Molecular Cloning, Recombinant Expression and Antifungal Activity of BnCPI, a Cystatin in Ramie (Boehmeria nivea L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth and Sampling
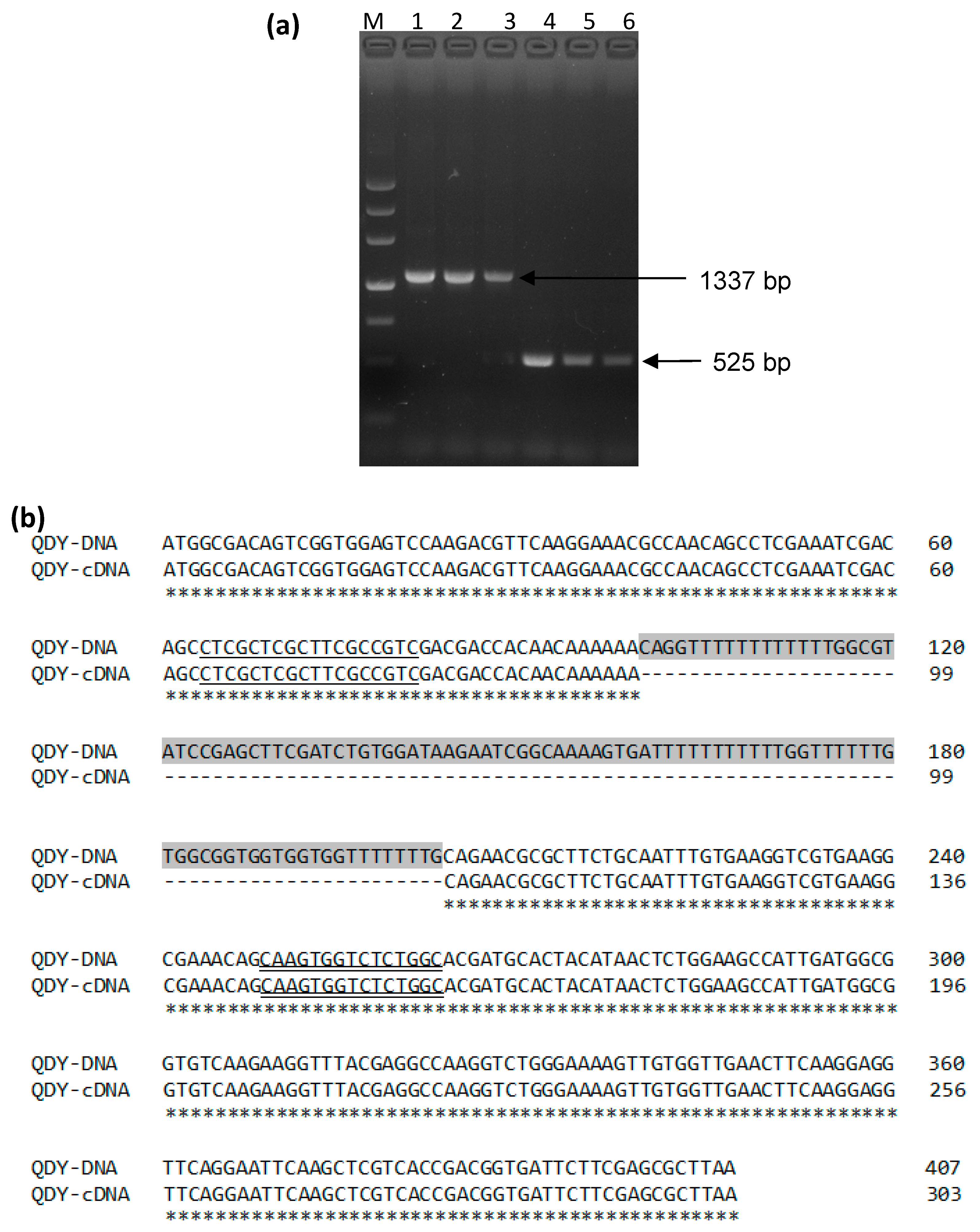
2.2. DNA and RNA Extraction and cDNA Synthesis
2.3. Molecular Cloning of the BnCPI Gene
2.4. Sequences Analysis and Phylogenetic Analysis
2.5. Expression of a Recombinant BnCPI in Escherichia coli
2.6. Protease Inhibitory Activity Assay
2.7. Antifungal Activity Assay of reBnCPI
3. Results
3.1. Sequence Analysis
3.2. Homology Analysis of BnCPI
3.3. Recombinant Expression of BnCPI
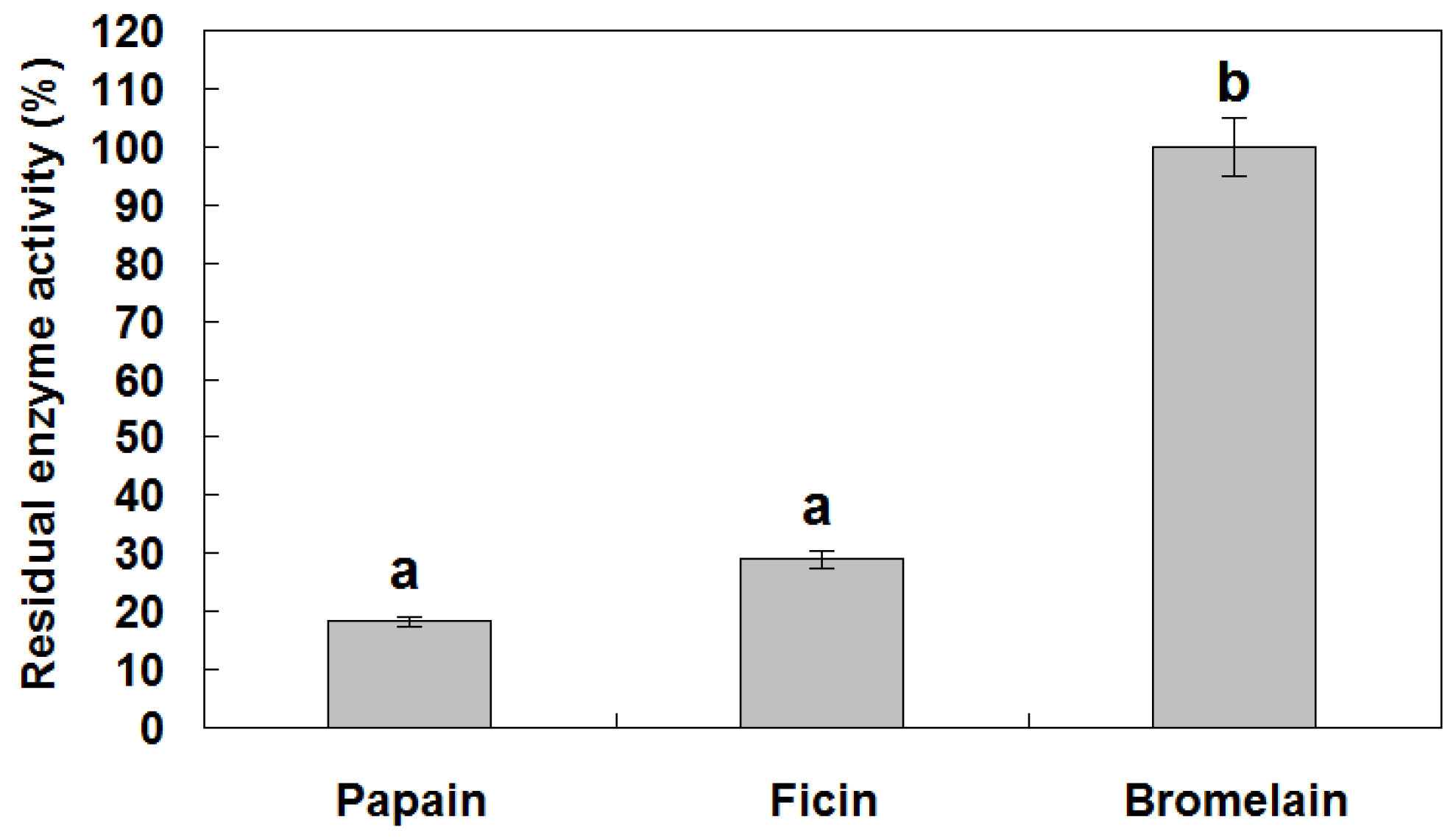
3.4. Protease Inhibitory Activity of reBnCPI
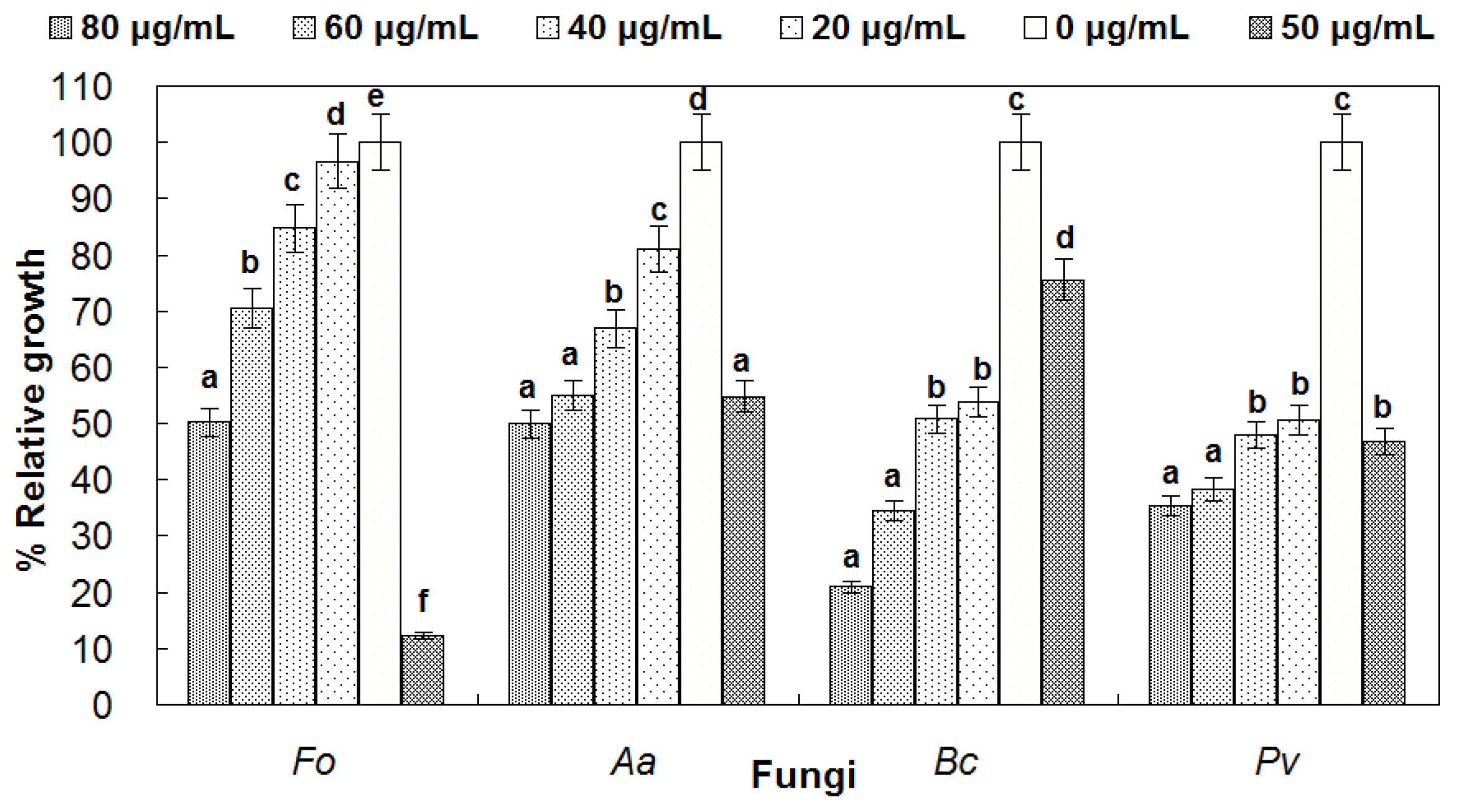
3.5. Antifungal Activity of reBnCPI
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kunert, K.J.; van Wyk, S.G.; Cullis, C.A.; Vorster, B.J.; Foyer, C.H. Potential use of phytocystatins in crop improvement, with a particular focus on legumes. J. Exp. Bot. 2015, 66, 3559–3570. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.M.; dos Reis, S.P.; de Souza, C.R. Phytocystatins and their potential to control plant diseases caused by fungi. Protein Pept. Lett. 2015, 22, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Santamaria, M.E.; Diaz-Mendoza, M.; Arnaiz, A.; Carrillo, L.; Ortego, F.; Diaz, I. Phytocystatins: Defense proteins against phytophagous insects and acari. Int. J. Mol. Sci. 2016, 17, 1747. [Google Scholar] [CrossRef] [PubMed]
- Oppert, B.; Morgan, T.D.; Hartzer, K.; Lenarcic, B.; Galesa, K.; Brzin, J.; Turk, V.; Yoza, K.; Ohtsubo, K.; Kramer, K.J. Effects of proteinase inhibitors on digestive proteinases and growth of the red flour beetle, Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2003, 134, 481–490. [Google Scholar] [CrossRef]
