Transcriptomic Analysis of the Porcine Endometrium during Embryo Implantation
Abstract
:1. Introduction
2. Experimental Section
2.1. Animals and Tissue Collection
2.2. mRNA Library Construction and Sequencing
2.3. RNA-Seq Data Analysis
2.4. Transcript Abundance Estimation and Differentially Expressed Testing
2.5. Functional Enrichment Analysis
2.6. Quantitative Real-Time PCR
| Gene Symbol | Gene Name | Primer Sequence | Target Accession No. | |
|---|---|---|---|---|
| ACTB | actin, beta | F: | CGAGCGCTTCCGGTGTCCAG | XM_003357928.1 |
| R: | GTGGTCCCGCCAGACAGCAC | |||
| RN18S | 18S ribosomal RNA | F: | GGGAGGAGGCTGACCGGGTT | NR_002170.3 |
| R: | ATACATGCCGACGGGCGCTG | |||
| FGF9 | fibroblast growth factor 9 (glia-activating factor) | F: | TTCCCAGGGGACCCGCAGTC | NM_213801.1 |
| R: | ATGCTGACCAAGCCCACGGC | |||
| IRF1 | interferon regulatory factor 1 | F: | CCTTGTGCACCGTAGGCGGG | NM_001097413.1 |
| R: | GGCTTGCCAGGCCCCAAGAG | |||
| PTGES | prostaglandin E synthase | F: | TGGTGAGCGGCCAGGTT | NM_001038631.1 |
| R: | TGGCCACTACGTACATCTTGATG | |||
| OSTN | osteocrin | F: | CCCCTGGACAGACTCTCAGCAGG | NM_001098597.1 |
| R: | GCCTCTGGAATTTGGAAGCCGGT | |||
| S100A9 | S100 calcium binding protein A9 | F: | ACCACATCCTGGAAGACCTG | NM_001177906.1 |
| R: | TCCTCGTGAGAAGCTACCGT | |||
| STAT1 | signal transducer and activator of transcription 1, 91 kDa | F: | AAATGCCGGCGCCAGAACCA | NM_213769.1 |
| R: | CGGGAGCTGGCTGACGTTGG | |||
| STC1 | stanniocalcin 1 | F: | ACACGCACCAGCGAGCTGAC | NM_001103212.1 |
| R: | GCTGTGAACACCTCGCCCCC | |||
3. Results
3.1. RNA-Seq Data for the Endometrium on Days 12, 18, and 25 of Pregnancy
| Sample | Raw Reads | Clean Reads | Clean Data | Mapped Reads | Unique Mapped Reads | Multi Mapped Reads | Pair-End Mapped Reads |
|---|---|---|---|---|---|---|---|
| D12-01 | 67169826 | 66077622 | 66077622 | 49833482 | 46142686 | 3690796 | 23127536 |
| D12-02 | 41784360 | 39324942 | 39324942 | 29499855 | 27337499 | 2162356 | 13706839 |
| D12-03 | 34275994 | 33688562 | 33688562 | 25394236 | 23553198 | 1841038 | 11998456 |
