.; Rocha-Sosa, M. Hormonal and stress induction of the gene encoding common bean acetyl-coenzyme A carboxylase. Plant Physiol. 2006, 142, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Kottapalli, K.R.; Rakwal, R.; Shibato, J.; Burow, G.; Tissues, D.; Burke, J.; Puppala, N.; Burow, M.; Payton, P. Physiology and proteomics of the water-deficit stress response in three contrasting peanut genotypes. Plant Cell Environ. 2009, 32, 380–407. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, H.; Kiyotaka, U.; Ishibashi, Y.; Yamaguchi, M.; Hirata, K.; Demura, T.; Kato, K. A short period of mannitol stress but not LiCl stress led to global translational repression in plants. Biosci. Biotechnol. Biochem. 2010, 74, 2110–2112. [Google Scholar] [CrossRef] [PubMed]
- Sormani, R.; Delannoy, E.; Lageix, S.; Bitton, F.; Lanet, E.; Saez-Vasquez, J.; Deragon, J.M.; Renou, J.P.; Robaglia, C. Sublethal cadmium intoxication in Arabidopsis thaliana impacts translation at multiple levels. Plant Cell Physiol. 2011, 52, 436–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; Momcilovic, I.; Prasad, P.V. Roles of protein synthesis elongation factor EF-Tu in heat tolerance in plants. J. Bot. 2012, 2012, 835836. [Google Scholar] [CrossRef]
- Bhadula, S.K.; Elthon, T.E.; Habben, J.E.; Helentjaris, T.G.; Jiao, S.; Ristic, Z. Heat-stress induced synthesis of chloroplast protein synthesis elongation factor (EF-Tu) in a heat-tolerant maize line. Planta 2001, 212, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Bukovnik, U.; Fu, J.; Bennett, M.; Prasad, P.V.V.; Ristic, Z. Heat tolerance and expression of protein synthesis elongation factors, EF-Tu and EF-1α, in spring wheat. Funct. Plant Biol. 2009, 36, 234–241. [Google Scholar] [CrossRef]
- Merewitz, E.B.; Gianfagna, T.; Huang, B. Protein accumulation in leaves and roots associated with improved drought tolerance in creeping bentgrass expressing an ipt gene for cytokinin synthesis. J. Exp. Bot. 2011, 2, 5311–5333. [Google Scholar] [CrossRef] [PubMed]
