Identification and Functional Analysis of Interleukin-1β in the Chinese Soft-Shelled Turtle Pelodiscus sinensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Tissue cDNA Samples
2.2. Cloning of the Genes Encoding Turtle Interleukin-1β (IL-1β) and Caspase-1
2.3. Sequence Analysis
2.4. Preparation of Polyclonal Antibodies Against tIL-1β
2.5. Quantitative PCR
2.6. tIL-1β Expression Analysis in Healthy or Aeromonas Hydrophila-Infected Turtles
2.7. Plasmid Construction and Transfection
2.8. Expression of Turtle IL-1β in the Baculovirus Expression System
2.9. Transcriptional Analysis of Inflammatory Cytokines in Turtle Peripheral Blood Monocytes Stimulated with tIL-1β
2.10. Western Blotting
2.11. Statistical Analysis
3. Results
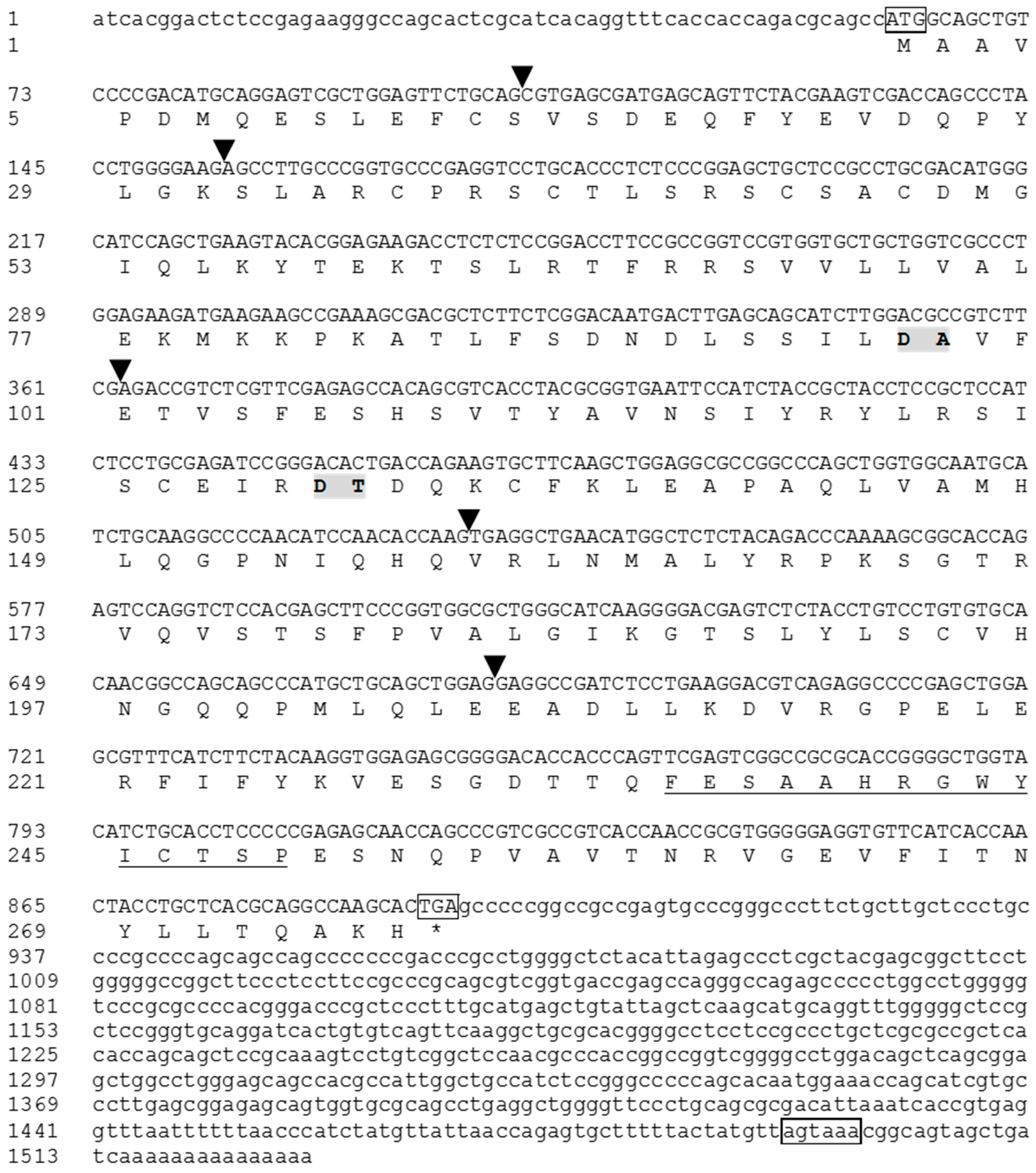
3.1. Cloning of the Full-Length tIL-1β cDNA
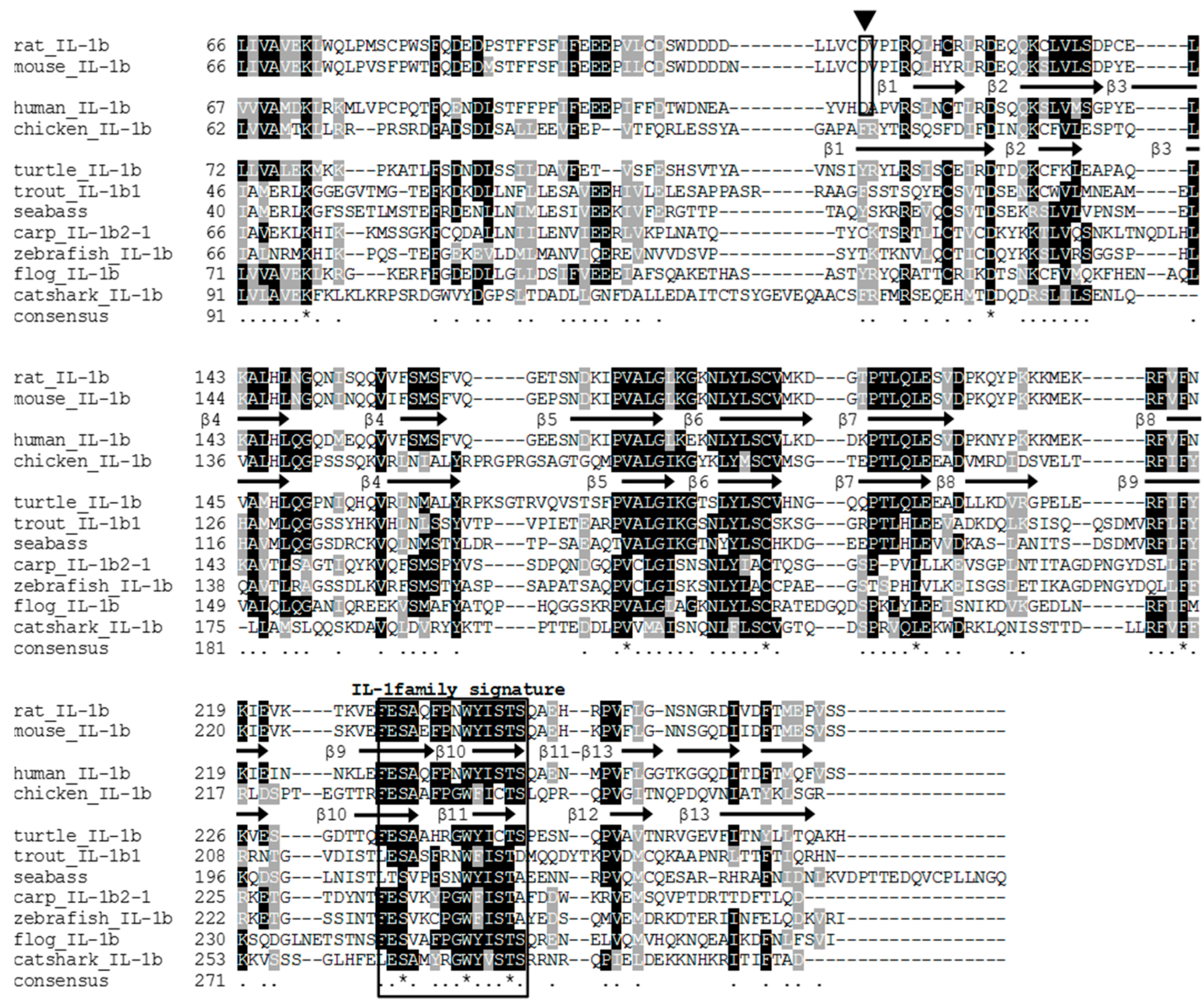
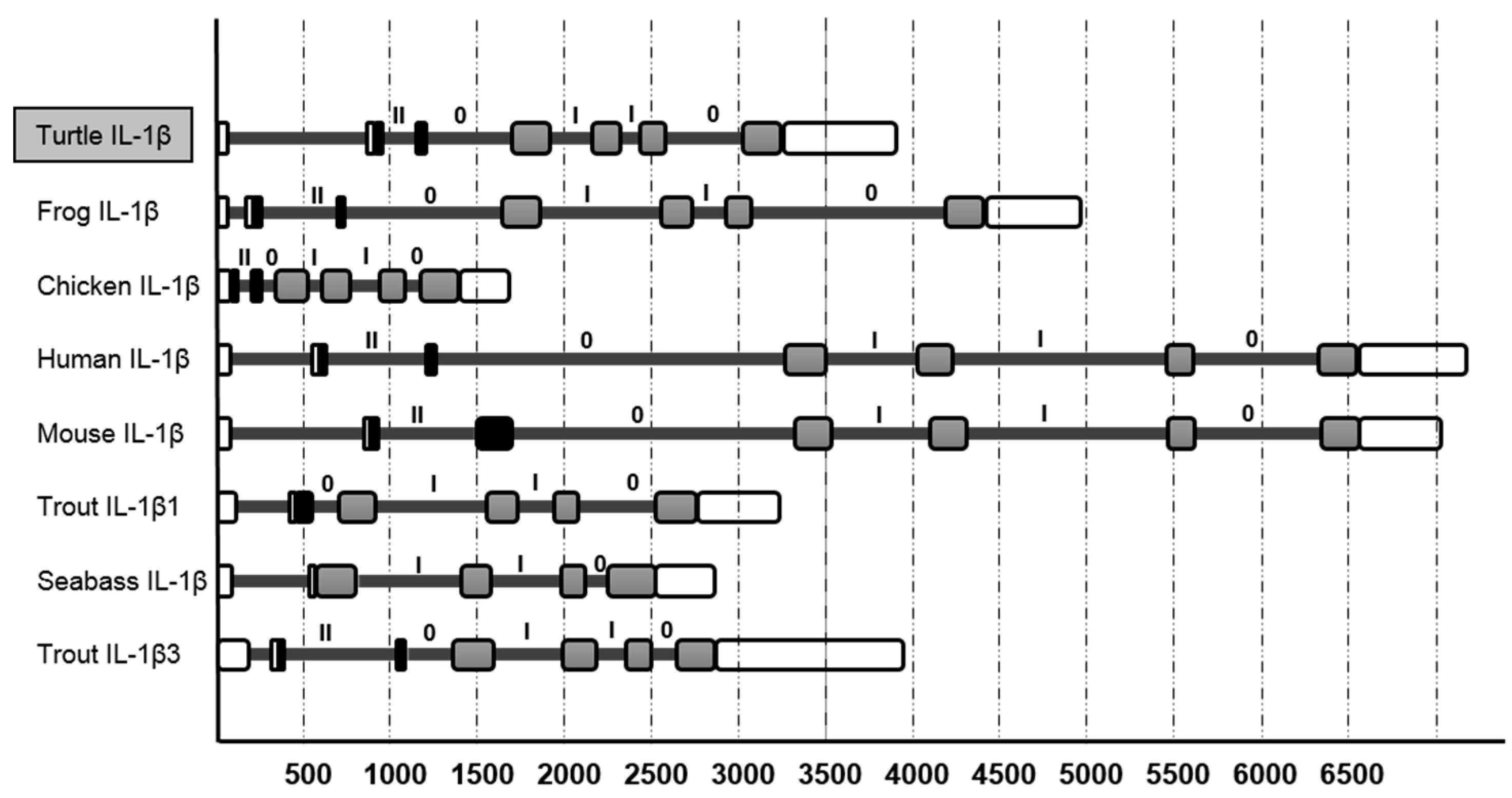
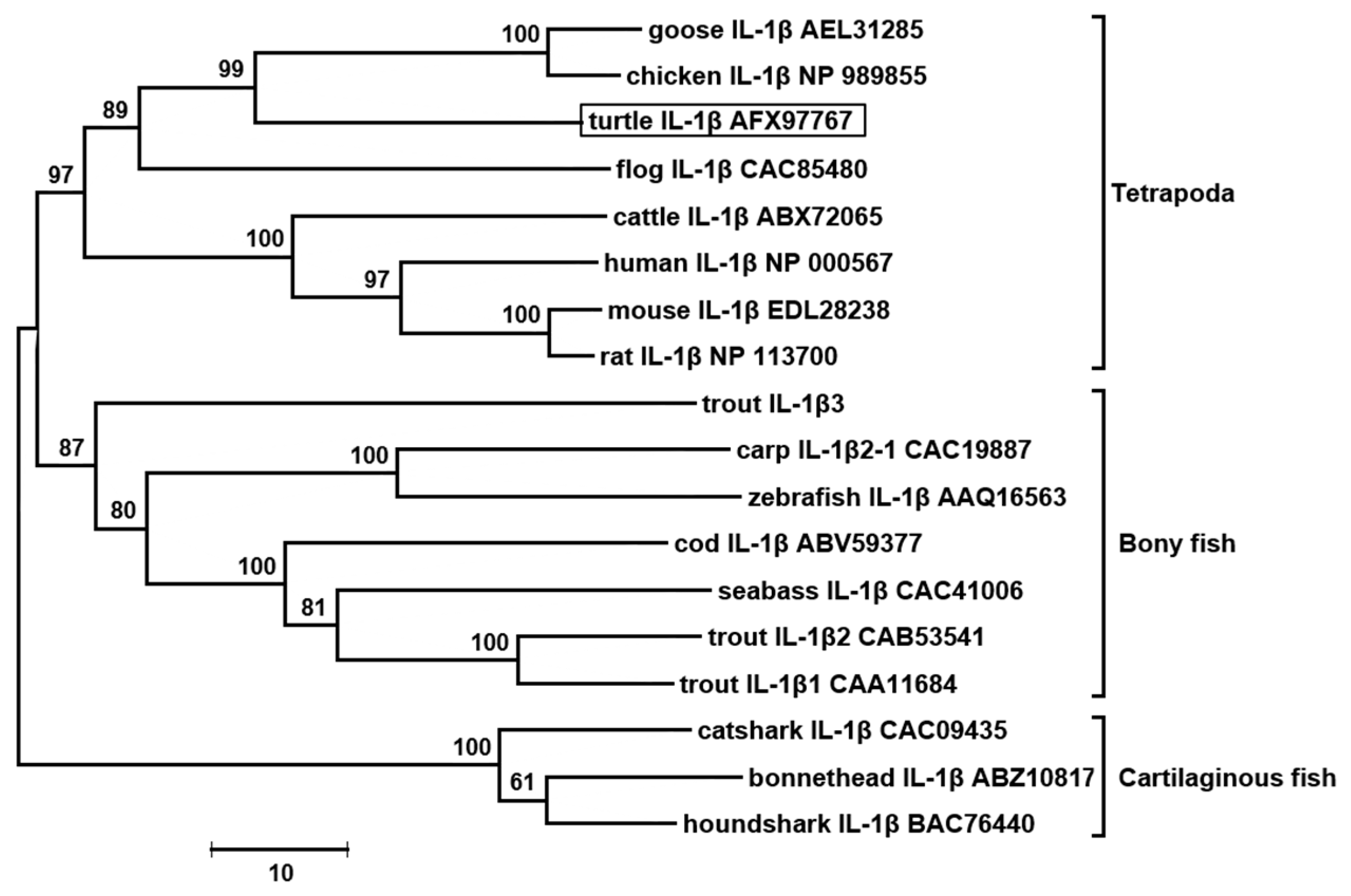
3.2. The tIL-1β Gene Organization and Phylogenetic Tree
3.3. Expression of tIL-1β in Different Tissues of Clinically Healthy Turtles or Aeromonas Hydrophila-Infected Turtles
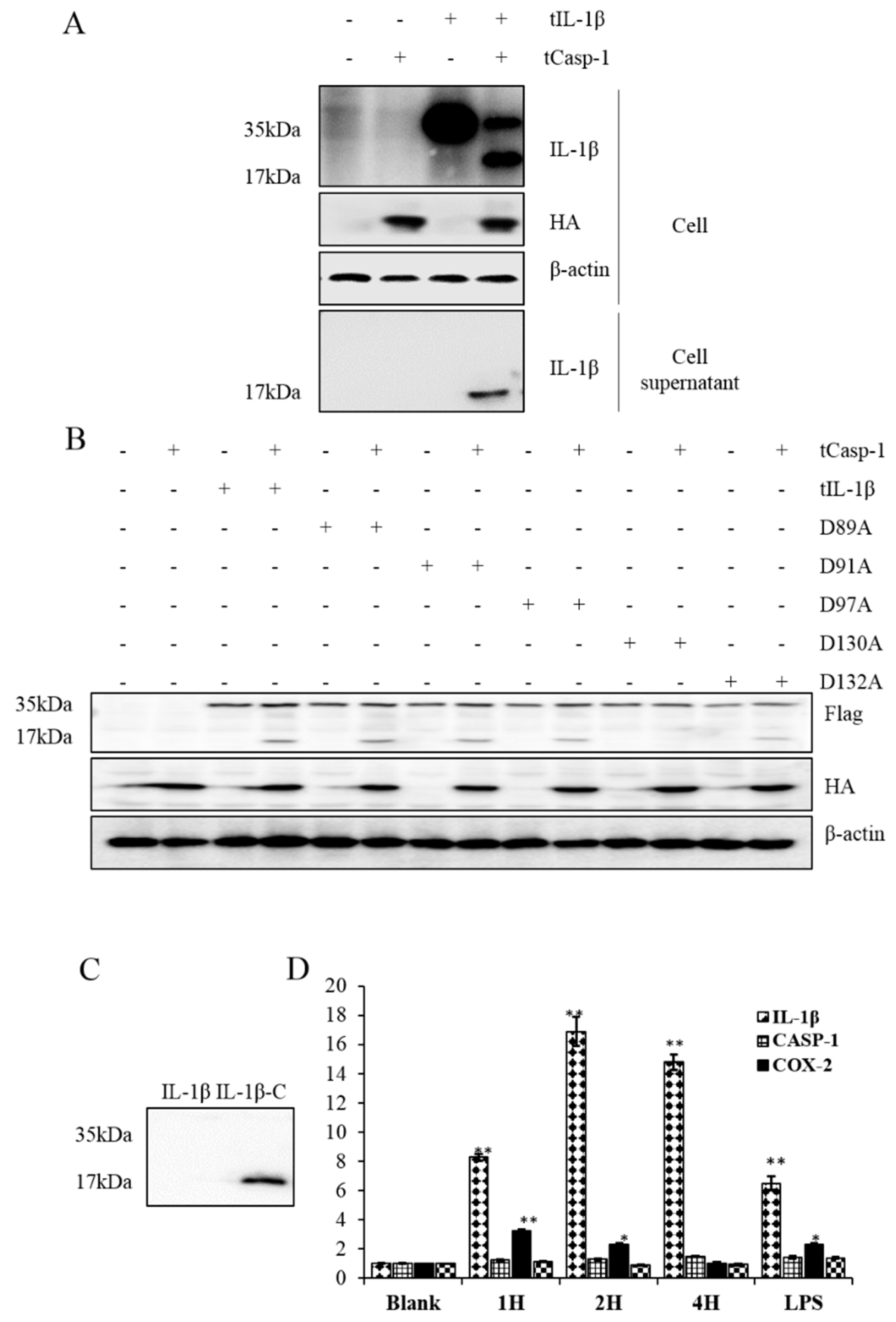
3.4. Processing of tIL-1β by tcaspase-1 in HEK293 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Husain, M.; Bird, S.; van Zwieten, R.; Secombes, C.J.; Wang, T. Cloning of the IL-1β3 gene and IL-1β4 pseudogene in salmonids uncovers a second type of IL-1β gene in teleost fish. Dev. Comp. Immunol. 2012, 38, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Bird, S.; Zou, J.; Wang, T.; Munday, B.; Cunningham, C.; Secombes, C.J. Evolution of interleukin-1beta. Cytokine Growth Factor Rev. 2002, 13, 483–502. [Google Scholar] [CrossRef]
