Modulation of Autophagy for Controlling Immunity
Abstract
:1. Introduction
2. Autophagy Signaling
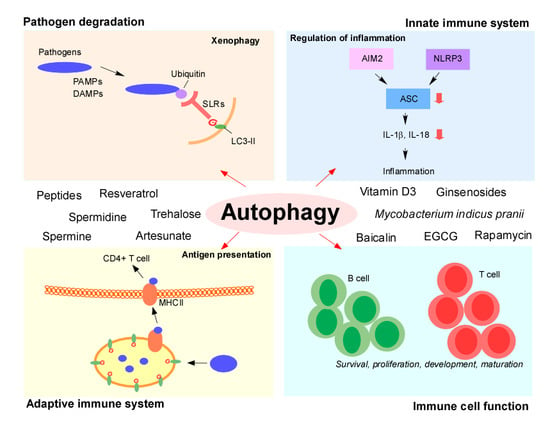
3. Pathogen Degradation: Xenophagy
4. The Proviral Role of Autophagy
5. Autophagy and Innate Immune Systems: Regulation of Inflammation
6. Autophagy in the Adaptive Immune System: Regulation of Antigen Presentation
7. Autophagy in Immune Cell Function
8. Modulators of Autophagy for Immune Control
8.1. Spermine
8.2. Spermidine
8.3. Resveratrol
8.4. Artesunate
8.5. Trehalose
8.6. Vitamin D3
8.7. Baicalin
8.8. Ginsenosides
8.9. Epigallocatechin-3-Ggallate (EGCG)
8.10. Rapamycin
8.11. Peptides and Mycobacterium
8.11.1. Peptides
8.11.2. Mycobacterium Indicus Pranii (MIP)
9. Conclusions
Funding
Conflicts of Interest
References
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Shibutani, S.T.; Saitoh, T.; Nowag, H.; Munz, C.; Yoshimori, T. Autophagy and autophagy-related proteins in the immune system. Nat. Immunol. 2015, 16, 1014–1024. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Galluzzi, L.; Zitvogel, L.; Kroemer, G. Autophagy and cellular immune responses. Immunity 2013, 39, 211–227. [Google Scholar] [CrossRef] [PubMed]
- Byun, S.; Lee, E.; Lee, K.W. Therapeutic Implications of Autophagy Inducers in Immunological Disorders, Infection, and Cancer. Int. J. Mol. Sci. 2017, 18, 1959. [Google Scholar] [CrossRef] [PubMed]
- Polson, H.E.; de Lartigue, J.; Rigden, D.J.; Reedijk, M.; Urbe, S.; Clague, M.J.; Tooze, S.A. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010, 6, 506–522. [Google Scholar] [CrossRef]
- Dooley, H.C.; Razi, M.; Polson, H.E.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell 2014, 55, 238–252. [Google Scholar] [CrossRef]
- Fujita, N.; Hayashi-Nishino, M.; Fukumoto, H.; Omori, H.; Yamamoto, A.; Noda, T.; Yoshimori, T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 2008, 19, 4651–4659. [Google Scholar] [CrossRef]
- Eskelinen, E.L.; Saftig, P. Autophagy: A lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta 2009, 1793, 664–673. [Google Scholar] [CrossRef]
- Sharma, V.; Verma, S.; Seranova, E.; Sarkar, S.; Kumar, D. Selective Autophagy and Xenophagy in Infection and Disease. Front. Cell Dev. Biol. 2018, 6, 147. [Google Scholar] [CrossRef] [PubMed]
- Bauckman, K.A.; Owusu-Boaitey, N.; Mysorekar, I.U. Selective autophagy: Xenophagy. Methods 2015, 75, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Sakowski, E.T.; Koster, S.; Portal Celhay, C.; Park, H.S.; Shrestha, E.; Hetzenecker, S.E.; Maurer, K.; Cadwell, K.; Philips, J.A. Ubiquilin 1 Promotes IFN-gamma-Induced Xenophagy of Mycobacterium tuberculosis. PLoS Pathog. 2015, 11, e1005076. [Google Scholar] [CrossRef] [PubMed]
- Castano-Rodriguez, N.; Kaakoush, N.O.; Goh, K.L.; Fock, K.M.; Mitchell, H.M. Autophagy in Helicobacter pylori Infection and Related Gastric Cancer. Helicobacter 2015, 20, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Yoshimori, T.; Suzuki, T.; Sagara, H.; Mizushima, N.; Sasakawa, C. Escape of intracellular Shigella from autophagy. Science 2005, 307, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.E.; Lee, H.K. Pattern recognition receptors and autophagy. Front. Immunol. 2014, 5, 300. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Coyne, C.B.; Zeh, H.J.; Lotze, M.T. PAMPs and DAMPs: Signal 0s that spur autophagy and immunity. Immunol. Rev. 2012, 249, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Travassos, L.H.; Carneiro, L.A.; Ramjeet, M.; Hussey, S.; Kim, Y.G.; Magalhaes, J.G.; Yuan, L.; Soares, F.; Chea, E.; Le Bourhis, L.; et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010, 11, 55–62. [Google Scholar] [CrossRef]