| D18-01 | 41684596 | 40824568 | 40824568 | 26668182 | 24658656 | 2009526 | 12636036 |
| D18-02 | 40426950 | 39987798 | 39987798 | 22327827 | 20562051 | 1765776 | 10550537 |
| D18-03 | 44795678 | 44415892 | 44415892 | 28210350 | 26213892 | 1996458 | 13001074 |
| D25-01 | 50047464 | 49009334 | 49009334 | 37099086 | 33754041 | 3345045 | 17284406 |
| D25-02 | 84890198 | 83601216 | 83601216 | 64826626 | 57999868 | 6826758 | 30054104 |
| D25-03 | 124850826 | 122914990 | 122914990 | 95857938 | 88801603 | 7056335 | 43719905 |
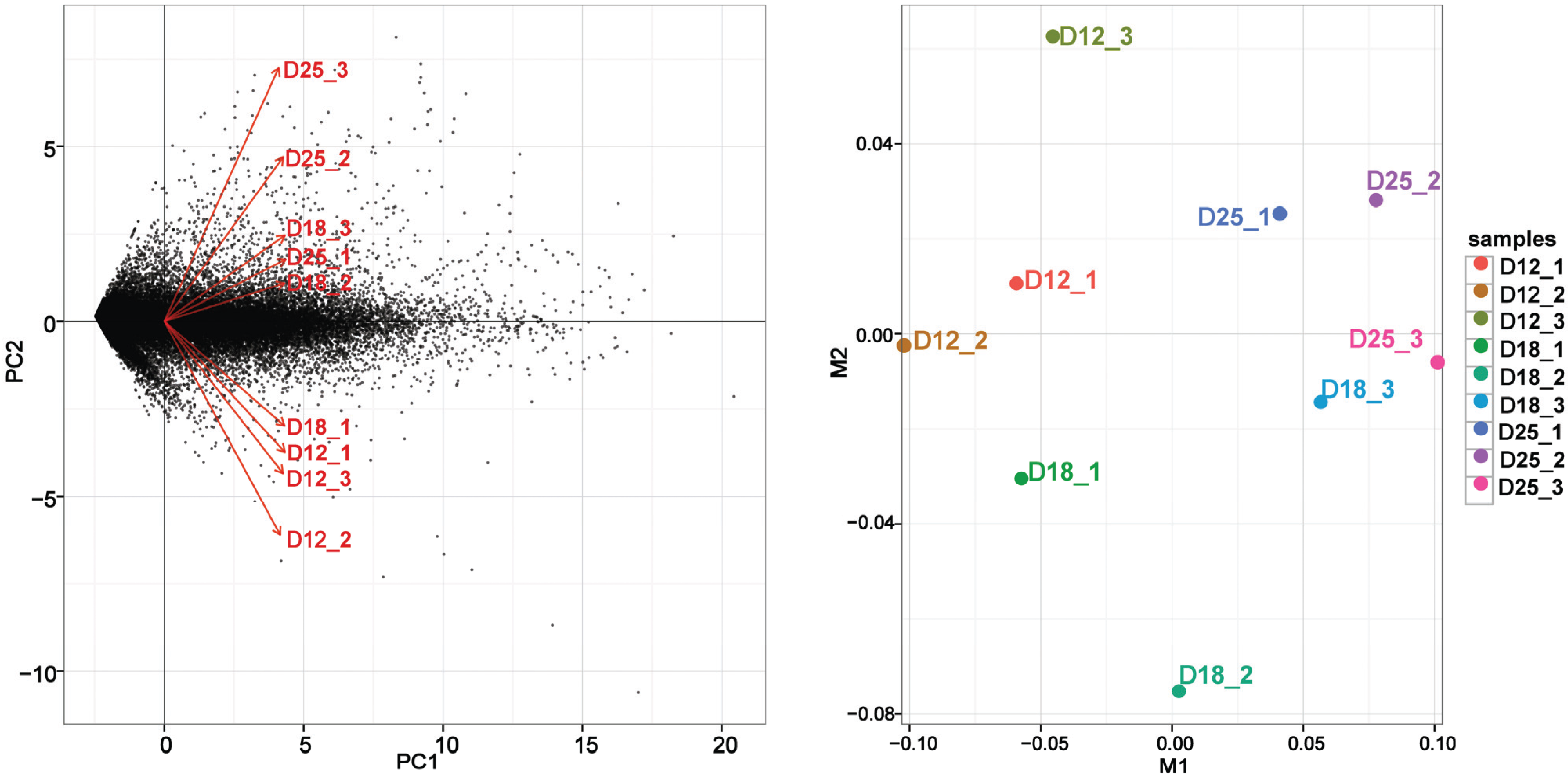
3.2. Gene Expression Variations between the Different Implantation Stages
| Sample | TB Gene Number | DEGs | Upregulation | Downregulation |
|---|---|---|---|---|
| D12 vs. D18 | 45828 | 1557 | 708 | 849 |
| D12 vs. D25 | 8951 | 4999 | 3952 | |
| D18 vs. D25 | 2345 | 1166 | 1179 |
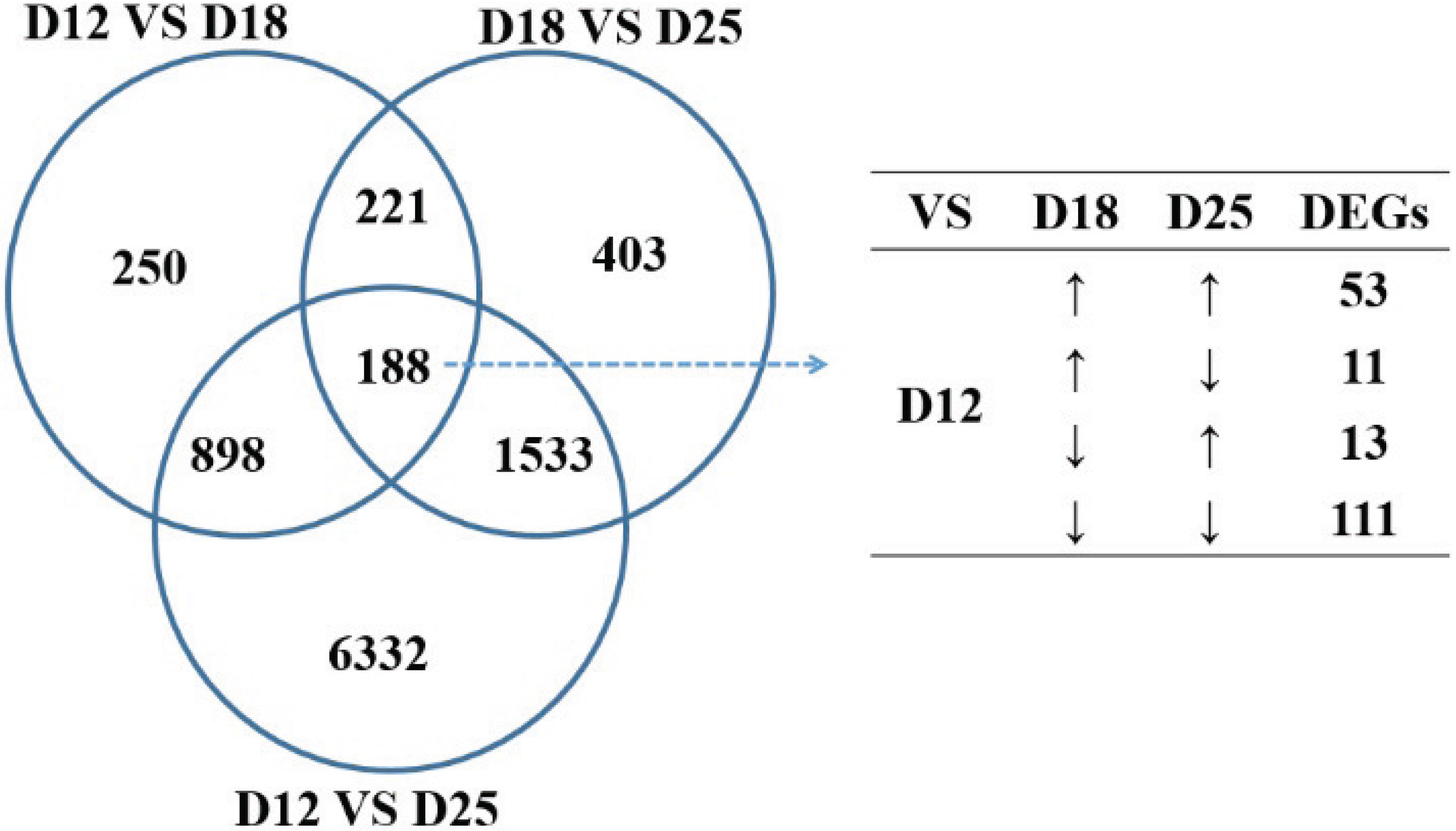
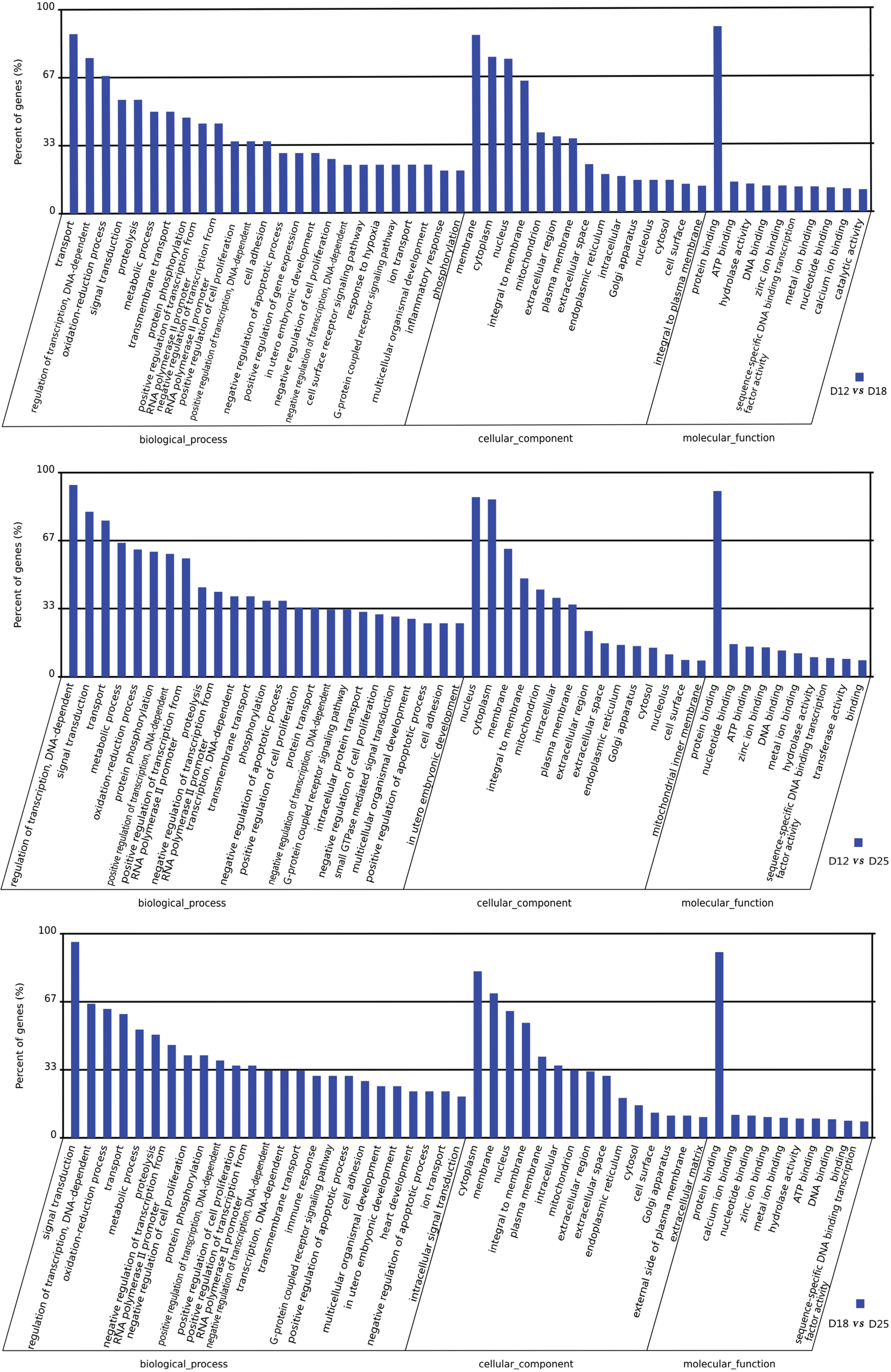
3.3. Gene Ontology (GO) and Genomes (KEGG) Ontology (KO) Classification (Up Arrow Indicates Upregulation, and Down Arrow Indicates Downregulation)
| Pathway ID | Pathway Name | Gene Number | p-Value | Gene List * | ||
|---|---|---|---|---|---|---|
| Day 12 vs. Day 18 | 1. | ssc04060 | Cytokine–cytokine receptor interaction | 13 | 1.79 × 10−4 | ACVR1, ACVR1B, FIGF, MET, CCR7, CSF1R, CSF2RB, CXCL14, CXCR3, IFNGR1, IL10RA, IL21R, IL2RG |
| 2. | ssc04610 | Complement and coagulation cascades | 10 | 2.26 × 10−7 | C7, DF, F5, C1QA, C2, CFH, F10, FGB, KNG1, PLAUR | |
| 3. | ssc04510 | Focal adhesion | 8 | 4.19 × 10−5 | COL5A3, ITGB8, COL11A1, ITGA4, LAMA3, MYL7, PARVG, SPP1 | |
| 4. | ssc04110 | Cell cycle | 8 | 7.39 × 10−4 | CDKN2B, CDKN2B, PLS1, SMAD2, CDC2, CDC6, SKP2, WEE1 | |
| 5. | ssc00270 | Cysteine and methionine metabolism | 6 | 1.18 × 10−6 | CDO1, AHCY, AMD1, DNMT3B, MAT1A, SMS | |
| 6. | ssc04142 | Lysosome | 6 | 1.66 × 10−3 | AP1B1, CTSC, CTSH, CTSZ, DNASE2, NPC2 | |
| 7. | ssc04360 | Axon guidance | 6 | 4.97 × 10−3 | EFNA1, SEMA6A, EPHA2, SEMA3F, SEMA4A, UNC5B | |