- Thornberry, N.A.; Bull, H.G.; Calaycay, J.R.; Chapman, K.T.; Howard, A.D.; Kostura, M.J.; Miller, D.K.; Molineaux, S.M.; Weidner, J.R.; Aunins, J. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 1992, 356, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Wasiliew, P.; Kracht, M. Interleukin-1beta (IL-1beta) processing pathway. Sci. Signal. 2010. [Google Scholar] [CrossRef]
- Irmler, M.; Hertig, S.; MacDonald, H.R.; Sadoul, R.; Becherer, J.; Proudfoot, A.; Solari, R.; Tschopp, J. Granzyme A is an interleukin 1 beta-converting enzyme. J. Exp. Med. 1995, 181, 1917–1922. [Google Scholar] [CrossRef] [PubMed]
- Black, R.A.; Kronheim, S.R.; Cantrell, M.; Deeley, M.C.; March, C.J.; Prickett, K.S.; Wignall, J.; Conlon, P.J.; Cosman, D.; Hopp, T.P. Generation of biologically active interleukin-1 beta by proteolytic cleavage of the inactive precursor. J. Biol. Chem. 1988, 263, 9437–9442. [Google Scholar] [PubMed]
- Lopez-Castejon, G.; Brough, D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011, 22, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Andrei, C.; Dazzi, C.; Lotti, L.; Torrisi, M.R.; Chimini, G.; Rubartelli, A. The secretory route of the leaderless protein interleukin 1β involves exocytosis of endolysosome-related vesicles. Mol. Boil. Cell 1999, 10, 1463–1475. [Google Scholar] [CrossRef]