- Deretic, V. Autophagy as an innate immunity paradigm: Expanding the scope and repertoire of pattern recognition receptors. Curr. Opin. Immunol. 2012, 24, 21–31. [Google Scholar] [CrossRef]
- Watson, R.O.; Manzanillo, P.S.; Cox, J.S.; Extracellular, M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 2012, 150, 803–815. [Google Scholar] [CrossRef]
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011, 333, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Iwasaki, A. Autophagy and antiviral immunity. Curr. Opin. Immunol. 2008, 20, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Kleeman, L.K.; Jiang, H.H.; Gordon, G.; Goldman, J.E.; Berry, G.; Herman, B.; Levine, B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 1998, 72, 8586–8596. [Google Scholar] [PubMed]
- Reed, M.; Morris, S.H.; Jang, S.; Mukherjee, S.; Yue, Z.; Lukacs, N.W. Autophagy-inducing protein beclin-1 in dendritic cells regulates CD4 T cell responses and disease severity during respiratory syncytial virus infection. J. Immunol. 2013, 191, 2526–2537. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Czymmek, K.; Talloczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Orvedahl, A.; MacPherson, S.; Sumpter, R., Jr.; Talloczy, Z.; Zou, Z.; Levine, B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 2010, 7, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Hafren, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.; Fang, R.; Sun, J. The role of autophagy in microbial infection and immunity. Immunotargets Ther. 2015, 4, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Li, J.K.; Liang, J.J.; Liao, C.L.; Lin, Y.L. Autophagy is involved in the early step of Japanese encephalitis virus infection. Microbes Infect. 2012, 14, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Dreux, M.; Gastaminza, P.; Wieland, S.F.; Chisari, F.V. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl Acad. Sci. USA 2009, 106, 14046–14051. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.W.; Ducroux, A.; Jeang, K.T.; Neuveut, C. Impact of cellular autophagy on viruses: Insights from hepatitis B virus and human retroviruses. J. Biomed. Sci. 2012, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Parnell, L.A.; Diamond, M.S.; Mysorekar, I.U. Inhibition of autophagy limits vertical transmission of Zika virus in pregnant mice. J. Exp. Med. 2017, 214, 2303–2313. [Google Scholar] [CrossRef]
- Campbell, G.R.; Spector, S.A. Inhibition of human immunodeficiency virus type-1 through autophagy. Curr. Opin. Microbiol. 2013, 16, 349–354. [Google Scholar] [CrossRef]
- Eekels, J.J.; Sagnier, S.; Geerts, D.; Jeeninga, R.E.; Biard-Piechaczyk, M.; Berkhout, B. Inhibition of HIV-1 replication with stable RNAi-mediated knockdown of autophagy factors. Virol. J. 2012, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Sagnier, S.; Daussy, C.F.; Borel, S.; Robert-Hebmann, V.; Faure, M.; Blanchet, F.P.; Beaumelle, B.; Biard-Piechaczyk, M.; Espert, L. Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes. J. Virol. 2015, 89, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Prentice, E.; Jerome, W.G.; Yoshimori, T.; Mizushima, N.; Denison, M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004, 279, 10136–10141. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Lund, J.M.; Ramanathan, B.; Mizushima, N.; Iwasaki, A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 2007, 315, 1398–1401. [Google Scholar] [CrossRef]