- Jedmowski, C.; Ashoub, A.; Beckhaus, T.; Berberich, T.; Karas, M.; Brüggemann, W. Comparative analysis of Sorghum bicolor proteome in response to drought stress and following recovery. Int. J. Proteom. 2014, 2014, 395905. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, M.E. Association of Rubisco activase with chaperonin-60β, a possible mechanism for protecting photosynthesis during heat stress. J. Exp. Bot. 2008, 59, 1923–1933. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, D.K.; Ansari, M.W.; Tuteja, N. Multiple abiotic stress responsive rice cyclophilin (OsCYP-25) mediates a wide range of cellular responses. Commun. Integr. Biol. 2013, 6, e25260. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.; Horton, P.; Gray, J.E. The Arabidopsis thaliana cyclophilin gene family. Plant Physiol. 2004, 134, 1268–1282. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 2014, 5, 135. [Google Scholar] [CrossRef] [PubMed]

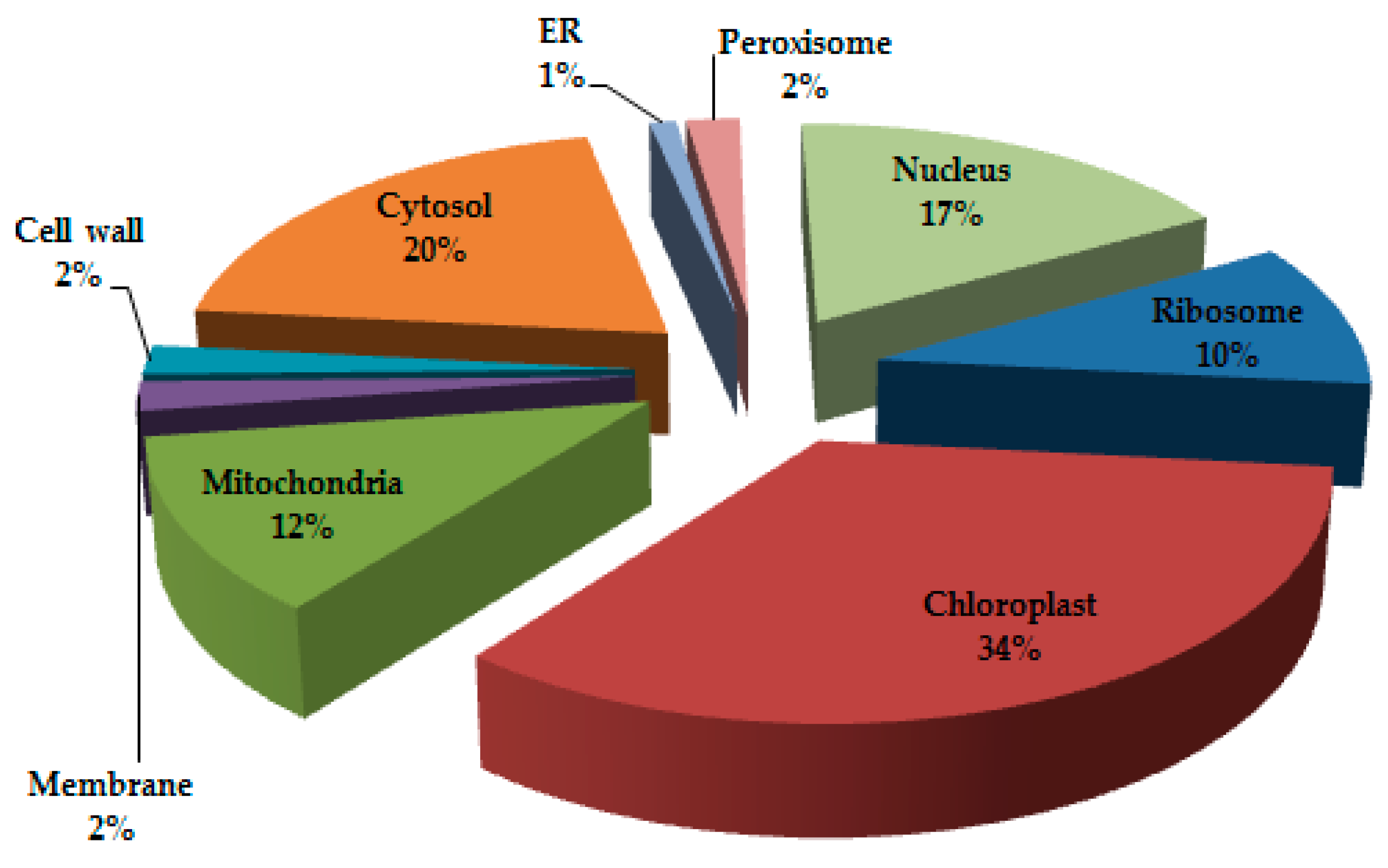


| S.N. | Accession No. | Name of Protein | Mr (Da) | Pi | M. SCO | No. of Matched Peptides | Location | Process | Mode of Regulation | Relative Spot Intensity | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UP2382 | VL616 | ||||||||||
| 1 | Q45NB2 | Glutamine synthetase | 46,852 | 5.96 | 120 | 9 | Chloroplast Mitochondria | Nitrogen metabolism | Upregulated |  | |
| 2 | P30110 | Glutathione transferase | 25,098 | 6.35 | 76 | 7 | Cytosol | Oxidative stress | Upregulated |  | |
| 3 | Q6EZE7 | Sucrose phosphate synthase | 12,874 | 6.81 | 84 | 11 | Cytosol | Carbohydrate metabolism | Upregulated |  | |
| 4 | M1NZ56 | Pyrroline-5-carboxylate synthetase | 30,883 | 5.74 | 122 | 6 | Cytosol | Osmo-regulation | Upregulated |  | |
| 5 | P27665 | Photosystem II oxygen-evolving complex protein 1 | 36,623 | 8.53 | 160 | 14 | Chloroplast | Photosynthesis | Upregulated |  | |
| 6 | Q6AUR2 | PII-like protein | 26,429 | 9.84 | 147 | 3 | Chloroplast | Nitrogen metabolism | Upregulated |  | |
| 7 | D8L9G7 | α-1,4-Glucan phosphorylase | 89,520 | 7.20 | 134 | 7 | Chloroplast | Osmo-regulation | Upregulated |  | |
| 8 | Q66NG6 | Plant histidine kinase (Cre 1) | 120,729 | 6.22 | 102 | 8 | Cytosol | Osmo-regulation | Upregulated |  | |
| 9 | P52589 | Protein disulfide isomerase | 56,921 | 5.01 | 149 | 3 | ER | Protein stabilization | Upregulated |  | |
| 10 | G9BRR4 | Heme oxygenase 4 | 32,415 | 5.81 | 62 | 6 | Chloroplast | Oxidative stress | Upregulated |  | |
| 11 | M8CY84 | Ferredoxin-dependent glutamate synthase 2 | 27,142 | 6.30 | 102 | 9 | Chloroplast | Nitrogen metabolism | Upregulated |  | |
| 12 | P12112 | ATPase F1 α subunit | 48,233 | 6.27 | 134 | 6 | Mitochondria | Energy metabolism | Upregulated |  | |