- Cingel, A.; Savic, J.; Lazarevic, J.; Cosic, T.; Raspor, M.; Smigocki, A.; Ninkovic, S. Co-expression of the proteinase inhibitors oryzacystatin I and oryzacystatin II in transgenic potato alters Colorado potato beetle larval development. Insect Sci. 2017, 24, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Schneider, V.K.; Soares-Costa, A.; Chakravarthi, M.; Ribeiro, C.; Chabregas, S.M.; Falco, M.C.; Henrique-Silva, F. Transgenic sugarcane overexpressing CaneCPI-1 negatively affects the growth and development of the sugarcane weevil Sphenophorus levis. Plant Cell Rep. 2017, 36, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.L.; Yang, A.H.; Chen, J.T.; Yeh, K.W.; Chan, M.T. Heterologouxpression of taro cystatin protects transgenic tomato against Meloidogyne incognita infection by means of interfering sex determination and suppressing gall formation. Plant Cell Rep. 2010, 29, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Roderick, H.; Tripathi, L.; Babirye, A.; Wang, D.; Tripathi, J.; Urwin, P.E.; Atkinson, H.J. Generation of transgenic plantain (Musa spp.) with resistance to plant pathogenic nematodes. Mol. Plant Pathol. 2012, 13, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Vieira, P.; Wantoch, S.; Lilley, C.J.; Chitwood, D.J.; Atkinson, H.J.; Kamo, K. Expression of a cystatin transgene can confer resistance to root lesion nematodes in Lilium longiflorum cv. ‘Nellie White’. Transgenic Res. 2015, 24, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Lopez-Solanilla, E.; Rodriguez-Palenzuela, P.; Carbonero, P.; Diaz, I. Inhibition of plant-pathogenic fungi by the barley cystatin Hv-CPI (gene Icy) is not associated with its cysteine-proteinase inhibitory properties. Mol. Plant Microbe Interact. 2003, 16, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.H.; Yeh, K.W. Molecular cloning, recombinant gene expression, and antifungal activity of cystatin from taro (Colocasia esculenta cv. Kaosiung No. 1). Planta 2005, 221, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, L.; Martinez, M.; Álvarez-Alfageme, F.; Castañera, P.; Smagghe, G.; Diaz, I.; Ortego, F. A barley cysteine-proteinase inhibitor reduces the performance of two aphid species in artificial diets and transgenic Arabidopsis plants. Transgenic Res. 2011, 20, 305–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senthilkumar, R.; Cheng, C.P.; Yeh, K.W. Genetically pyramiding protease-inhibitor genes for dual broad-spectrum resistance against insect and phytopathogens in transgenic tobacco. Plant Biotechnol. J. 2010, 8, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; McKerrow, J.H. Cysteine proteases of parasitic organisms. Mol. Biochem. Parasitol. 