RNA-Seq Analysis of Plant Maturity in Crested Wheatgrass (Agropyron cristatum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, RNA Extraction, and Illumina Sequencing
2.2. De Novo Assembly, Differential Gene Expression Analysis and Sequence Annotation
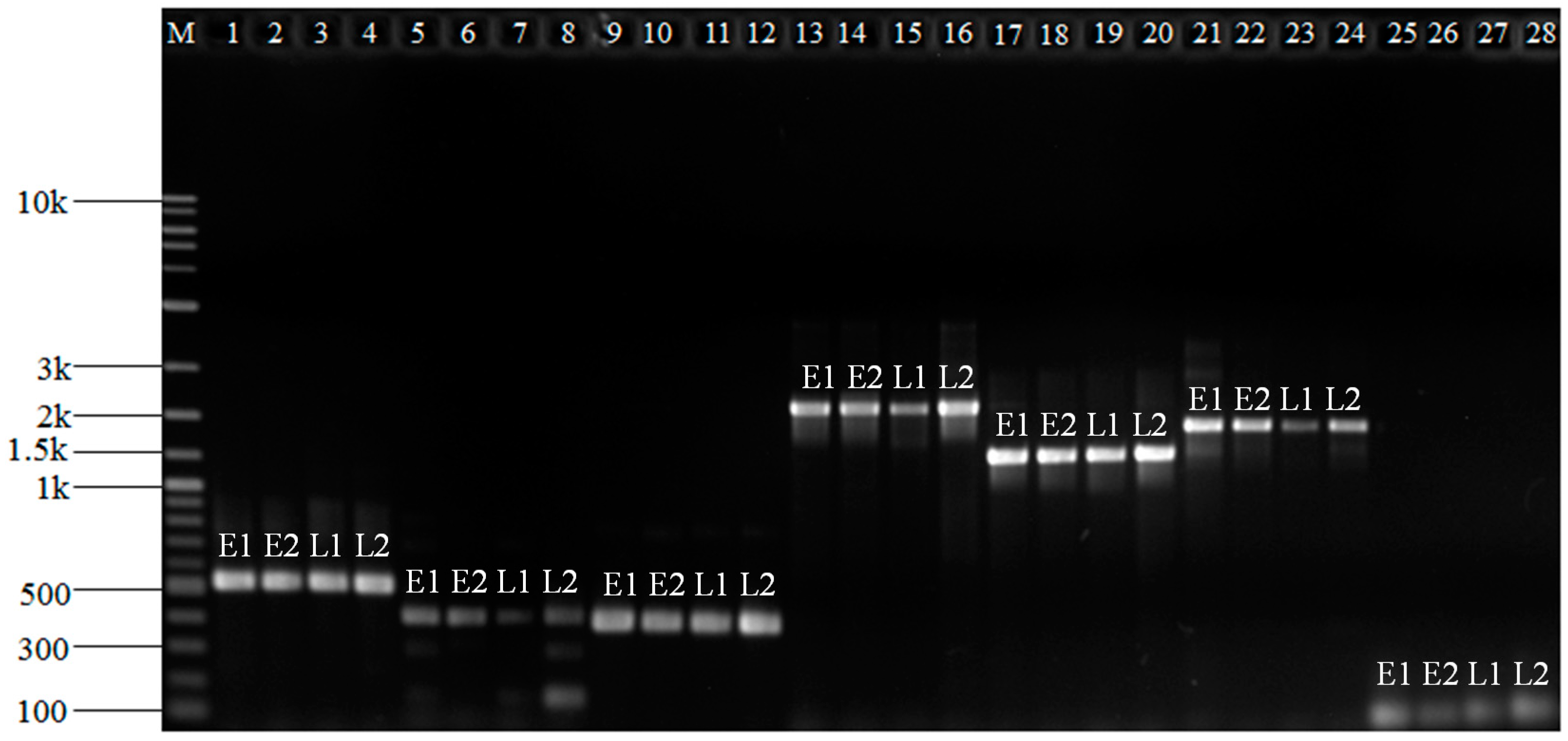
2.3. The Validation of Differentially Expressed Genes
3. Results and Discussion
3.1. De Novo Assembly
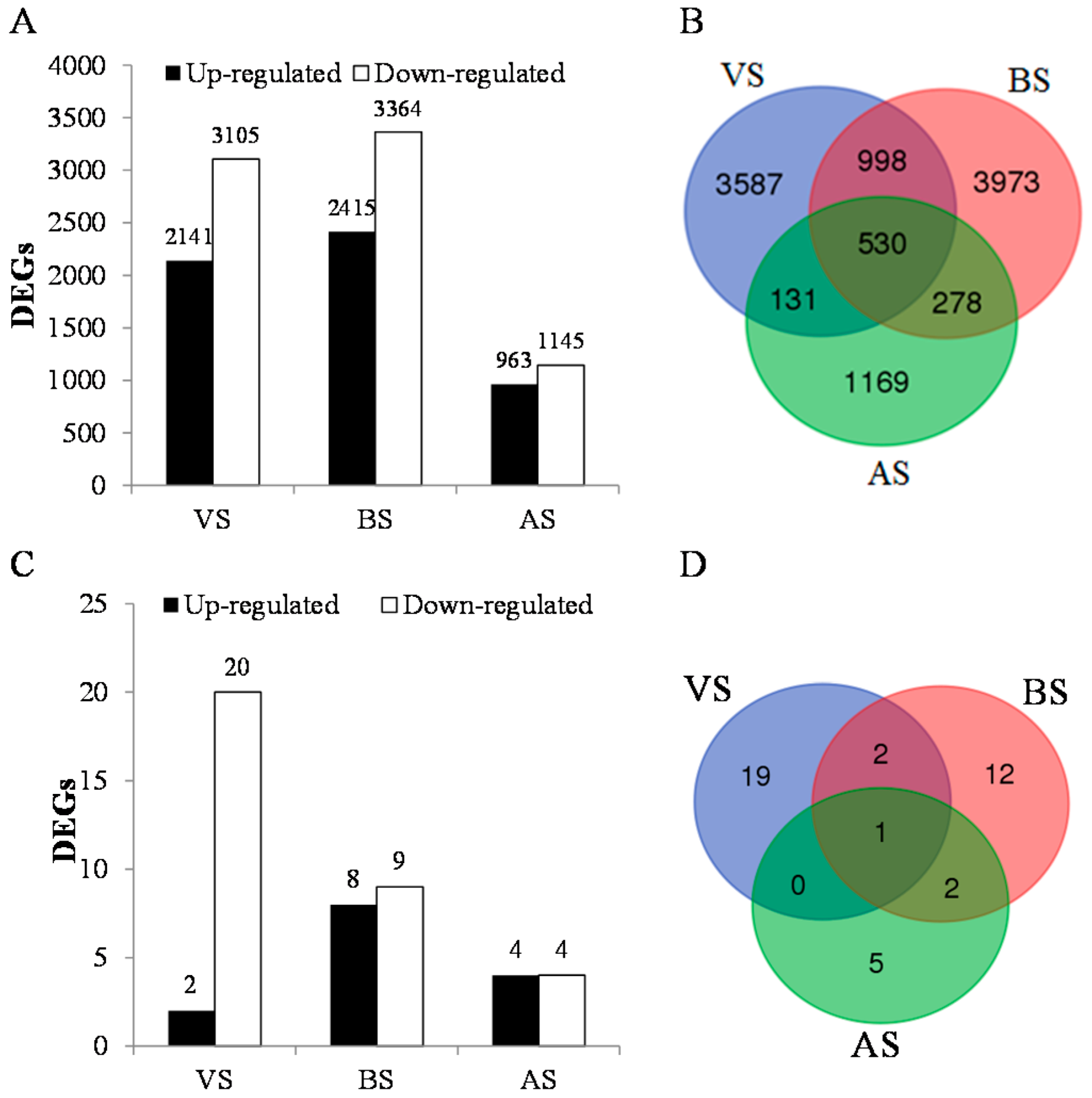
3.2. Differentially Expressed Genes Detection and Annotation
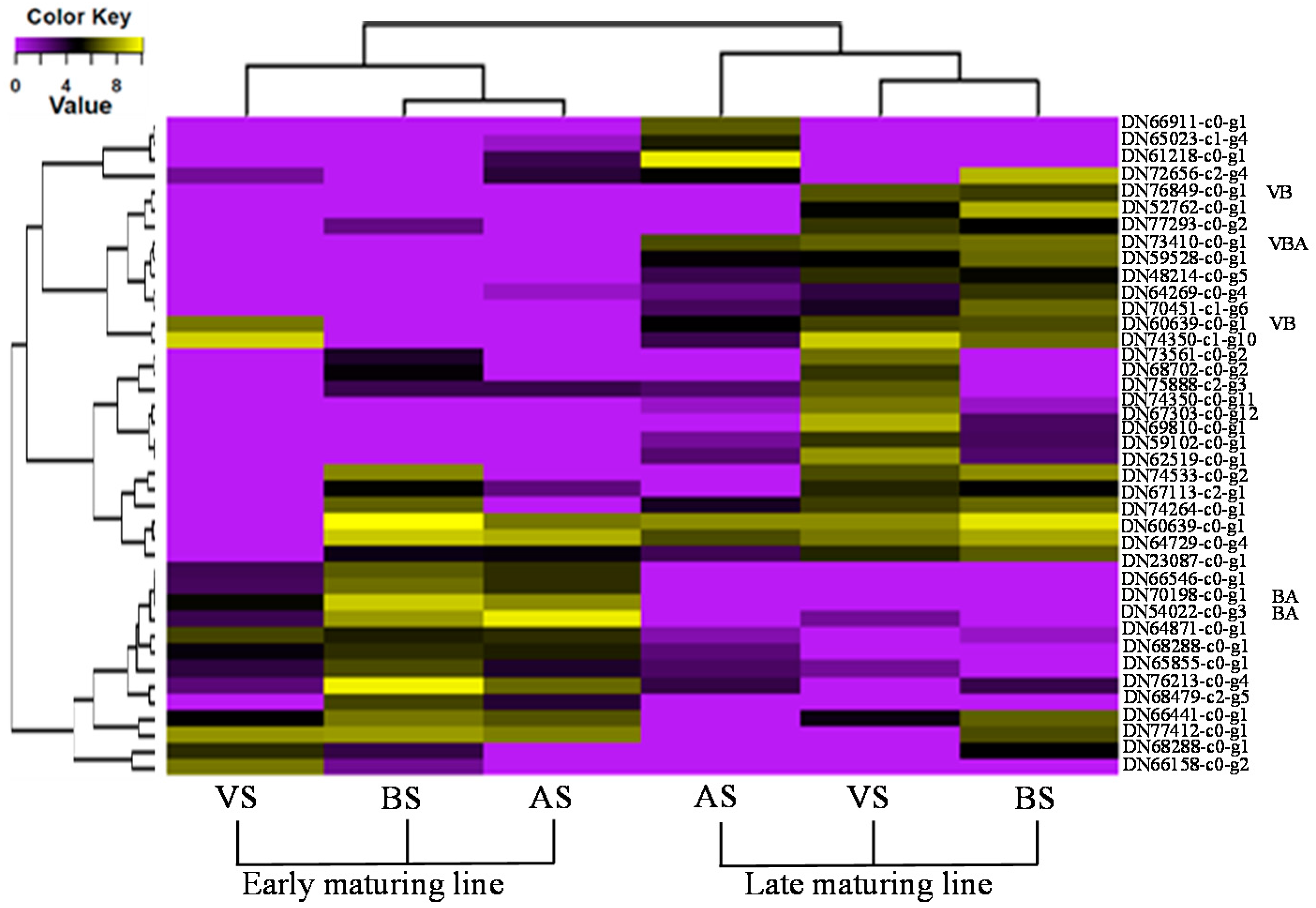
3.3. Flowering Related Differentially Expressed Genes
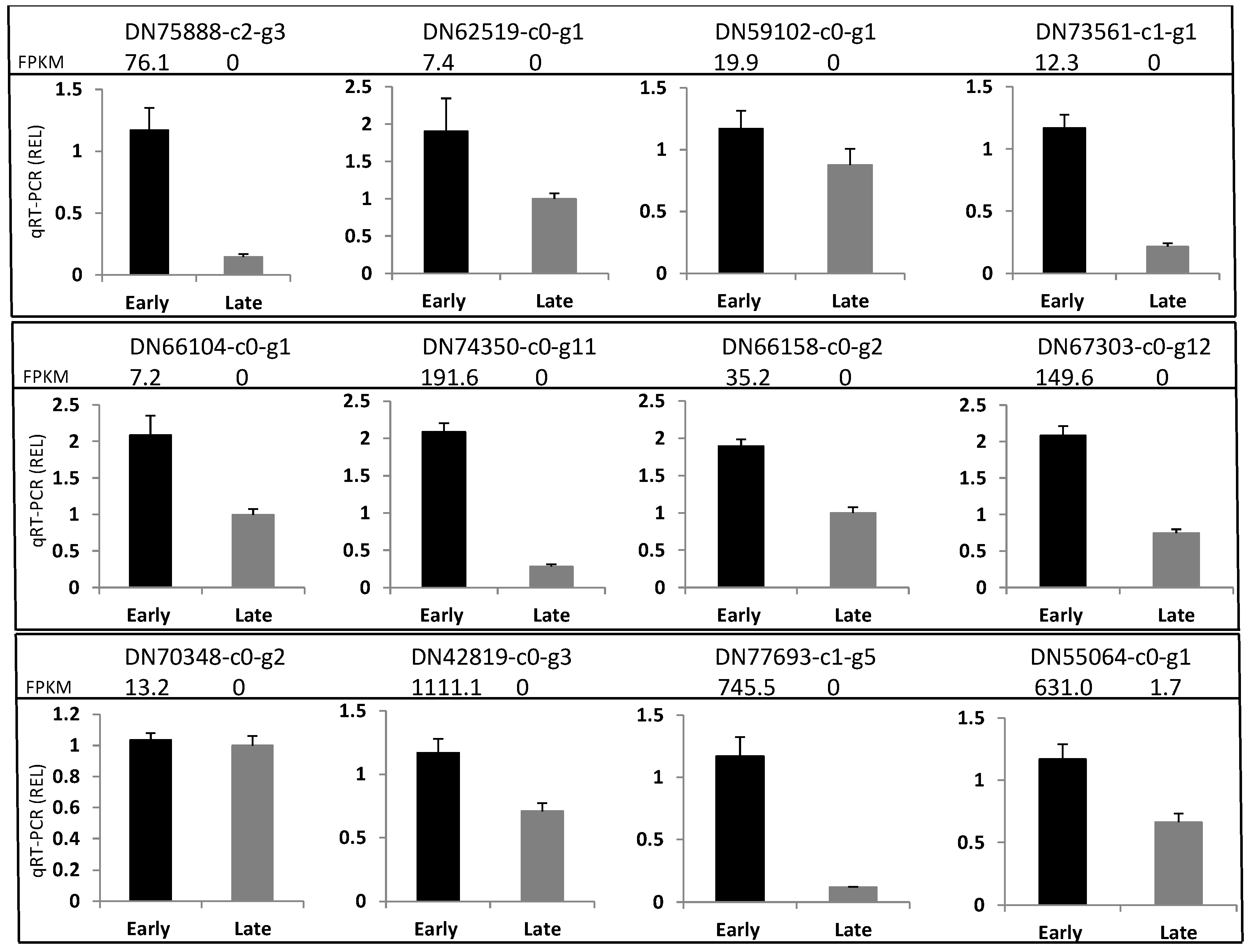
3.4. Differentially Expressed Genes Validation
3.5. Implications for RNA-Seq Analysis and Breeding
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contribution
Conflicts of Interest
References
- Daugherty, D.A.; Britton, C.M.; Turner, H.A. Grazing management of crested wheatgrass range for yearling steers. J. Range Manag. 1982, 35, 347–350. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, W.; Han, H.; Song, L.; Bai, L.; Gao, Z.; Zhang, Y.; Yang, X.; Li, X.; Gao, A.; et al. De novo transcriptome sequencing of Agropyron cristatum to identify available gene resources for the enhancement of wheat. Genomics 2015, 106, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Nagatani, A. Flowering regulation by tissue specific functions of photoreceptors. Plant Signal. Behav. 2008, 3, 47–48. [Google Scholar] [CrossRef] [PubMed]