- Yang, C.S.; Rodgers, M.; Min, C.K.; Lee, J.S.; Kingeter, L.; Lee, J.Y.; Jong, A.; Kramnik, I.; Lin, X.; Jung, J.U. The autophagy regulator Rubicon is a feedback inhibitor of CARD9-mediated host innate immunity. Cell Host Microbe 2012, 11, 277–289. [Google Scholar] [CrossRef]
- Netea, M.G.; Simon, A.; van de Veerdonk, F.; Kullberg, B.J.; Van der Meer, J.W.; Joosten, L.A. IL-1beta processing in host defense: Beyond the inflammasomes. PLoS Pathog. 2010, 6, e1000661. [Google Scholar] [CrossRef]
- Rathinam, V.A.; Vanaja, S.K.; Fitzgerald, K.A. Regulation of inflammasome signaling. Nat. Immunol. 2012, 13, 333–342. [Google Scholar] [CrossRef]
- Won, J.H.; Park, S.; Hong, S.; Son, S.; Yu, J.W. Rotenone-induced Impairment of Mitochondrial Electron Transport Chain Confers a Selective Priming Signal for NLRP3 Inflammasome Activation. J. Biol. Chem. 2015, 290, 27425–27437. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Nakahira, K.; Haspel, J.A.; Rathinam, V.A.; Lee, S.J.; Dolinay, T.; Lam, H.C.; Englert, J.A.; Rabinovitch, M.; Cernadas, M.; Kim, H.P.; et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011, 12, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Lupfer, C.; Thomas, P.G.; Anand, P.K.; Vogel, P.; Milasta, S.; Martinez, J.; Huang, G.; Green, M.; Kundu, M.; Chi, H.; et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 2013, 14, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.; Hartman, M.; Roche, C.; Zeng, S.G.; O’Shea, A.; Sharp, F.A.; Lambe, E.M.; Creagh, E.M.; Golenbock, D.T.; Tschopp, J.; et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J. Biol. Chem. 2011, 286, 9587–9597. [Google Scholar] [CrossRef]
- Paul, S.; Kashyap, A.K.; Jia, W.; He, Y.W.; Schaefer, B.C. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity 2012, 36, 947–958. [Google Scholar] [CrossRef]
- Shibata, Y.; Oyama, M.; Kozuka-Hata, H.; Han, X.; Tanaka, Y.; Gohda, J.; Inoue, J. p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO. Nat. Commun. 2012, 3, 1061. [Google Scholar] [CrossRef]
- Fliss, P.M.; Jowers, T.P.; Brinkmann, M.M.; Holstermann, B.; Mack, C.; Dickinson, P.; Hohenberg, H.; Ghazal, P.; Brune, W. Viral mediated redirection of NEMO/IKKgamma to autophagosomes curtails the inflammatory cascade. PLoS Pathog. 2012, 8, e1002517. [Google Scholar] [CrossRef]
- Munz, C. Antigen Processing for MHC Class II Presentation via Autophagy. Front. Immunol. 2012, 3, 9. [Google Scholar] [CrossRef]
- Van Kaer, L.; Parekh, V.V.; Postoak, J.L.; Wu, L. Role of autophagy in MHC class I-restricted antigen presentation. Mol. Immunol. 2017. [Google Scholar] [CrossRef]
- Romao, S.; Gannage, M.; Munz, C. Checking the garbage bin for problems in the house, or how autophagy assists in antigen presentation to the immune system. Semin Cancer Biol. 2013, 23, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Dengjel, J.; Schoor, O.; Fischer, R.; Reich, M.; Kraus, M.; Muller, M.; Kreymborg, K.; Altenberend, F.; Brandenburg, J.; Kalbacher, H.; et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 7922–7927. [Google Scholar] [CrossRef] [PubMed]
- Cooney, R.; Baker, J.; Brain, O.; Danis, B.; Pichulik, T.; Allan, P.; Ferguson, D.J.; Campbell, B.J.; Jewell, D.; Simmons, A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 2010, 16, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Jagannath, C.; Lindsey, D.R.; Dhandayuthapani, S.; Xu, Y.; Hunter, R.L., Jr.; Eissa, N.T. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 2009, 15, 267–276. [Google Scholar] [CrossRef] [PubMed]