2002, 120, 1–21. [Google Scholar] [CrossRef]
- Li, Y.; Wang, K.; Xie, H.; Wang, D.W.; Xu, C.L.; Huang, X.; Wu, W.J.; Li, D.L. Cathepsin B cysteine proteinase issential for the development and pathogenesis of the plant parasitic nematode Radopholus similis. Int. J. Biol. Sci. 2015, 11, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, Y.; Huang, X.; Wang, D.W.; Xu, C.L.; Xie, H. The cathepsin S cysteine proteinase of the burrowing nematode Radopholus similis is essential for the reproduction and invasion. Cell Biosci. 2016, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Shingles, J.; Lilley, C.J.; Atkinson, H.J.; Urwin, P.E. Meloidogyne incognita: Molecular and biochemical characterisation of a cathepsin L cysteine proteinase and the effect on parasitism following RNAi. Exp. Parasitol. 2007, 115, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Rodriguez, S.; Cedro-Tanda, A.; Aguilar-Hernandez, V.; Cortes-Onofre, E.; Blanco-Labra, A.; Guerrero-Rangel, A. Recombinant amaranth cystatin (AhCPI) inhibits the growth of phytopathogenic fungi. Plant Physiol. Biochem. 2010, 48, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Luan, M.B.; Chen, B.F.; Zou, Z.Z.; Zhu, J.J.; Wang, X.F.; Xu, Y.; Sun, Z.M.; Chen, J.H. Molecular identity of ramie germplasm using simple sequence repeat markers. Genet. Mol. Res. 2015, 14, 2302–2311. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhu, S.; Tang, Q.; Chen, P.; Yu, Y.; Tang, S. De novo assembly and characterization of transcriptome using Illumina paired-end sequencing and identification of CesA gene in ramie (Boehmeria nivea L. Gaud). BMC Genom. 2013, 14, 125. [Google Scholar] [CrossRef] [PubMed]
- Kipriotis, E.; Xiong, H.P.; Vafeiadakis, T.; Kiprioti, M.; Alexopoulou, E. Ramie and kenaf as feed crops. Ind. Crop. Prod. 2015, 68, 126–130. [Google Scholar] [CrossRef]