- O’Neill, L.A. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol. Rev. 2008, 226, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, B.; Chen, N.; Hu, H.; Chen, J.; Zhang, X.; Li, J.; Fang, W. Molecular characterization and functional analysis of MyD88 in Chinese soft-shelled turtle Trionyx sinensis. Fish Shellfish Immunol. 2011, 30, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Osterbur, K.; Yu, D.H.; DeClue, A.E. Interleukin-1β, tumour necrosis factor-α and lipopolysaccharide induce C-type natriuretic peptide from canine aortic endothelial cells. Res. Vet. Sci. 2013, 94, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Roca, F.J.; Cayuela, M.L.; Secombes, C.J.; Meseguer, J.; Mulero, V. Post-transcriptional regulation of cytokine genes in fish: A role for conserved AU-rich elements located in the 3’-untranslated region of their mRNAs. Mol. Immunol. 2007, 44, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Bufler, P.; Gamboni-Robertson, F.; Azam, T.; Kim, S.H.; Dinarello, C.A. Interleukin-1 homologues IL-1F7b and IL-18 contain functional mRNA instability elements within the coding region responsive to lipopolysaccharide. Biochem. J. 2004, 381, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Greten, F.R.; Arkan, M.C.; Bollrath, J.; Hsu, L.C.; Goode, J.; Miething, C.; Göktuna, S.I.; Neuenhahn, M.; Fierer, J.; Paxian, S. NF-κB is a negative regulator of IL-1β secretion as revealed by genetic and pharmacological inhibition of IKKβ. Cell 2007, 130, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Nishida, T.; Nishino, N.; Takano, M.; Kawai, K.; Bando, K.; Masui, Y.; Nakai, S.; Hirai, Y. cDNA cloning of IL-1 alpha and IL-1 beta from mRNA of U937 cell line. Biochem. Biophys. Res. Commun. 1987, 143, 345–352. [Google Scholar] [CrossRef]