- Amasino, R. Seasonal and developmental timing of flowering. Plant J. 2010, 61, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Amasino, R. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999, 11, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Trevaskis, B.; Hemming, M.N.; Dennis, E.S.; Peacock, W.J. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007, 12, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Kellogg, E.A. Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 2006, 174, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Loukoianov, A.; Blechl, A.; Tranquilli, G.; Ramakrishna, W.; SanMiguel, P.; Bennetzen, J.L.; Echenique, V.; Dubcovsky, J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 2004, 303, 1640–1644. [Google Scholar] [CrossRef] [PubMed]
- Dubcovsky, J.; Chen, C.; Yan, L. Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol. Breed. 2005, 15, 395–407. [Google Scholar] [CrossRef]
- Karsai, I.; Szűcs, P.; Mészáros, K.; Filichkina, T.; Hayes, P.; Skinner, J.; Láng, L.; Bedo, Z. The Vrn-H2 locus is a major determinant of flowering time in a facultative × winter growth habit barley (Hordeum vulgare L.) mapping population. Theor. Appl. Genet. 2005, 110, 1458–1466. [Google Scholar] [CrossRef] [PubMed]
- Distelfeld, A.; Tranquilli, G.; Li, C.; Yan, L.; Dubcovsky, J. Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 2009, 149, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Fu, D.; Li, C.; Blechl, A.; Tranquilli, G.; Bonafede, M.; Sanchez, A.; Valarik, M.; Yasuda, S.; Dubcovsky, J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 2006, 103, 19581–19586. [Google Scholar] [CrossRef] [PubMed]
- Sasani, S.; Hemming, M.N.; Oliver, S.N.; Greenup, A.; Tavakkol-Afshari, R.; Mahfoozi, S.; Poustini, K.; Sharifi, H.; Dennis, E.S.; Peacock, W.J.; et al. The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare). J. Exp. Bot. 2009, 60, 2169–2178. [Google Scholar] [CrossRef] [PubMed]