- Yu, Y.T.; Liu, H.L.; Zhu, A.G.; Zhang, G.; Zeng, L.B.; Xue, S.D. A review of root lesion nematode: Identification and plant resistance. Adv. Microb. 2012, 2, 411–416. [Google Scholar] [CrossRef]
- Yu, Y.; Zeng, L.; Yan, Z.; Liu, T.; Sun, K.; Zhu, T.; Zhu, A. Identification of ramie genes in response to Pratylenchus coffeae infection challenge by digital gene expression analysis. Int. J. Mol. Sci. 2015, 16, 21989–22007. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Panchenko, A.R.; Shoemaker, B.A.; Thiessen, P.A.; Geer, L.Y.; Bryant, S.H. CDD: A database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002, 30, 281–283. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.T.; Chen, J.; Gao, C.S.; Zeng, L.B.; Li, Z.M.; Zhu, T.T.; Sun, K.; Cheng, Y.; Sun, X.P.; Yan, L.; et al. First report of brown root rot caused by Pythium vexans on ramie in Hunan, China. Can. J. Plant Pathol. 2016, 38, 405–410. [Google Scholar] [CrossRef]
- Yu, Y.T.; Zeng, L.B.; Huang, L.L.; Yan, Z.; Sun, K.; Zhu, T.T.; Zhu, A.G. First report of black leaf spot caused by Alternaria alternata on ramie in China. J. Phytopathol. 2016, 164, 358–361. [Google Scholar] [CrossRef]
- Pernas, M.; López-Solanilla, E.; Sánchez-Monge, R.; Salcedo, G.; Rodríguez-Palenzuela, P. Antifungal activity of a plant cystatin. Mol. Plant Microbe Interact. 1999, 12, 624–627. [Google Scholar] [CrossRef]
- Dutt, S.; Gaur, V.S.; Taj, G.; Kumar, A. Differential induction of two different cystatin genes during pathogenesis of Karnal bunt (Tilletia indica) in wheat under the influence of jasmonic acid. Gene 2012, 506, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.J.; Irene, D.; Lo, C.J.; Cai, Y.L.; Tzen, T.C.; Lin, T.H.; Chyan, C.L. Resonance assignments and secondary structure of a phytocystatin from Sesamum indicum. Biomol. NMR Assign. 2015, 9, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Domoto, C.; Watanabe, H.; Abe, K.; Arai, S. Structural organization of the gene encoding corn cystatin. Biosci. Biotechnol. Biochem. 1996, 60, 1173–1175. [Google Scholar] [PubMed]