| 8. | ssc00190 | Oxidative phosphorylation | 6 | 8.96 × 10−3 | ATP6V1C2, ATP5G1, ATP6V1G3, NDUFA10, NDUFS3, UQCRC1 | |
| 9. | ssc04514 | Cell adhesion molecules (CAMs) | 6 | 8.96 × 10−3 | SELL, CD2, CD4, CD8A, ITGAL, PTPRC | |
| 10. | ssc00860 | Porphyrin and chlorophyll metabolism | 5 | 6.97 × 10−5 | CP, CPOX, EPRS, HMBS, UROD | |
| 11. | ssc00512 | Mucin type O-Glycan biosynthesis | 5 | 1.98 × 10−4 | GALNT11, GALNTL5, GCNT1, B4GALT5, GALNT9 | |
| 12. | ssc04115 | p53 signaling pathway | 4 | 1.98 × 10−4 | SESN3, CYCS, GTSE1, SERPINB5 | |
| 13. | ssc04650 | Natural killer cell mediated cytotoxicity | 4 | 5.28 × 10−4 | CD244, CD48, FCER1G, PRF1 | |
| 14. | ssc03320 | PPAR signaling pathway | 4 | 5.00 × 10−3 | CD36, APOA1, FABP5, PPARG | |
| 15. | ssc02010 | ABC transporters | 4 | 5.95 × 10−3 | ABCA1, ABCB10, ABCC2, CFTR | |
| 16. | ssc04614 | Renin–angiotensin system | 3 | 1.88 × 10−5 | ACE2, LNPEP, MME | |
| 17. | ssc04350 | TGF-beta signaling pathway | 3 | 3.72 × 10−4 | FST, ID1, ID2 | |
| 18. | ssc05010 | Alzheimer’s disease | 2 | 6.84 × 10−3 | APOE, BACE2 | |
| Day 12 vs. Day 25 | 1. | ssc04141 | Protein processing in endoplasmic reticulum | 30 | 6.70 × 10−6 | ATF6B, BAK1, DNAJB2, HSPA4L, UBXN6, CALR, CRYAB, DERL3, DNAJA1, DNAJB11, DNAJB12, DNAJC10, ERLEC1, ERP29, GRP-58, HSP90AA1, HSP90B1, HSPH1, LMAN1, NGLY1, PDIA4, PLAA, SAR1A, SAR1B, SEC23B, SEC31A, SSR1, SSR4, TRAM1, VIMP |
| 2. | ssc04110 | Cell cycle | 29 | 1.88 × 10−9 | ANAPC5, ATM, CDC14A, CDKN2B, EP300, FZR1, PLS1, SMAD2, ATM, BUB3, CCNB2, CDC2, CDC23, CDC25C, CDC25C, CDC6, CDK6, CDKN2C, E2F1, E2F3, HDAC2, MAD2L1, ORC3, PTTG1, SKP2, SMAD4, TTK, WEE1, YWHAH | |
| 3. | ssc00190 | Oxidative phosphorylation | 25 | 5.06 × 10−6 | ATP6V0D2, TCIRG1, ATP5A1, ATP5G1, ATP5G3, ATP6V0D1, ATP6V0E1, ATP6V1C1, ATP6V1E1, ATP6V1G3, ATP6V1H, COX5B, COX7C, NDUFA5, NDUFAB1, NDUFB3, NDUFB5, NDUFB6, NDUFS1, NDUFS3, NDUFS4, NDUFS5, NDUFV1, NDUFV2, UQCRH | |
| 4. | ssc04145 | Phagosome | 19 | 5.71 × 10−4 | CD61, ITGAV, MRC2, SCARB1, SFTPD, SLA, SLA-DMA, TUBA4A, CH242-21O2.1, DYNC1LI1, FCGR1A, ITGA5, ITGB1, MRC2, MSR1, NCF2, THBS1, THBS3, TLR2 | |