- Gray, P.W.; Glaister, D.; Chen, E.; Goeddel, D.V.; Pennica, D. Two interleukin 1 genes in the mouse: Cloning and expression of the cDNA for murine interleukin 1 beta. J. Immunol. 1986, 137, 3644–3648. [Google Scholar] [PubMed]
- Leong, S.R.; Flaggs, G.M.; Lawman, M.; Gray, P.W. The nucleotide sequence for the cDNA of bovine interleukin-1 beta. Nucleic Acids Res. 1988, 16, 9054–9054. [Google Scholar] [CrossRef] [PubMed]
- Fiskerstrand, C.; Sargan, D. Nucleotide sequence of ovine interleukin-1 beta. Nucleic Acids Res. 1990. [Google Scholar] [CrossRef]
- Huether, M.J.; Lin, G.; Smith, D.M.; Murtaugh, M.P.; Molitor, T.W. Cloning, sequencing and regulation of an mRNA encoding porcine interleukin-1 beta. Gene 1993, 129, 285–289. [Google Scholar] [CrossRef]
- Weining, K.C.; Sick, C.; Kaspers, B.; Staeheli, P. A chicken homolog of mammalian interleukin-1 beta: cDNA cloning and purification of active recombinant protein. Eur. J. Biochem. 1998, 258, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Grabowski, P.S.; Cunningham, C.; Secombes, C.J. Molecular cloning of interleukin 1beta from rainbow trout Oncorhynchus mykiss reveals no evidence of an ice cut site. Cytokine 1999, 11, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Scapigliati, G.; Buonocore, F.; Bird, S.; Zou, J.; Pelegrin, P.; Falasca, C.; Prugnoli, D.; Secombes, C.J. Phylogeny of cytokines: Molecular cloning and expression analysis of sea bass Dicentrarchus labrax interleukin-1beta. Fish Shellfish Immunol. 