- Misaka, T.; Kuroda, M.; Iwabuchi, K.; Abe, K.; Arai, S. Soyacystatin, a novel cysteine proteinase inhibitor in soybean, is distinct in protein structure and gene organization from other cystatins of animal and plant origin. FEBS J. 1996, 240, 609–614. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, P.; Zhou, X.M.; Xiong, H.X.; Sun, M.X. Genome-wide identification and characterization of cystatin family genes in rice (Oryza sativa L.). Plant Cell Rep. 2015, 34, 1579–1592. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.L.; Li, R.; Wang, L.; Chen, H.F.; Zhang, C.J.; Chen, L.M.; Hao, Q.N.; Shan, Z.H.; Zhang, X.J.; Chen, S.L.; et al. Search for nodulation and nodule development-related cystatin genes in the genome of Soybean (Glycine max). Front. Plant Sci. 2016, 7, 1595. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.; Abraham, Z.; Carbonero, P.; Díaz, I. Comparative phylogenetic analysis of cystatin gene families from Arabidopsis, rice and barley. Mol. Genet. Genom. 2005, 273, 423–432. [Google Scholar]
- Pernas, M.; Sánchez-Monge, R.; Gómez, L.; Salcedo, G. A chestnut seed cystatin differentially effective against cysteine proteinases from closely related pests. Plant Mol. Biol. 1998, 38, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Gaddour, K.; Vicente-Carbajosa, J.; Lara, P.; Isabel-Lamoneda, I.; Díaz, I.; Carbonero, P. A constitutive cystatin-encoding gene from barley (Icy) responds differentially to abiotic stimuli. Plant Mol. Biol. 2001, 45, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Neuteboom, L.W.; Matsumoto, K.O.; Christopher, D.A. An extended AE-rich N-terminal trunk in secreted pineapple cystatin enhances inhibition of fruit bromelain and is posttranslationally removed during ripening. Plant Physiol. 2009, 151, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Shyu, D.J.; Chyan, C.L.; Tzen, J.T.; Chou, W.M. Molecular cloning, expression, and functional characterization of a cystatin from pineapple stem. Biosci. Biotechnol. Biochem. 