| 5. | ssc03008 | Ribosome biogenesis in eukaryotes | 18 | 1.49 ×10 −4 | RPP25L, XRN1, DKC1, GNL3, LSG1, MPHOSPH10, NHP2L1, NXT2, POP5, RAN, REXO2, RIOK1, RPP30, RPP40, SBDS, UTP18, UTP6, WDR75 | |
| 6. | ssc04360 | Axon guidance | 17 | 5.74 × 10−3 | ABLIM1, EFNA1, EPHB4, FYN, L1CAM, LIMK2, SEMA3G, SEMA6A, SEMA6C, SEMA6D, CFL2, EPHA2, EPHA4, EPHB2, SEMA3F, SEMA4F, UNC5B | |
| 7. | ssc04310 | Wnt signaling pathway | 16 | 4.82 × 10−3 | CHD8, CSNK1E, DVL1, FZD10, FZD5, NFATC4, SOX17, TCF7L2, VANGL1, VANGL2, CACYBP, FOSL1, SFRP2, SFRP5, TCF7L1, WNT2B | |
| 8. | ssc04510 | Focal adhesion | 15 | 1.66 × 10−3 | ITGA3, ITGA6, ITGB4, ITGB8, VWF, COL11A1, COL5A2, FN1, ITGA1, ITGA11, ITGA4, ITGA8, LAMA3, MYL7, VCL | |
| 9. | ssc04610 | Complement and coagulation cascades | 13 | 5.65 × 10−3 | DF, F5, SERPINC1, C2, C7, CFH, CFI, FGB, MASP1, PLAU, PLAUR, SERPINA5, TFPI | |
| 10. | ssc04115 | p53 signaling pathway | 9 | 4.02 × 10−4 | CCNG2, SESN3, SHISA5, BID, CYCS, EI24, GTSE1, SERPINB5, TP53I3 | |
| 11. | ssc04530 | Tight junction | 8 | 6.02 × 10−3 | CGN, EPB41L1, MAGI3, MPP5, AMOTL1, CRB3, EPB41L3, MYH1 | |
| 12. | ssc00860 | Porphyrin and chlorophyll metabolism | 8 | 2.04 × 10−3 | HMOX2, BLVRA, CPOX, EPRS, HMBS, MMAB, UROD, UROS | |
| 13. | ssc00480 | Glutathione metabolism | 7 | 2.04 × 10-3 | GCLC, GSTM3, GCLM, GPX2, GSTT1, MGST3, TXNDC12 | |
| 14. | ssc00260 | Glycine, serine and threonine metabolism | 7 | 6.02 × 10−3 | CHDH, GATM, GLYCTK, GNMT, PHGDH, PSAT1, PSPH | |
| 15. | ssc00512 | Mucin type O-Glycan biosynthesis | 7 | 9.44 × 10−3 | GALNT11, GALNTL5, GCNT1, B4GALT5, GALNT10, GALNT3, GALNT9 | |
| 16. | ssc00100 | Steroid biosynthesis | 7 | 2.68 × 10−4 | LIPA, CYP51, HSD17B7, NSDHL, SC4MOL, SQLE, TM7SF2 | |
| 17. | ssc05322 | Systemic lupus erythematosus | 7 | 1.70 × 10−3 | H2AFV, TRIM21, H2AFZ, H3F3A, H3F3C, HIST2H2AC, SSB | |
| 18. | ssc00520 | Amino sugar and nucleotide sugar metabolism | 7 | 6.10 × 10−3 | CYB5R3, GNPDA1, CHI3L1, CMAS, GNPDA2, NANS, UAP1 | |
| 19. | ssc03060 | Protein export | 7 | 6.10 × 10−3 | HSPA5, SEC11C, SEC61B, SEC61G, SPCS1, SPCS3, SRPRB | |