- Hemming, M.N.; Peacock, W.J.; Dennis, E.S.; Trevaskis, B. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol. 2008, 147, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Helguera, M.; Kato, K.; Fukuyama, S.; Sherman, J.; Dubcovsky, J. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor. Appl. Genet. 2004, 109, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.; Himelblau, E.; Kim, S.; Schomburg, F.; Amasino, R. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 2005, 137, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Fowler, S.; Lee, K.; Onouchi, H.; Samach, A.; Richardson, K.; Morris, B.; Coupland, G.; Putterill, J. GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999, 18, 4679–4688. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.; Kubota, A.; Imaizumi, T. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 2017, 173, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Green, R.M.; Tobin, E.M. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. USA 1999, 96, 4176–4179. [Google Scholar] [CrossRef] [PubMed]
- Hicks, K.; Millar, A.J.; Carré, I.A.; Somers, D.E.; Straume, M.; Meeks-Wagner, D.R.; Kay, S.A. Conditional circadian dysfunction of the Arabidopsis early-flowering-3 mutant. Science 1996, 274, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Park, D.H.; Somers, D.E.; Kim, Y.S.; Choy, Y.H.; Lim, H.K.; Soh, M.S.; Kim, H.J.; Kay, S.A.; Nam, H.G. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 1999, 285, 1579–1582. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, R.; Ramsay, N.; Samach, A.; Corden, S.; Putterill, J.; Carré, I.A.; Coupland, G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998, 93, 1219–1229. [Google Scholar] [CrossRef]
