Textile Hemp vs. Salinity: Insights from a Targeted Gene Expression Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Set-Up, Plant Material and Growth Conditions
2.2. Soil Electric Conductivity and pH Measurement
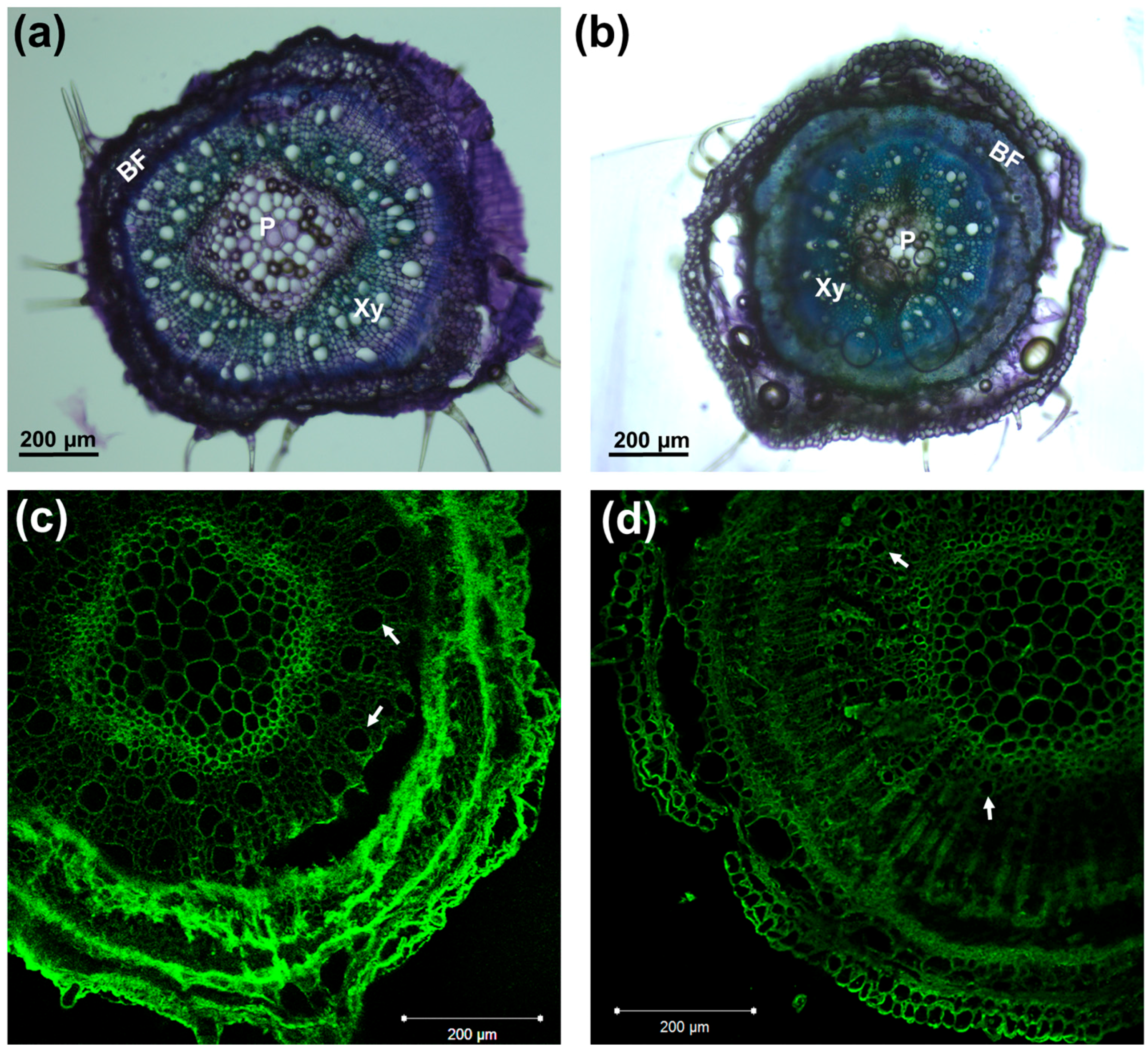
2.3. Toluidine Blue Staining and Immunohistochemistry
2.4. RNA Extraction and Reverse Transcription-quantitative PCR (RT-qPCR)
2.5. Primer Design
3. Results and Discussion
3.1. Phenotype of Hemp Plantlets Exposed to NaCl
3.2. Gene Expression Pattern in the Leaves
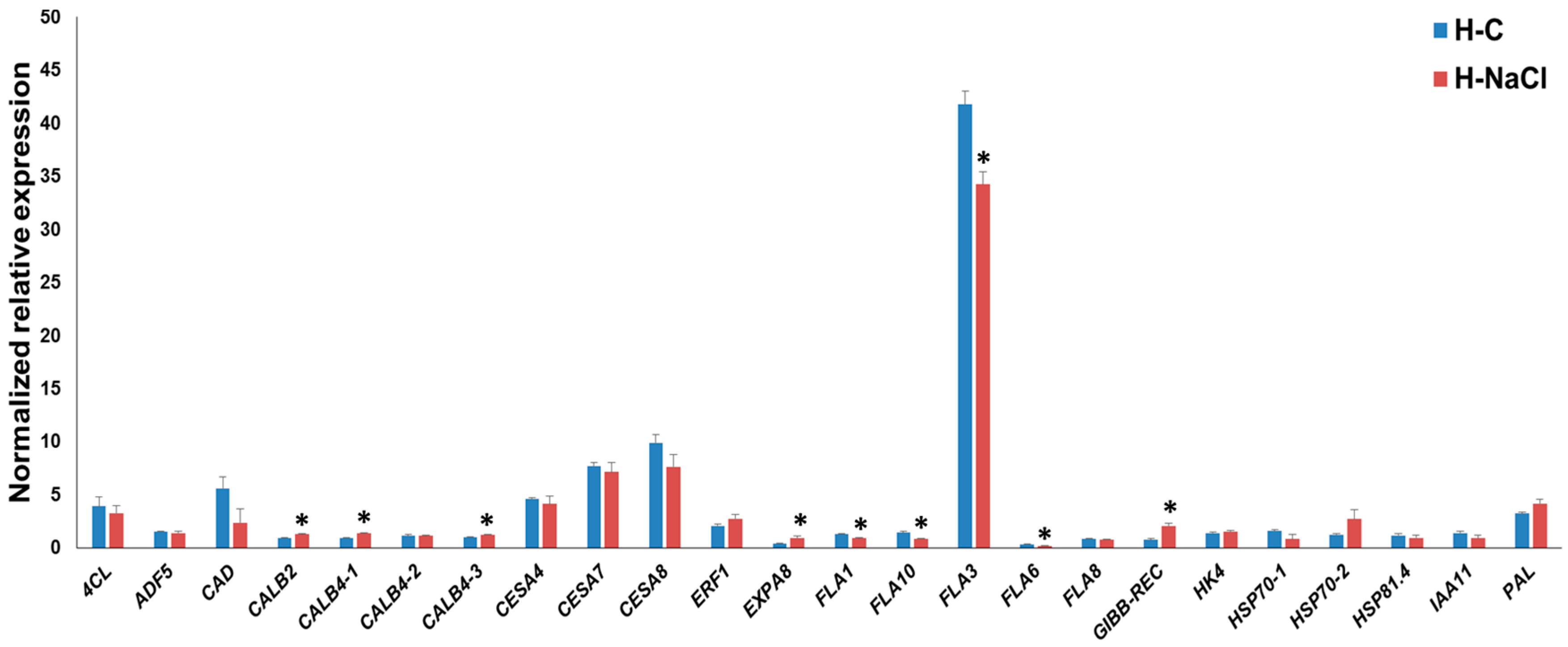
3.3. Gene Expression Pattern in the Hypocotyls
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cardi, M.; Castiglia, D.; Ferrara, M.; Guerriero, G.; Chiurazzi, M.; Esposito, S. The effects of salt stress cause a diversion of basal metabolism in barley roots: Possible different roles for glucose-6-phosphate dehydrogenase isoforms. Plant Physiol. Biochem. 2015, 86, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Legay, S.; Hausman, J.-F. Alfalfa Cellulose synthase gene expression under abiotic stress: A Hitchhiker’s guide to RT-qPCR normalization. PLoS ONE 2014, 9, e103808. [Google Scholar] [CrossRef] [PubMed]
- Behr, M.; Legay, S.; Hausman, J.-F.; Guerriero, G. Analysis of Cell Wall-Related Genes in Organs of Medicago sativa L. under Different Abiotic Stresses. Int. J. Mol. Sci. 2015, 16, 16104–16124. [Google Scholar] [CrossRef] [PubMed]
- Woodrow, P.; Ciarmiello, L.F.; Annunziata, M.G.; Pacifico, S.; Iannuzzi, F.; Mirto, A.; D’Amelia, L.; Dell’Aversana, E.; Piccolella, S.; Fuggi, A.; et al. Durum wheat seedling responses to simultaneous high light and salinity involve a fine reconfiguration of amino acids and carbohydrate metabolism. Physiol. Plant. 2017, 159, 290–312. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Maximova, E.; Fuggi, A.; Carillo, P. Durum Wheat Roots Adapt to Salinity Remodeling the Cellular Content of Nitrogen Metabolites and Sucrose. Front. Plant Sci. 2016, 7, 2035. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Cabot, C.; Sibole, J.V.; Barceló, J.; Poschenrieder, C. Lessons from crop plants struggling with salinity. Plant Sci. Int. J. Exp. Plant Biol. 2014, 226, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Landi, S.; Hausman, J.