| 13 | P08823 | Chaperonin 60 β precursor | 54,321 | 4.61 | 158 | 5 | Mitochondria | Protein stabilization | Upregulated |  | |
| 14 | Q8LD97 | Glyoxylase I 7 | 32,564 | 5.82 | 74 | 2 | Peroxisomes | Oxidative stress | Upregulated |  | |
| 15 | M8B2C5 | l-Ascorbate peroxidase 5 | 34,852 | 6.84 | 94 | 4 | Chloroplast | Oxidative stress | Upregulated |  | |
| 16 | M8A9B6 | Abscisic acid-inducible protein kinase | 37,023 | 5.51 | 54 | 6 | Cytosol | Osmotic adjustment | Upregulated |  | |
| 17 | Q43349 | 29 kDa ribonucleoprotein A, chloroplast precursor | 28,326 | 4.75 | 165 | 5 | Nucleus | Protein synthesis | Downregulated |  | |
| 18 | P34791 | Chloroplast-localised cyclophilin | 26,527 | 8.48 | 146 | 6 | Chloroplast | Protein stability | Upregulated |  | |
| 19 | Q95H51 | Ribosomal protein L14 | 13,716 | 9.11 | 54 | 8 | Ribosome | Protein synthesis | Downregulated |  | |
| 20 | X5CQE2 | Flavonol synthase | 36,401 | 5.33 | 94 | 8 | Cytoplasm, Nucleus | Oxidative stress | Upregulated |  | |
| 21 | M8BQR6 | Osmotin like protein | 27,148 | 7.87 | 135 | 6 | Cell wall | Cell wall synthesis | Upregulated |  | |
| 22 | M7Z1M4 | Triosephosphate isomerase | 31,955 | 6.00 | 148 | 14 | Chloroplast, Cytosol | Glycolysis | Downregulated |  | |
| 23 | P60577 | Ribosomal protein S19 | 10,589 | 11.0 | 50 | 15 | Ribosome, Chloroplast, Mitochondria | Protein synthesis | Downregulated |  | |
| 24 | A0A1D5ZWW7 | Translation elongation factor-Tu | 52,275 | 5.09 | 99 | 9 | Plastid, Mitochondria, Cytosol | Protein synthesis | Upregulated |  | |
| 25 | -- | Unknown | 9,371 | 6.25 | 58 | 3 | - | - | Upregulated |  | |
| 26 | -- | Predicted protein | 6,649 | 4.62 | 42 | 2 | - | - | Downregulated |  | |
| 27 | P46285 | Sedoheptulose-1,7-bisphosphatase | 42,068 | 6.26 | 153 | 15 | Chloroplast | Calvin cycle | Downregulated |  | |
| 28 | M7ZTK2 | Glucosamine-fructose-6-phosphate aminotransferase | 73,834 | 6.98 | 47 | 6 | Cytosol | Nitrogen metabolism | Upregulated |  | |
| 29 | Q9ZSR6 | Small heat shock protein | 25,622 | 9.20 | 61 | 7 | Nucleus | Protein stabilization | Upregulated |  | |
| 30 | C1K2Q2 | Heat shock responsive transcription factor | 38,362 | 5.21 | 54 | 5 | Nucleus | Protein stabilization | Upregulated |  | |
| 31 | A5YVV3 | Glyceraldehyde 3-phosphate dehydrogenase | 42,766 | 7.62 | 141 | 4 | Plastid, Mitochondria | Glycolysis | Downregulated |  | |