- Wang, Z.; Tobin, E. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 1998, 93, 1207–1217. [Google Scholar] [CrossRef]
- Strayer, C.; Oyama, T.; Schultz, T.F.; Raman, R.; Somers, D.E.; Más, P.; Panda, S.; Kreps, J.A.; Kay, S.A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, M.; Hanhart, C.; van der Veen, J. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 1991, 229, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Suárez-López, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Biligetu, B.; Coulman, B.; Schellenberg, M.P.; Fu, Y.B. RNA-Seq analysis of gene expression for floral development in crested wheatgrass (Agropyron cristatum L.). PLoS ONE 2017, 12, e0177417. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.; Moser, L.; Vogel, K.; Waller, S.; Johnson, B.E.; Pedersen, J.F. Describing and quantifying growth stages of perennial forage grasses. Agron. J. 1991, 83, 1073–1077. [Google Scholar] [CrossRef]
- Grabherr, M.; Haas, B.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mao, X.; Cai, T.; Luo, J.; Wei, L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006, 34 (Suppl. 2), W720–W724. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.G. Evolution of flowering in response to day length flipping the CONSTANS switch. Bioessays 2003, 25, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Yanovsky, M.J.; Kay, S.A. Molecular basis of seasonal time measurement in Arabidopsis. Nature 2002, 419, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Turck, F.; Fornara, F.; Coupland, G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 2008, 59, 573–594. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.B.; Yang, M.; Zeng, F.; Biligetu, B. Searching for an accurate marker-based prediction of an individual quantitative trait in molecular plant breeding. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
| Year | Accession Name | Ploidy | Row Number | Plant Number | Heading Date a | Number of Heads/Plant | Plant Height (cm) |
|---|---|---|---|---|---|---|---|
| 2016 | W625134 (Early) | tetraploid | 18 | 8 | 26-May-2016 | >20 | 71 |
| 2016 | S9516 (Late) | tetraploid | 40 | 30 | 3-June-2016 | 2 | 85 |
| 2015 | W625134 (Early) | tetraploid | 18 | 8 | 3-June-2015 | >20 | 70 |
| 2015 | S9516 (Late) | tetraploid | 40 | 30 | 8-June-2015 | 11 | 90 |
| W625134 | S9516 | |||||
|---|---|---|---|---|---|---|
| VS | BS | AS | VS | BS | AS | |
| Raw reads | 23,073,394 | 29,312,848 | 25,603,776 | 25,948,120 | 28,249,736 | 22,494,862 |
| Clean reads | 22,064,163 | 28,810,091 | 24,343,976 | 24,294,860 | 27,763,034 | 21,957,198 |
| Total clean nucleotides | 2,727,558,541 | 3,563,181,662 | 2,966,838,914 | 3,005,892,104 | 3,432,700,188 | 2,714,079,492 |
| Average read length | 123.6 | 123.7 | 121.9 | 123.7 | 123.6 | 123.6 |
| All six libraries | ||||||
| Total trinity transcripts | 401,587 | |||||
| Mean length of contigs | 546 | |||||
| N50 of contigs | 691 | |||||
| Total assembled bases | 219,458,456 | |||||
| “Genes” with TPM > 5 a | 94,461 | |||||
| “Genes” with TPM > 10 | 45,689 | |||||
| Gene ID a | Putative Function | Nr ID | log2FC | FDR |
|---|---|---|---|---|