-F.; Guerriero, G.; Esposito, S. Poaceae vs. Abiotic Stress: Focus on Drought and Salt Stress, Recent Insights and Perspectives. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Carillo, P.; Parisi, D.; Woodrow, P.; Pontecorvo, G.; Massaro, G.; Annunziata, M.G.; Fuggi, A.; Sulpice, R. Salt-induced accumulation of glycine betaine is inhibited by high light in durum wheat. Funct. Plant Biol. 2011, 38, 139–150. [Google Scholar] [CrossRef]
- Gao, H.-J.; Yang, H.-Y.; Bai, J.-P.; Liang, X.-Y.; Lou, Y.; Zhang, J.-L.; Wang, D.; Zhang, J.-L.; Niu, S.-Q.; Chen, Y.-L. Ultrastructural and physiological responses of potato (Solanum tuberosum L.) plantlets to gradient saline stress. Front. Plant Sci. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, H.; Chen, T.; Pen, J.; Yu, S.; Zhao, X. Morphological and Physiological Responses of Cotton (Gossypium hirsutum L.) Plants to Salinity. PLoS ONE 2014, 9, e112807. [Google Scholar] [CrossRef] [PubMed]
- Katsuhara, M.; Kawasaki, T. Salt Stress Induced Nuclear and DNA Degradation in Meristematic Cells of Barley Roots. Plant Cell Physiol. 1996, 37, 169–173. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.; Wang, D.; Gong, Q. Autophagy Is Rapidly Induced by Salt Stress and Is Required for Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1459. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, P.; Mohanty, B.; Wijaya, E.; Lee, D.-Y.; Lim, T.-M.; Lin, Q.; Xu, J.; Loh, C.-S.; Kumar, P.P. Transcriptomics analysis of salt stress tolerance in the roots of the mangrove Avicennia officinalis. Sci. Rep. 2017, 7, 10031. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cui, G.; Hu, G.; Wang, M.; Zhang, P.; Qin, L.; Shang, C.; Zhang, H.; Zhu, X.; Qu, M. Proteome dynamics and physiological responses to short-term salt stress in Leymus chinensis leaves. PLoS ONE 2017, 12, e0183615. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yang, A.; Zhang, W.-H. Comparative studies on tolerance of rice genotypes differing in their tolerance to moderate salt stress. BMC Plant Biol. 2017, 17, 141. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Huang, W.; Chen, H.; Wu, G.; Yuan, H.; Song, X.; Kang, Q.; Zhao, D.; Jiang, W.; Liu, Y.; et al. Identification of differentially expressed genes in flax (Linum usitatissimum L.) under saline-alkaline stress by digital gene expression. Gene 2014, 549, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qiao, Q.; Cheng, X.; Du, G.; Deng, G.; Zhao, M.; Liu, F. Transcriptome differences between fiber-type and seed-type Cannabis sativa variety exposed to salinity. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2016, 22, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Behr, M.; Legay, S.; Žižková, E.; Motyka, V.; Dobrev, P.I.; Hausman, J.-F.; Lutts, S.; Guerriero, G. Studying Secondary Growth and Bast Fiber Development: The Hemp Hypocotyl Peeks behind the Wall. Front. Plant Sci. 2016, 7, 1733. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Behr, M.; Legay, S.; Mangeot-Peter, L.; Zorzan, S.; Ghoniem, M.; Hausman, J.-F. Transcriptomic profiling of hemp bast fibres at different developmental stages. Sci. Rep. 2017, 7, 4961. [Google Scholar] [CrossRef] [PubMed]
- Shavrukov, Y. Salt stress or salt shock: Which genes are we studying? J. Exp. Bot. 2013, 64, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Legay, S.; Lamoureux, D.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Monitoring gene expression of potato under salinity using cDNA microarrays. Plant Cell Rep. 2009, 28, 1799–1816. [Google Scholar] [CrossRef] [PubMed]
- Mangeot-Peter, L.; Legay, S.; Hausman, J.-F.; Esposito, S.; Guerriero, G. Identification of Reference Genes for RT-qPCR Data Normalization in Cannabis sativa Stem Tissues. Int. J. Mol. Sci. 2016, 17, 1556. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Marschner, P.; Cao, W.; Zuo, C.; Qin, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef]
- Carillo, P.; Annunziata, M.G.; Pontecorvo, G.; Fuggi, A.; Woodrow, P. Salinity Stress and Salt Tolerance. In Abiotic Stress in Plants-Mechanisms and Adaptations; InTech: Rijeka, Croatia, 2011. [Google Scholar] [CrossRef]