- English, L.; Chemali, M.; Duron, J.; Rondeau, C.; Laplante, A.; Gingras, D.; Alexander, D.; Leib, D.; Norbury, C.; Lippe, R.; et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 2009, 10, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Munz, C. Antigen processing for MHC presentation by autophagy. F1000 Biol. Rep. 2010, 2, 61. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Kaveri, S.V.; Bayry, J. Cross-presentation of antigens by dendritic cells: Role of autophagy. Oncotarget 2015, 6, 28527–28528. [Google Scholar] [CrossRef]
- Kovacs, J.R.; Li, C.; Yang, Q.; Li, G.; Garcia, I.G.; Ju, S.; Roodman, D.G.; Windle, J.J.; Zhang, X.; Lu, B. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ. 2012, 19, 144–152. [Google Scholar] [CrossRef]
- Stephenson, L.M.; Miller, B.C.; Ng, A.; Eisenberg, J.; Zhao, Z.; Cadwell, K.; Graham, D.B.; Mizushima, N.N.; Xavier, R.; Virgin, H.W.; et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy 2009, 5, 625–635. [Google Scholar] [CrossRef]
- Henson, S.M.; Lanna, A.; Riddell, N.E.; Franzese, O.; Macaulay, R.; Griffiths, S.J.; Puleston, D.J.; Watson, A.S.; Simon, A.K.; Tooze, S.A.; et al. p38 signaling inhibits mTORC1-independent autophagy in senescent human CD8(+) T cells. J. Clin. Investig. 2014, 124, 4004–4016. [Google Scholar] [CrossRef]
- Miller, B.C.; Zhao, Z.; Stephenson, L.M.; Cadwell, K.; Pua, H.H.; Lee, H.K.; Mizushima, N.N.; Iwasaki, A.; He, Y.W.; Swat, W.; et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy 2008, 4, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Araki, K.; Li, S.; Han, J.H.; Ye, L.; Tan, W.G.; Konieczny, B.T.; Bruinsma, M.W.; Martinez, J.; Pearce, E.L.; et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat. Immunol. 2014, 15, 1152–1161. [Google Scholar] [CrossRef] [PubMed]
- Atiya Ali, M.; Poortvliet, E.; Stromberg, R.; Yngve, A. Polyamines in foods: Development of a food database. Food Nutr. Res. 2011, 55. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.B.; Kim, M.M. Activation of p53 by spermine mediates induction of autophagy in HT1080 cells. Int. J. Biol. Macromol. 2014, 63, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Yang, W.; Jiang, D.; Tao, K.; Dong, A.; Cheng, H. Spermine ameliorates ischemia/reperfusion injury in cardiomyocytes via regulation of autophagy. Am. J. Transl. Res. 2016, 8, 3976–3985. [Google Scholar] [PubMed]
- Han, L.P.; Yuan, L.B.; Shentu, Y.P.; Shao, J.D. Spermine reduced no-reflow size induced by ischemia-reperfusion through regulating autophagy. Int. J. Cardiol. 2013, 168, 3145–3147. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Wu, X.; Jia, G.; Zhao, H.; Chen, X.; Wu, C.; Tang, J.; Wang, J.; Cai, J.; Liu, G. New insights into the role of dietary spermine on inflammation, immune function and related-signalling molecules in the thymus and spleen of piglets. Arch. Anim Nutr. 2017, 71, 175–191. [Google Scholar] [CrossRef]
- Zhou, S.; Gu, J.; Liu, R.; Wei, S.; Wang, Q.; Shen, H.; Dai, Y.; Zhou, H.; Zhang, F.; Lu, L. Spermine Alleviates Acute Liver Injury by Inhibiting Liver-Resident Macrophage Pro-Inflammatory Response Through ATG5-Dependent Autophagy. Front. Immunol. 2018, 9, 948. [Google Scholar] [CrossRef]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Buttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef]
- Eisenberg, T.; Abdellatif, M.; Schroeder, S.; Primessnig, U.; Stekovic, S.; Pendl, T.; Harger, A.; Schipke, J.; Zimmermann, A.; Schmidt, A.; et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 2016, 22, 1428–1438. [Google Scholar] [CrossRef]