| 20. | ssc05010 | Alzheimer’s disease | 6 | 4.90 × 10−4 | ADAM10, CDK5R1, SNCA, APOE, BACE2, IDE | |
| 21. | ssc04972 | Pancreatic secretion | 4 | 9.37 × 10−3 | RAB27B, RAB3D, TPCN2, RAB8A | |
| 22. | ssc00740 | Riboflavin metabolism | 4 | 6.14 × 10−3 | FLAD1, RFK, ACP1, ACP6 | |
| Day 18 vs. Day 25 | 1. | ssc04060 | Cytokine–cytokine receptor interaction | 19 | 7.75 × 10−6 | CCR7, CD27, CD40, CSF1R, CXCL10, CXCL16, CXCR3, FLT3LG, IFNE, IL10RA, IL21R, IL2RG, LIFR, LTB, MET, TNFRSF21, CSF3, IL13RA1, IL18 |
| 2. | ssc04145 | Phagosome | 13 | 3.31 × 10−7 | CD61, DMB, SLA, SLA-DMA, TUBA4A, CD209, CH242-21O2.1, ITGB1, MRC2, MSR1, THBS1, THBS2, THBS3 | |
| 3. | ssc00190 | Oxidative phosphorylation | 12 | 1.18 × 10−5 | ATP6V0C, ATP6V0D2, NDUFS6, TCIRG1, ATP6V1C2, ATP6V1E1, NDUFA5, NDUFB3, NDUFB6, NDUFS4, NDUFS5, NDUFV2 | |
| 4. | ssc04610 | Complement and coagulation cascades | 11 | 1.12 × 10−6 | C1QA, C8G, DF, FGB, KNG1, SERPINA1, SERPINA5, SERPINC1, C7, CFH, CFI | |
| 5. | ssc04510 | Focal adhesion | 10 | 1.96 × 10−5 | COL11A1, ITGA3, ITGA4, LAMB3, PARVG, COL5A2, ITGA11, ITGA8, MYL7, VCL | |
| 6. | ssc04514 | Cell adhesion molecules (CAMs) | 8 | 5.26 × 10−3 | CD8A, CLDN8, ITGAL, PTPRC, PECAM1, SELL, VCAM1, VCAN | |
| 7. | ssc00260 | Glycine, serine, and threonine metabolism | 5 | 9.15 × 10−5 | GLYCTK, GNMT, PHGDH, PSAT1, AOC3 | |
| 8. | ssc03320 | PPAR signaling pathway | 5 | 3.57 × 10−3 | APOA1, APOA2, APOC3, PPARD, PPARG | |
| 9. | ssc04270 | Vascular smooth muscle contraction | 4 | 2.04 × 10−4 | ACTA2, CALD1, KCNMB1, PPP1R12B | |
| 10. | ssc04650 | Natural killer cell-mediated cytotoxicity | 4 | 2.43 × 10−3 | CD3Z, CD48, PRF1, ZAP70 | |
| 11. | ssc00480 | Glutathione metabolism | 4 | 4.53 × 10−3 | GCLC, GCLM, GPX2, TXNDC12 | |
| 12. | ssc00590 | Arachidonic acid metabolism | 4 | 7.58 × 10−3 | ALOX12, ALOX15B, LTA4H, PTGIS | |
| 13. | ssc00340 | Histidine metabolism | 3 | 4.01 × 10−3 | HAL, HDC, UROC1 | |
| 14. | ssc00100 | Steroid biosynthesis | 3 | 8.91 × 10−3 | CYP51, HSD17B7, SC4MOL |