2001, 11, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Bird, S.; Minter, R.; Horton, J.; Cunningham, C.; Secombes, C.J. Molecular cloning of the gene for interleukin-1β from Xenopus laevis and analysis of expression in vivo and in vitro. Immunogenetics 2000, 51, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Bird, S.; Wang, T.; Zou, J.; Cunningham, C.; Secombes, C.J. The first cytokine sequence within cartilaginous fish: IL-1β in the small spotted catshark (Scyliorhinus canicula). J. Immunol. 2002, 168, 3329–3340. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, L.M.; Vogel, L.A.; Bowden, R.M. Understanding the vertebrate immune system: Insights from the reptilian perspective. J. Exp. Biol. 2010, 213, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, C.; Fang, W.; Lin, F. White-spot disease of Chinese soft-shelled turtles (Trionyx sinens) caused by Paecilomyces lilacinus. J. Zhejiang Univ. Sci. B 2008, 9, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhu, N.; Kong, L.; Bei, Y.; Zheng, T.; Ding, X.; He, Z. First case of soft shell disease in Chinese soft-shelled turtle (Trionyx sinens) associated with Aeromonas sobria—A. veronii complex. Aquaculture 2013, 406, 62–67. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, N.; Kong, L.; Bei, Y.; Zheng, T.; Ding, X.; He, Z. First reported fatal Bacillus thuringiensis infections in Chinese soft-shelled turtles (Trionyx sinensis). Aquaculture 2014, 428, 16–20. [Google Scholar] [CrossRef]
- Florea, L.; Hartzell, G.; Zhang, Z.; Rubin, G.M.; Miller, W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998, 8, 967–974. [Google Scholar] [PubMed]