| DN67303-c0-g12 | flowering locus T [Lolium perenne] | AIE58042.1 | −11.5 | 1.90 × 10−6 |
| DN59102-c0-g1 | timing of cab expression 1 | AMK48976.1 | −10.9 | 7.50 × 10−6 |
| DN74264-c0-g1 | SYD isoform X1 | EMT16433.1 | −10.7 | 1.10 × 10−5 |
| DN74350-c1-g11 | flowering locus T [Lolium perenne] | AAW23034.1 | −10.4 | 2.40 × 10−5 |
| DN60639-c0-g1 | transcriptional corepressor SEUSS [Oryza sativa] | BAJ98061.1 | −10.3 | 2.70 × 10−5 |
| DN73561-c0-g2 | GIGANTEA [Oryza sativa] | CDM81775.1 | −10.3 | 3.30 × 10−5 |
| DN73410-c0-g1 | nucleic acid binding [Zea mays] | BAJ87586.1 | −10.0 | 6.50 × 10−5 |
| DN75888-c2-g3 | cryptochrome 2 | ABX58030.1 | −9.9 | 8.10 × 10−5 |
| DN66158-c0-g2 | HD3A_ORYSJ | BAH30246.1 | −9.9 | 8.20 × 10−5 |
| DN76849-c0-g1 | phosphatidylinositol 4-phosphate 5-kinase 1-like | EMS61702.1 | −9.7 | 1.23 × 10−4 |
| DN62519-c0-g1 | spotted leaf 11 | BAJ85648.1 | −9.6 | 1.61 × 10−4 |
| DN67113-c2-g1 | auxin response factor | EMT32630.1 | −9.2 | 3.60 × 10−4 |
| DN66104-c0-g1 | COL10_ARATH | Q9LUA9.1 | −9.2 | 1.12 × 10−5 |
| DN68702-c0-g2 | SWI SNF complex subunit SWI3B | EMT14023.1 | −9.1 | 4.87 × 10−4 |
| DN69810-c0-g1 | CONSTANS CO6 [Zea mays] | BAJ98422.1 | −9.1 | 4.74 × 10−4 |
| DN77293-c0-g2 | probable serine threonine- kinase vps15 isoform | EMT17455.1 | −9.1 | 4.74 × 10−4 |
| DN70348-c0-g2 | MADS-box transcription factor 18 | XP_006657934 | −9.1 | 1.28 × 10−5 |
| DN48214-c0-g5 | constans-like 1 [Picea abies] | EMT25416.1 | −9.0 | 5.74 × 10−4 |
| DN64729-c0-g4 | MADS-domain transcription factor [Zea mays] | ABF57916.1 | −8.8 | 9.36 × 10−4 |
| DN74533-c0-g2 | phragmoplast-associated kinesin [Oryza sativa] | EMT21759.1 | −8.8 | 8.80 × 10−4 |