- Morselli, E.; Marino, G.; Bennetzen, M.V.; Eisenberg, T.; Megalou, E.; Schroeder, S.; Cabrera, S.; Benit, P.; Rustin, P.; Criollo, A.; et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 2011, 192, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Puleston, D.J.; Zhang, H.L.; Powell, T.J.; Lipina, E.; Sims, S.; Panse, I.; Watson, A.S.; Cerundolo, V.; Townsend, A.R.M.; Klenerman, P.; et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife 2014, 3. [Google Scholar] [CrossRef]
- Araki, K.; Turner, A.P.; Shaffer, V.O.; Gangappa, S.; Keller, S.A.; Bachmann, M.F.; Larsen, C.P.; Ahmed, R. mTOR regulates memory CD8 T-cell differentiation. Nature 2009, 460, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Song, X.; Cao, W.; Lu, J.; Wang, X.; Wang, G.; Wang, Z.; Chen, X. Autophagy and mitochondrial dysfunction in adjuvant-arthritis rats treatment with resveratrol. Sci. Rep. 2016, 6, 32928. [Google Scholar] [CrossRef]
- Qin, N.; Wei, L.; Li, W.; Yang, W.; Cai, L.; Qian, Z.; Wu, S. Local intra-articular injection of resveratrol delays cartilage degeneration in C57BL/6 mice by inducing autophagy via AMPK/mTOR pathway. J. Pharmacol. Sci. 2017, 134, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Jiang, J.; Zhang, G.; Bu, Y.; Zhang, G.; Zhao, X. Resveratrol and caloric restriction prevent hepatic steatosis by regulating SIRT1-autophagy pathway and alleviating endoplasmic reticulum stress in high-fat diet-fed rats. PLoS ONE 2017, 12, e0183541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, M.L.; Zhou, Y.; Yi, L.; Gao, Y.X.; Ran, L.; Chen, S.H.; Zhang, T.; Zhou, X.; Zou, D.; et al. Resveratrol improves hepatic steatosis by inducing autophagy through the cAMP signaling pathway. Mol. Nutr. Food Res. 2015, 59, 1443–1457. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.J.; Chen, N. Resveratrol as a Natural Autophagy Regulator for Prevention and Treatment of Alzheimer’s Disease. Nutrients 2017, 9, 927. [Google Scholar] [CrossRef]
- Hu, J.; Han, H.; Cao, P.; Yu, W.; Yang, C.; Gao, Y.; Yuan, W. Resveratrol improves neuron protection and functional recovery through enhancement of autophagy after spinal cord injury in mice. Am. J. Transl. Res. 2017, 9, 4607–4616. [Google Scholar]
- Vitale, N.; Kisslinger, A.; Paladino, S.; Procaccini, C.; Matarese, G.; Pierantoni, G.M.; Mancini, F.P.; Tramontano, D. Resveratrol couples apoptosis with autophagy in UVB-irradiated HaCaT cells. PLoS ONE 2013, 8, e80728. [Google Scholar] [CrossRef]
- Ferraresi, A.; Phadngam, S.; Morani, F.; Galetto, A.; Alabiso, O.; Chiorino, G.; Isidoro, C. Resveratrol inhibits IL-6-induced ovarian cancer cell migration through epigenetic up-regulation of autophagy. Mol. Carcinog. 2017, 56, 1164–1181. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Lee, C.Y.; Lu, C.C.; Tsai, F.J.; Hsu, Y.M.; Tsao, J.W.; Juan, Y.N.; Chiu, H.Y.; Yang, J.S.; Wang, C.C. Resveratrol-induced autophagy and apoptosis in cisplatin-resistant human oral cancer CAR cells: A key role of AMPK and Akt/mTOR signaling. Int. J. Oncol. 2017, 50, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Luyten, T.; Welkenhuyzen, K.; Roest, G.; Kania, E.; Wang, L.W.; Bittremieux, M.; Yule, D.I.; Parys, J.B.; Bultynck, G. Resveratrol-induced autophagy is dependent on IP(3)Rs and on cytosolic Ca2+. Bba-Mol. Cell Res. 2017, 1864, 947–956. [Google Scholar] [CrossRef]