- Li, X.; Kang, Y.; Zhang, X.; Zhu, B.; Fang, W. Identification of a heat shock cognate protein 70 gene in Chinese soft-shell turtle (Pelodiscus sinensis) and its expression profiles under thermal stress. J. Zhejiang Univ. Sci. B 2012, 13, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G. Caspases: the executioners of apoptosis. Biochem. J. 1997, 326, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Thummabancha, K.; Onparn, N.; Srisapoome, P. Molecular characterization and expression analyses of cDNAs encoding the thioredoxin-interacting protein and selenoprotein P genes and histological changes in Nile tilapia (Oreochromis niloticus) in response to silver nanoparticle exposure. Gene 2016, 577, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Lüthi, A.; Martin, S. The CASBAH: A searchable database of caspase substrates. Cell Death Differ. 2007, 14, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Caput, D.; Beutler, B.; Hartog, K.; Thayer, R.; Brown-Shimer, S.; Cerami, A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Nat. Acad. Sci. USA 1986, 83, 1670–1674. [Google Scholar] [CrossRef] [PubMed]
- Pesole, G.; Mignone, F.; Gissi, C.; Grillo, G.; Licciulli, F.; Liuni, S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene 2001, 276, 73–81. [Google Scholar] [CrossRef]
- Sims, J.E.; Smith, D.E. The IL-1 family: regulators of immunity. Nat. Rev. Immunol. 2010, 10, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Yang, W.; Dai, Q.; Fu, J. Using genes as characters and a parsimony analysis to explore the phylogenetic position of turtles. PLoS ONE 2013, 8, e79348. [Google Scholar] [CrossRef] [PubMed]