| DN66288-c0-g1 | Os01g0687700 [Oryza sativa] | CDM83875.1 | 8.9 | 7.84 × 10−4 |
| DN64871-c0-g1 | gamma-glutamylcysteine synthetase | BAJ84988.1 | 9.3 | 2.75 × 10−4 |
| BS (Early vs. Late) | ||||
|---|---|---|---|---|
| Gene ID a | Putative Function | Nr ID | log2FC | FDR |
| DN52762-c0-g1 | mediator complex subunit 25 | AMW92183.1 | −11.4 | 2.59 × 10−6 |
| DN59528-c0-g1 | pentatricopeptide repeat-containing mitochondrial | EMT01957.1 | −10.1 | 5.38 × 10−5 |
| DN60639-c0-g1 | transcriptional corepressor SEUSS [Oryza sativa] | BAJ98061.1 | −9.6 | 1.90 × 10−4 |
| DN64269-c0-g4 | Cullin-associated NEDD8-dissociated 1 | BAJ94036.1 | −9.2 | 5.57 × 10−4 |
| DN70451-c1-g6 | polycomb group EMBRYONIC FLOWER 2 isoform X1 | BAJ97315.1 | −10.0 | 7.29 × 10−5 |
| DN72656-c2-g4 | glycerol-3-phosphate 2-O-acyltransferase 6 | AKL71379.1 | −11.5 | 2.02 × 10−6 |
| DN73410-c0-g1 | nucleic acid binding [Zea mays] | BAJ87586.1 | −10.2 | 4.27 × 10−5 |
| DN74350-c1-g10 | flowering locus T [Lolium perenne] | AAW23034.1 | −10.1 | 5.38 × 10−5 |
| DN76849-c0-g1 | phosphatidylinositol 4-phosphate 5-kinase 1-like | EMS61702.1 | −9.3 | 3.78 × 10−4 |
| DN65855-c0-g1 | CONSTANS CO5 | AAL99264.1 | 9.5 | 2.28 × 10−4 |
| DN66546-c0-g1 | Eukaryotic translation initiation factor 3 subunit E | EMT06719.1 | 10.2 | 5.00 × 10−5 |
| DN68288-c0-g1 | Peptide chain release factor 2 | EMT30094.1 | 8.9 | 9.55 × 10−4 |
| DN68479-c2-g5 | LRR receptor-like serine threonine-kinase FEI 1 | EMS63993.1 | 9.3 | 4.00 × 10−4 |
| DN70198-c0-g1 | WD-40 repeat-containing MSI1 | ABB92268.1 | 11.9 | 9.36 × 10−7 |
| DN76213-c0-g4 | minichromosome maintenance 5 partial | BAJ93067.1 | 6.3 | 3.26 × 10−5 |