2004, 68, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Abraham, Z.; Martinez, M.; Carbonero, P.; Diaz, I. Structural and functional diversity within the cystatin gene family of Hordeum vulgare. J. Exp. Bot. 2006, 57, 4245–4255. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Abraham, Z.; Gambardella, M.; Echaide, M.; Carbonero, P.; Diaz, I. The strawberry gene Cyf1 encodes a phytocystatin with antifungal properties. J. Exp. Bot. 2005, 56, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M.; Andjelkovic, U.; Burazer, L.; Lindner, B.; Petersen, A.; Gavrovic-Jankulovic, M. Biochemical and immunological characterization of a recombinantly-produced antifungal cysteine proteinase inhibitor from green kiwifruit (Actinidia deliciosa). Phytochemistry 2013, 94, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Porruana, R.; Chundetb, R.; Anuntalabhochaia, S. Characterization of a cDNA encoding cystatin with antifungal activity from Siam tulip Curcuma alismatifolia. Sci. Asia 2013, 39, 596–604. [Google Scholar] [CrossRef]
- Carrillo, L.; Herrero, I.; Cambra, I.; Sánchez-Monge, R.; Diaz, I.; Martinez, M. Differential in vitro and in effect of barley cysteine and serine protease inhibitors on phytopathogenic microorganisms. Plant Physiol. Biochem. 2011, 49, 1191–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Zhang, G.; Li, Z.; Cheng, Y.; Gao, C.; Zeng, L.; Chen, J.; Yan, L.; Sun, X.; Guo, L.; et al. Molecular Cloning, Recombinant Expression and Antifungal Activity of BnCPI, a Cystatin in Ramie (Boehmeria nivea L.). Genes 2017, 8, 265. https://doi.org/10.3390/genes8100265
Yu Y, Zhang G, Li Z, Cheng Y, Gao C, Zeng L, Chen J, Yan L, Sun X, Guo L, et al. Molecular Cloning, Recombinant Expression and Antifungal Activity of BnCPI, a Cystatin in Ramie (Boehmeria nivea L.). Genes. 2017; 8(10):265. https://doi.org/10.3390/genes8100265
Chicago/Turabian StyleYu, Yongting, Gang Zhang, Zhimin Li, Yi Cheng, Chunsheng Gao, Liangbin Zeng, Jia Chen, Li Yan, Xiangping Sun, Litao Guo, and et al. 2017. "Molecular Cloning, Recombinant Expression and Antifungal Activity of BnCPI, a Cystatin in Ramie (Boehmeria nivea L.)" Genes 8, no. 10: 265. https://doi.org/10.3390/genes8100265