- Junghans, U.; Polle, A.; Düchting, P.; Weiler, E.; Kuhlman, B.; Gruber, F.; Teichmann, T. Adaptation to high salinity in poplar involves changes in xylem anatomy and auxin physiology. Plant Cell Environ. 2006, 29, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Zang, L.; Zheng, T.; Chu, Y.; Ding, C.; Zhang, W.; Huang, Q.; Su, X. Genome-Wide Analysis of the Fasciclin-Like Arabinogalactan Protein Gene Family Reveals Differential Expression Patterns, Localization, and Salt Stress Response in Populus. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zhao, J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J. Exp. Bot. 2010, 61, 2647–2668. [Google Scholar] [CrossRef] [PubMed]
- Faik, A.; Abouzouhair, J.; Sarhan, F. Putative fasciclin-like arabinogalactan-proteins (FLA) in wheat (Triticum aestivum) and rice (Oryza sativa): Identification and bioinformatic analyses. Mol. Genet. Genom. MGG 2006, 276, 478–494. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Kim, Y.; Guo, Y.; Stevenson, B.; Zhu, J.-K. The Arabidopsis SOS5 Locus Encodes a Putative Cell Surface Adhesion Protein and Is Required for Normal Cell Expansion. Plant Cell 2003, 15, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Mangeot-Peter, L.; Legay, S.; Behr, M.; Lutts, S.; Siddiqui, K.S.; Hausman, J.-F. Identification of fasciclin-like arabinogalactan proteins in textile hemp (Cannabis sativa L.): In silico analyses and gene expression patterns in different tissues. BMC Genom. 2017, 18, 741. [Google Scholar] [CrossRef] [PubMed]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef] [PubMed]
- Moura, J.C.M.S.; Bonine, C.A.V.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Zörb, C.; Mühling, K.H.; Kutschera, U.; Geilfus, C.-M. Salinity stiffens the epidermal cell walls of salt-stressed maize leaves: Is the epidermis growth-restricting? PLoS ONE 2015, 10, e0118406. [Google Scholar] [CrossRef] [PubMed]
- Inada, N. Plant actin depolymerizing factor: Actin microfilament disassembly and more. J. Plant Res. 2017, 130, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.-C.; Liao, P.-M.; Kuo, W.-W.; Lin, T.-P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 Regulates Abiotic Stress-Responsive Gene Expression by Binding to Different cis-Acting Elements in Response to Different Stress Signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Deng, G.; Su, Y.; Liu, J.J.; Yang, Y.; Du, G.H.; Chen, Z.Y.; Liu, F.H. Protein mechanisms in response to NaCl-stress of salt-tolerant and salt-sensitive industrial hemp based on iTRAQ technology. Ind. Crops Prod. 2016, 83, 444–452. [Google Scholar] [CrossRef]