3.4. Validation of the RNA-Seq Data
| Gene | vs. | Fold-Change | p-Value | Correlation | ||
|---|---|---|---|---|---|---|
| RNA-Seq | Real-time PCR | RNA-Seq | Real-time PCR | |||
| FGF9 | D12 vs. D18 | 1.01 | 0.89 | 0.9076 | 0.7736 | 0.8825 |
| D12 vs. D25 | 2.92 | 2.87 | < 0.0001 | < 0.0001 | 0.9939 | |
| D18 vs. D25 | 2.82 | 3.32 | 0.0002 | < 0.0001 | 0.9692 | |
| IRF1 | D12 vs. D18 | 1.16 | 1.83 | 0.7325 | 0.2655 | 0.9800 |
| D12 vs. D25 | 5.62 | 7.54 | < 0.0001 | < 0.0001 | 0.8467 | |
| D18 vs. D25 | 4.72 | 4.23 | 0.0005 | 0.0136 | 0.9092 | |
| OSTN | D12 vs. D18 | −17.40 | −15.82 | < 0.0001 | < 0.0001 | 0.9732 |
| D12 vs. D25 | −530.57 | −1015.42 | < 0.0001 | < 0.0001 | 0.7383 | |
| D18 vs. D25 | −30.93 | −63.69 | 0.0003 | 0.0009 | 0.9799 | |
| PTGES | D12 vs. D18 | −3.45 | −5.37 | < 0.0001 | < 0.0001 | 0.9606 |
| D12 vs. D25 | −4.05 | −13.64 | < 0.0001 | < 0.0001 | 0.9807 | |
| D18 vs. D25 | −1.15 | −2.55 | 0.2223 | 0.0912 | 0.9201 | |
| S100A9 | D12 vs. D18 | 610.76 | 329.82 | < 0.0001 | < 0.0001 | 0.7967 |
| D12 vs. D25 | 372.60 | 266.37 | < 0.0001 | 0.0002 | 0.8823 | |
| D18 vs. D25 | 0.60 | 0.83 | 0.0685 | 0.0502 | 0.9734 | |
| STAT1 | D12 vs. D18 | 0.91 | 0.62 | 0.9542 | 0.3827 | 0.9987 |
| D12 vs. D25 | 1.98 | 0.87 | 0.0001 | 0.0106 | 0.9336 | |
| D18 vs. D25 | 2.22 | 1.19 | < 0.0001 | 0.0114 | 0.9725 | |
| STC1 | D12 vs. D18 | 9.40 | 8.39 | < 0.0001 | < 0.0001 | 0.9480 |
| D12 vs. D25 | 51.34 | 38.83 | < 0.0001 | < 0.0001 | 0.8834 | |
| D18 vs. D25 | 5.91 | 4.43 | < 0.0001 | 0.0057 | 0.9508 | |
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interests
References
- Morris, D.; Diskin, M. Effect of progesterone on embryo survival. Animal 2008, 2, 1112–1119. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Johnson, G.A. Pig blastocyst-uterine interactions. Differentiation 2014, 87, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Spencer, T.E.; Johnson, G.A.; Burghardt, R.C.; Bazer, F.W. Progesterone and placental hormone actions on the uterus: Insights from domestic animals. Biol. Reprod. 2004, 71, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Thatcher, W.W. Theory of maternal recognition of pregnancy in swine based on estrogen controlled endocrine versus exocrine secretion of prostaglandin F2alpha by the uterine endometrium. Prostaglandins 1977, 14, 397–400. [Google Scholar] [CrossRef]