- Secombes, C.J.; Wang, T.; Bird, S. The interleukins of fish. Dev. Comp. Immunol. 2011, 35, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Lynch, M. Ubiquitous internal gene duplication and intron creation in eukaryotes. Proc. Nat. Acad. Sci. USA 2009, 106, 20818–20823. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, D.; Li, J.; Liu, Z. Molecular characterization, recombinant expression and bioactivity analysis of the interleukin-1β from the yellowfin sea bream, Acanthopagrus latus (Houttuyn). Fish Shellfish Immunol. 2008, 24, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Lepen Pleić, I.; Secombes, C.; Bird, S.; Mladineo, I. Characterization of three major cytokines: IL1, TNF1 and TNF2 in reared Atlantic bluefin tuna Thunnus thynnus. Fish Shellfish Immunol. 2013. [Google Scholar] [CrossRef]
- Lu, X.J.; Chen, J.; He, Y.Q.; Shi, Y.H. Molecular characterization of an IL-1β gene from ayu, Plecoglossus altivelis. Fish Shellfish Immunol. 2013, 34, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Lepen Pleić, I.; Secombes, C.J.; Bird, S.; Mladineo, I. Characterization of three pro-inflammatory cytokines, TNFα1, TNFα2 and IL-1β, in cage-reared Atlantic bluefin tuna Thunnus thynnus. Fish Shellfish Immunol. 2014, 36, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Bird, S.; Koussounadis, A.; Holland, J.W.; Carrington, A.; Zou, J.; Secombes, C.J. Identification of a novel IL-1 cytokine family member in teleost fish. J. Immunol. 2009, 183, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Biologic basis for interleukin-1 in disease. Blood 1996, 87, 2095–2147. [Google Scholar] [PubMed]