| 32 | Q9FKW6 | RecName: Full=Ferredoxin--NADP reductase, chloroplastic | 41,322 | 8.54 | 139 | 7 | Chloroplast | Photosynthesis | Upregulated |  | |
| 33 | P11383 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 53,901 | 6.22 | 351 | 18 | Chloroplast | Photosynthesis | Downregulated |  | |
| 34 | W5G9W6 | Chlorophyll a-b binding protein 1 | 24,836 | 5.11 | 112 | 6 | Chloroplast | Photosynthesis | Upregulated |  | |
| 35 | A0A1D5TQL0 | Cu/Zn superoxide dismutase | 20,352 | 5.35 | 130 | 4 | Cytosol | Oxidative stress | Upregulated |  | |
| 36 | Q9XPS8 | RNA polymerase β chain | 79,908 | 9.00 | 217 | 8 | Nucleus | Transcription | Downregulated |  | |
| 37 | P56765 | Acetyl-CoA carboxylase β subunit | 40,207 | 9.02 | 53 | 5 | Cytoplasm, plastid | Fatty acid metabolism | Upregulated |  | |
| 38 | P10896 | Rubisco activase chloroplast precursor | 51,786 | 5.43 | 138 | 4 | Chloroplast | Photosynthesis | Upregulated |  | |
| 39 | P83970 | ATPase | 110,868 | 6.50 | 169 | 9 | Plasma membrane, tonoplast | Energy metabolism | Upregulated |  | |
| 40 | P11383 | Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit | 52,235 | 6.41 | 45 | 8 | Chloroplast | Photosynthesis | Downregulated |  | |
| 41 | A5BMQ9 | Hypothetical protein VITISV_041859 | 50,670 | 6.25 | 38 | 5 | - | - | Downregulated |  | |
| 42 | D0TZF0 | Isoamylase precursor | 75,052 | 6.40 | 52 | 2 | Cytosol | Carbohydrate metabolism | Upregulated |  | |
| 43 | P69326 | Ubiquitin | 4,693 | 8.90 | 72 | 6 | Cytosol | Protein degradation | Upregulated |  | |
| 44 | W5B4J0 | Histone acetyl-transferase complex component | 16,392 | 5.41 | 48 | 4 | Nucleus | Transcription | Upregulated |  | |
| 45 | Q2QWY8 | Transposon protein, putative, CACTA, En/Spm sub-class | 55,850 | 8.34 | 41 | 7 | Nucleus | - | Upregulated |  | |
| 46 | Q9SAX6 | Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit, partial | 13,215 | 6.60 | 87 | 10 | Chloroplast | Photosynthesis | Downregulated |  | |
| 47 | B6TJX4 | SnRK1-interacting protein 1 | 14,800 | 7.23 | 82 | 6 | Nucleus | Transcription | Upregulated |  | |
| 48 | M7ZIN5 | Translation factor 5A1 | 17,532 | 5.70 | 114 | 5 | Ribosome | Protein synthesis | Downregulated |  | |
| 49 | D7LVK3 | ABA-responsive element binding protein 3 | 32,674 | 6.53 | 43 | 9 | Nucleus | Transcription | Upregulated |  | |