- Geisert, R.D.; Renegar, R.H.; Thatcher, W.W.; Roberts, R.M.; Bazer, F.W. Establishment of pregnancy in the pig: I. Interrelationships between preimplantation development of the pig blastocyst and uterine endometrial secretions. Biol. Reprod. 1982, 27, 925–939. [Google Scholar] [CrossRef] [PubMed]
- Ziecik, A.J.; Waclawik, A.; Kaczmarek, M.M.; Blitek, A.; Jalali, B.M.; Andronowska, A. Mechanisms for the establishment of pregnancy in the pig. Reprod. Domest. Anim. 2011, 46, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Thatcher, W.W.; Martinat-Botte, F.; Terqui, M. Conceptus development in large white and prolific Chinese Meishan pigs. J. Reprod. Fertil. 1988, 84, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Østrup, E.; Bauersachs, S.; Blum, H.; Wolf, E.; Hyttel, P. Differential endometrial gene expression in pregnant and nonpregnant sows. Biol. Reprod. 2010, 83, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Samborski, A.; Graf, A.; Krebs, S.; Kessler, B.; Bauersachs, S. Deep sequencing of the porcine endometrial transcriptome on day 14 of pregnancy. Biol. Reprod. 2013. [Google Scholar] [CrossRef] [PubMed]
- Samborski, A.; Graf, A.; Krebs, S.; Kessler, B.; Reichenbach, M.; Reichenbach, H.D.; Ulbrich, S.E.; Bauersachs, S. Transcriptome changes in the porcine endometrium during the preattachment phase. Biol. Reprod. 2013, 89, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Franczak, A.; Wojciechowicz, B.; Kotwica, G. Transcriptomic analysis of the porcine endometrium during early pregnancy and the estrous cycle. Reprod. Biol. 2013, 13, 229–237. [Google Scholar] [CrossRef] [PubMed]
- UCSC Genome Bioinformatics. Swine Genome Sequencing Consortium Sscrofa10.2. Available online: http://genome.ucsc.edu (accessed on 12 January 2015).