- Dinarello, C.A. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 2009, 27, 519–550. [Google Scholar] [CrossRef] [PubMed]
- Swaan, P.W.; Knoell, D.L.; Helsper, F.; Wewers, M.D. Sequential processing of human ProIL-1beta by caspase-1 and subsequent folding determined by a combined in vitro and in silico approach. Pharm. Res. 2001, 18, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Vojtech, L.N.; Scharping, N.; Woodson, J.C.; Hansen, J.D. Roles of Inflammatory caspases during processing of zebrafish interleukin-1beta in Francisella noatunensis infection. Infect. Immun. 2012, 80, 2878–2885. [Google Scholar] [CrossRef] [PubMed]
- Garlanda, C.; Dinarello, C.A.; Mantovani, A. The interleukin-1 family: Back to the future. Immunity 2013, 39, 1003–1018. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Q.; Li, W.; Guo, N.; Tong, C.; Zhou, Y.; Fang, W.; Li, X. Identification and Functional Analysis of Interleukin-1β in the Chinese Soft-Shelled Turtle Pelodiscus sinensis. Genes 2016, 7, 18. https://doi.org/10.3390/genes7050018
Liang Q, Li W, Guo N, Tong C, Zhou Y, Fang W, Li X. Identification and Functional Analysis of Interleukin-1β in the Chinese Soft-Shelled Turtle Pelodiscus sinensis. Genes. 2016; 7(5):18. https://doi.org/10.3390/genes7050018
Chicago/Turabian StyleLiang, Quan, Weifen Li, Ningning Guo, Chao Tong, Yingshan Zhou, Weihuan Fang, and Xiaoliang Li. 2016. "Identification and Functional Analysis of Interleukin-1β in the Chinese Soft-Shelled Turtle Pelodiscus sinensis" Genes 7, no. 5: 18. https://doi.org/10.3390/genes7050018