- De Silva, K.; Laska, B.; Brown, C.; Sederoff, H.W.; Khodakovskaya, M. Arabidopsis thaliana calcium-dependent lipid-binding protein (AtCLB): A novel repressor of abiotic stress response. J. Exp. Bot. 2011, 62, 2679–2689. [Google Scholar] [CrossRef] [PubMed]

| Primer Name | Sequence (5′ → 3′) | Amplicon Size (bp) | Amplification Efficiency (%) |
|---|---|---|---|
| IAA11 Fwd | GTGGCCTCCAATCAGAACTTAC | 79 | 110.7 |
| IAA11 Rev | GCTGAATCCTTCATTGAGTGG | ||
| ADF5 Fwd | TGGATGTGATTCAGGACAGG | 116 | 105.5 |
| ADF5 Rev | TGGATAAAAGCATAGCCCTTG | ||
| ERF1 Fwd | TACTTCAATGGCAGCAGCAC | 96 | 105.3 |
| ERF1 Rev | TTTGGTGGTGGGTCGTTTAG | ||
| CALB2 Fwd | GCTTCCTCCAAGTTTTCGTG | 128 | 93.8 |
| CALB2 Rev | CAACCACAACCGTCGATATG | ||
| CALB4-1 Fwd | TGTTCTTGCTCATCCTGCTC | 114 | 92.8 |
| CALB4-1 Rev | GCTTCAAGGGTTGCTGATTC | ||
| CALB4-2 Fwd | GGCTGGTAGGCAAGTTTTTG | 90 | 94.2 |
| CALB4-2 Rev | TGAAGCTCCAATCCCCTATG | ||
| CALB4-3 Fwd | ATCTGGGCGAGATATTGCTG | 85 | 95.7 |
| CALB4-3 Rev | CGAGTCATAACTTGGCACAAC | ||
| HSP70-1 Fwd | TCAACCATTTCGTCCAGGAG | 98 | 91.6 |
| HSP70-1 Rev | CGCTCTCTCACAAGATGTTCTC | ||
| HSP70-2 Fwd | GGGATTCTGAATGTGTCTGC | 92 | 104.1 |
| HSP70-2 Rev | TCTTCCTTGCTCAACCTTCC | ||
| HSP81.4 Fwd | AGGAGTTCATGGAGGCATTG | 94 | 92.2 |
| HSP81.4 Rev | TTCTCGGCAACAAGGTAAGC | ||
| GibbRec Fwd | TAATCTTCTTCGGCGGTCTG | 102 | 102.6 |
| GibbRec Rev | GAGAAAACCCCATCAACAGG | ||
| HK4 Fwd | TGCTGAGGTGGGTTTTTCTC | 139 | 93.5 |
| HK4 Rev | TTGTGCTGCATCTTCAGACC |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerriero, G.; Behr, M.; Hausman, J.-F.; Legay, S. Textile Hemp vs. Salinity: Insights from a Targeted Gene Expression Analysis. Genes 2017, 8, 242. https://doi.org/10.3390/genes8100242
Guerriero G, Behr M, Hausman J-F, Legay S. Textile Hemp vs. Salinity: Insights from a Targeted Gene Expression Analysis. Genes. 2017; 8(10):242. https://doi.org/10.3390/genes8100242
Chicago/Turabian StyleGuerriero, Gea, Marc Behr, Jean-Francois Hausman, and Sylvain Legay. 2017. "Textile Hemp vs. Salinity: Insights from a Targeted Gene Expression Analysis" Genes 8, no. 10: 242. https://doi.org/10.3390/genes8100242