| DN54022-c0-g3 | MADS-box transcription factor 7 | CAM59055.1 | 10.9 | 8.63 × 10−6 |
| DN23087-c0-g1 | Polyglutamine-binding 1 | EMT21570.1 | 9.7 | 1.55 × 10−4 |
| AS (Early vs. Late) | ||||
|---|---|---|---|---|
| Gene ID | Putative Function | Nr ID | log2FC | FDR |
| DN61281-c0-g1 | Aldose reductase | CDM85015.1 | −7.2 | 7.90 × 10−6 |
| DN65023-c1-g4 | flower-specific gamma-thionin precursor [Zea mays] | BAK07823.1 | −6.5 | 8.11 × 10−4 |
| DN66911-c0-g1 | flower-specific gamma-thionin precursor [Zea mays] | BAK07823.1 | −10.4 | 5.20 × 10−5 |
| DN73410-c0-g1 | nucleic acid binding [Zea mays] | BAJ87586.1 | −10.1 | 1.04 × 10−4 |
| DN77412-c0-g1 | SYD isoform X1 | EMT16433.1 | 10.0 | 1.40 × 10−4 |
| DN54022-c0-g3 | MADS-box transcription factor 7 | CAM59055.1 | 12.1 | 2.85 × 10−6 |
| DN66441-c0-g1 | flowering time control FPA isoform X1 | BAJ91078.1 | 9.2 | 8.54 × 10−4 |
| DN70198-c0-g1 | WD-40 repeat-containing MSI1 | ABB92268.1 | 10.3 | 6.58 × 10−5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, F.; Biligetu, B.; Coulman, B.; Schellenberg, M.P.; Fu, Y.-B. RNA-Seq Analysis of Plant Maturity in Crested Wheatgrass (Agropyron cristatum L.). Genes 2017, 8, 291. https://doi.org/10.3390/genes8110291
Zeng F, Biligetu B, Coulman B, Schellenberg MP, Fu Y-B. RNA-Seq Analysis of Plant Maturity in Crested Wheatgrass (Agropyron cristatum L.). Genes. 2017; 8(11):291. https://doi.org/10.3390/genes8110291
Chicago/Turabian StyleZeng, Fangqin, Bill Biligetu, Bruce Coulman, Michael P. Schellenberg, and Yong-Bi Fu. 2017. "RNA-Seq Analysis of Plant Maturity in Crested Wheatgrass (Agropyron cristatum L.)" Genes 8, no. 11: 291. https://doi.org/10.3390/genes8110291