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Database for Annotation, Visualization and Integrated Discovery. DAVID Bioinformatics Resources 6.7. Available online: http://david.abcc.ncifcrf.gov (accessed on 18 March 2015).
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Anderson, L.L. Growth, protein content and distribution of early pig embryos. Anat. Rec. 1978, 190, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Geisert, R.D.; Schmitt, R.A.M. Early embryonic survival in the pig: Can it be improved? J. Anim. Sci. 2002, 80, 54–65. [Google Scholar]
- Kayser, J.P.; Kim, J.G.; Cerny, R.L.; Vallet, J.L. Global characterization of porcine intrauterine proteins during early pregnancy. Reproduction 2006, 131, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Jalali, B.M.; Bogacki, M.; Dietrich, M.; Likszo, P.; Wasielak, M. Proteomic analysis of porcine endometrial tissue during peri-implantation period reveals altered protein abundance. J. Proteomics 2015, 125, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, H.; Croy, B.A.; King, G.J. Role of uterine immune cells in early pregnancy in pigs. J. Reprod. Fertil. Suppl. 1997, 52, 115–131. [Google Scholar] [PubMed]
- Engelhardt, H.; Croy, B.A.; King, G.J. Conceptus influences the distribution of uterine leukocytes during early porcine pregnancy. Biol. Reprod. 2002, 66, 1875–1880. [Google Scholar] [CrossRef] [PubMed]
- Passey, R.J.; Williams, E.; Lichanska, A.M.; Wells, C.; Hu, S.; Geczy, C.L.; Little, M.H.; Hume, D.A. A null mutation in the inflammation-associated S100 protein S100A8 causes early resorption of the mouse embryo. J. Immunol. 1999, 163, 2209–2216. [Google Scholar] [PubMed]
- Nair, R.R.; Khanna, A.; Singh, K. Role of inflammatory proteins S100A8 and S100A9 in pathophysiology of recurrent early pregnancy loss. Placenta 2013, 34, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Bowen, J.A.; Bazer, F.W.; Burghardt, R.C. Spatial and temporal analysis of integrin and Muc-1 expression in porcine uterine epithelium and trophectoderm in vivo. Biol. Reprod. 1996, 55, 1098–1106. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.; Chard, T. Cytokines in implantation. Cytokine Growth Factor Rev. 1998, 9, 287–296. [Google Scholar] [CrossRef]
- Dey, S.K.; Lim, H.; Das, S.K.; Reese, J.; Paria, B.C.; Daikoku, T.; Wang, H. Molecular cues to implantation. Endocr. Rev. 2004, 25, 341–373. [Google Scholar]
- Spencer, T.E.; Bazer, F.W. Uterine and placental factors regulating conceptus growth in domestic animals. J. Anim. Sci. 2004, 82, 4–13. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.; Wang, H.; Wang, Y.; Liu, C.; Wang, C.; Guo, J. Transcriptomic Analysis of the Porcine Endometrium during Embryo Implantation. Genes 2015, 6, 1330-1346. https://doi.org/10.3390/genes6041330
Lin H, Wang H, Wang Y, Liu C, Wang C, Guo J. Transcriptomic Analysis of the Porcine Endometrium during Embryo Implantation. Genes. 2015; 6(4):1330-1346. https://doi.org/10.3390/genes6041330
Chicago/Turabian StyleLin, Haichao, Huaizhong Wang, Yanping Wang, Chang Liu, Cheng Wang, and Jianfeng Guo. 2015. "Transcriptomic Analysis of the Porcine Endometrium during Embryo Implantation" Genes 6, no. 4: 1330-1346. https://doi.org/10.3390/genes6041330