- Park, D.; Jeong, H.; Lee, M.N.; Koh, A.; Kwon, O.; Yang, Y.R.; Noh, J.; Suh, P.G.; Park, H.; Ryu, S.H. Resveratrol induces autophagy by directly inhibiting mTOR through ATP competition. Sci. Rep. 2016, 6, 21772. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Iqbal, M.A.; Singh, R.K.; Bamezai, R.N. Resveratrol inhibits TIGAR to promote ROS induced apoptosis and autophagy. Biochimie 2015, 118, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, S.; Gao, K.; Zhou, Z.; Wang, C.; Shen, Z.; Guo, Y.; Li, Z.; Wan, Z.; Liu, C.; et al. Resveratrol protects against spinal cord injury by activating autophagy and inhibiting apoptosis mediated by the SIRT1/AMPK signaling pathway. Neuroscience 2017, 348, 241–251. [Google Scholar] [CrossRef]
- Duan, W.J.; Li, Y.F.; Liu, F.L.; Deng, J.; Wu, Y.P.; Yuan, W.L.; Tsoi, B.; Chen, J.L.; Wang, Q.; Cai, S.H.; et al. A SIRT3/AMPK/autophagy network orchestrates the protective effects of trans-resveratrol in stressed peritoneal macrophages and RAW 264.7 macrophages. Free Radic Biol. Med. 2016, 95, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.J.; Liu, F.L.; He, R.R.; Yuan, W.L.; Li, Y.F.; Tsoi, B.; Su, W.W.; Yao, X.S.; Kurihara, H. Autophagy is involved in the effects of resveratrol on prevention of splenocyte apoptosis caused by oxidative stress in restrained mice. Mol. Nutr. Food Res. 2013, 57, 1145–1157. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, X.; Zhu, G.; Zhang, Y.; He, M.; Zhang, J. The role of Resveratrol-induced mitophagy/autophagy in peritoneal mesothelial cells inflammatory injury via NLRP3 inflammasome activation triggered by mitochondrial ROS. Exp. Cell Res. 2016, 341, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.L.; Yi, L.; Jin, X.; Liang, X.Y.; Zhou, Y.; Zhang, T.; Xie, Q.; Zhou, X.; Chang, H.; Fu, Y.J.; et al. Resveratrol attenuates vascular endothelial inflammation by inducing autophagy through the cAMP signaling pathway. Autophagy 2013, 9, 2033–2045. [Google Scholar] [CrossRef] [PubMed]
- He, S.P.; Tan, G.Y.; Li, G.; Tan, W.M.; Nan, T.G.; Wang, B.M.; Li, Z.H.; Li, Q.X. Development of a sensitive monoclonal antibody-based enzyme-linked immunosorbent assay for the antimalaria active ingredient artemisinin in the Chinese herb Artemisia annua L. Anal. Bioanal. Chem. 2009, 393, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Putalun, W.; De-Eknamkul, W.; Matangkasombut, O.; Shoyama, Y. Preparation of a novel monoclonal antibody against the antimalarial drugs, artemisinin and artesunate. Planta Med. 2007, 73, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Ayede, I.A.; Falade, A.G.; Sowunmi, A.; Jansen, F.H. An open randomized clinical trial in comparing two artesunate-based combination treatments on Plasmodium falciparum malaria in Nigerian children: Artesunate/sulphamethoxypyrazine/pyrimethamine (fixed dose over 24 hours) versus artesunate/amodiaquine (fixed dose over 48 hours). Malar. J. 2010, 9, 378. [Google Scholar] [CrossRef] [PubMed]
- Guruprasad, B.; Chaudhary, P.; Choedon, T.; Kumar, V.L. Artesunate Ameliorates Functional Limitations in Freund’s Complete Adjuvant-Induced Monoarthritis in Rat by Maintaining Oxidative Homeostasis and Inhibiting COX-2 Expression. Inflammation 2015, 38, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.B.; Qiu, H.Y. Effects of Artesunate on chondrocyte proliferation, apoptosis and autophagy through the PI3K/AKT/mTOR signaling pathway in rat models with rheumatoid arthritis. Biomed. Pharmacother. 2018, 102, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.W.; Cen, Y.Y.; Song, Y.; Li, P.; Qin, R.X.; Liu, C.; Zhao, Y.B.; Zheng, J.; Zhou, H. Artesunate attenuated progression of atherosclerosis lesion formation alone or combined with rosuvastatin through inhibition of pro-inflammatory cytokines and pro-inflammatory chemokines. Phytomedicine 2016, 23, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Shen, Y.; Sun, H.J.; Meng, D.L.; Huo, W.; Qi, X. Protectiveness of Artesunate Given Prior Ischemic Cerebral Infarction Is Mediated by Increased Autophagy. Front. Neurol. 2018, 9. [Google Scholar] [CrossRef]