| 50 | B5B3P8 | PR-10 protein | 16,464 | 7.04 | 22 | 8 | Nucleus | - | Downregulated |  | |
| 51 | A0A1D5WC57 | Zinc finger family protein | 38,647 | 8.01 | 47 | 9 | Nucleus | Transcription | Upregulated |  | |
| 52 | W5ZRX4 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 52,563 | 6.05 | 212 | 5 | Chloroplast | Photosynthesis | Downregulated |  | |
| 53 | -- | Predicted protein | 11,607 | 10.46 | 125 | 7 | - | - | Upregulated |  | |
| 54 | S4Z9E7 | Ribosomal protein S19 | 6,795 | 9.12 | 110 | 6 | Ribosome | Protein synthesis | Downregulated |  | |
| 55 | -- | Unnamed protein product | 4,414 | 7.90 | 24 | 8 | - | - | Downregulated |  | |
| 56 | Q95H61 | Ribosomal protein S4 | 23,540 | 10.80 | 47 | 6 | Ribosome | Protein synthesis | Downregulated |  | |
| 57 | Q9LXC9 | Inorganic pyrophosphatase family protein | 32,567 | 5.72 | 112 | 10 | Chloroplast | Phosphorus metabolism | Upregulated |  | |
| 58 | A2ZLN9 | Hypothetical protein OsI_38734 | 15,336 | 4.25 | 41 | 11 | - | Chloroplast | Upregulated |  | |
| 59 | A0A1D5ZI60 | Methionine synthase | 54,794 | 5.79 | 101 | 8 | Mitochondria | Amino-acid metabolism | Upregulated |  | |
| 60 | P69667 | Ribosomal protein L23 | 9,563 | 10.02 | 71 | 6 | Ribosome | Protein synthesis | Downregulated |  | |
| 61 | A0A1D5ST37 | Malate dehydrogenase | 38,104 | 5.61 | 58 | 8 | Mitochondria | Citric acid cycle | Downregulated |  | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yousuf, P.Y.; Abd_Allah, E.F.; Nauman, M.; Asif, A.; Hashem, A.; Alqarawi, A.A.; Ahmad, A. Responsive Proteins in Wheat Cultivars with Contrasting Nitrogen Efficiencies under the Combined Stress of High Temperature and Low Nitrogen. Genes 2017, 8, 356. https://doi.org/10.3390/genes8120356
Yousuf PY, Abd_Allah EF, Nauman M, Asif A, Hashem A, Alqarawi AA, Ahmad A. Responsive Proteins in Wheat Cultivars with Contrasting Nitrogen Efficiencies under the Combined Stress of High Temperature and Low Nitrogen. Genes. 2017; 8(12):356. https://doi.org/10.3390/genes8120356
Chicago/Turabian StyleYousuf, Peerzada Yasir, Elsayed Fathi Abd_Allah, Mohd Nauman, Ambreen Asif, Abeer Hashem, Abdulaziz A. Alqarawi, and Altaf Ahmad. 2017. "Responsive Proteins in Wheat Cultivars with Contrasting Nitrogen Efficiencies under the Combined Stress of High Temperature and Low Nitrogen" Genes 8, no. 12: 356. https://doi.org/10.3390/genes8120356