- Qin, G.; Wu, L.; Liu, H.; Pang, Y.; Zhao, C.; Wu, S.; Wang, X.; Chen, T. Artesunate induces apoptosis via a ROS-independent and Bax-mediated intrinsic pathway in HepG2 cells. Exp. Cell Res. 2015, 336, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhou, J.Y.; Zhang, D.; Liu, M.H.; Chen, Y.G. Artesunate induces apoptosis and autophagy in HCT116 colon cancer cells, and autophagy inhibition enhances the artesunateinduced apoptosis. Int. J. Mol. Med. 2018, 42, 1295–1304. [Google Scholar] [CrossRef]
- Chen, K.; Shou, L.M.; Lin, F.; Duan, W.M.; Wu, M.Y.; Xie, X.; Xie, Y.F.; Li, W.; Tao, M. Artesunate induces G2/M cell cycle arrest through autophagy induction in breast cancer cells. Anti-Cancer Drug 2014, 25, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.C.; Liu, Y.; Wang, H.; Han, Q.K.; Lu, C. Research on the relationship between artesunate and Raji cell autophagy and apoptosis of Burkitt’s lymphoma and its mechanism. Eur. Rev. Med. Pharmaco. 2017, 21, 2238–2243. [Google Scholar]
- Wan, Q.; Chen, H.; Li, X.; Yan, L.; Sun, Y.; Wang, J. Artesunate inhibits fibroblasts proliferation and reduces surgery-induced epidural fibrosis via the autophagy-mediated p53/p21(waf1/cip1) pathway. Eur. J. Pharmacol. 2019, 842, 197–207. [Google Scholar] [CrossRef]
- Kuang, M.; Cen, Y.; Qin, R.; Shang, S.; Zhai, Z.; Liu, C.; Pan, X.; Zhou, H. Artesunate Attenuates Pro-Inflammatory Cytokine Release from Macrophages by Inhibiting TLR4-Mediated Autophagic Activation via the TRAF6-Beclin1-PI3KC3 Pathway. Cell Physiol. Biochem. 2018, 47, 475–488. [Google Scholar] [CrossRef]
- Li, B.; Zhang, R.; Li, J.; Zhang, L.; Ding, G.; Luo, P.; He, S.; Dong, Y.; Jiang, W.; Lu, Y.; et al. Antimalarial artesunate protects sepsis model mice against heat-killed Escherichia coli challenge by decreasing TLR4, TLR9 mRNA expressions and transcription factor NF-kappa B activation. Int. Immunopharmacol. 2008, 8, 379–389. [Google Scholar] [CrossRef]
- Benaroudj, N.; Lee, D.H.; Goldberg, A.L. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 2001, 276, 24261–24267. [Google Scholar] [CrossRef]
- Echigo, R.; Shimohata, N.; Karatsu, K.; Yano, F.; Kayasuga-Kariya, Y.; Fujisawa, A.; Ohto, T.; Kita, Y.; Nakamura, M.; Suzuki, S.; et al. Trehalose treatment suppresses inflammation, oxidative stress, and vasospasm induced by experimental subarachnoid hemorrhage. J. Transl. Med. 2012, 10, 80. [Google Scholar] [CrossRef]
- Rodriguez-Navarro, J.A.; Rodriguez, L.; Casarejos, M.J.; Solano, R.M.; Gomez, A.; Perucho, J.; Cuervo, A.M.; Garcia de Yebenes, J.; Mena, M.A. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol. Dis. 2010, 39, 423–438. [Google Scholar] [CrossRef]
- Du, J.; Liang, Y.; Xu, F.; Sun, B.; Wang, Z. Trehalose rescues Alzheimer’s disease phenotypes in APP/PS1 transgenic mice. J. Pharm. Pharmacol. 2013, 65, 1753–1756. [Google Scholar] [CrossRef]
- Williams, B.; Njaci, I.; Moghaddam, L.; Long, H.; Dickman, M.B.; Zhang, X.; Mundree, S. Trehalose Accumulation Triggers Autophagy during Plant Desiccation. PLoS Genet. 2015, 11, e1005705. [Google Scholar] [CrossRef]
- Chen, X.; Li, M.; Li, L.; Xu, S.; Huang, D.; Ju, M.; Huang, J.; Chen, K.; Gu, H. Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mTOR-independent pathway. Sci. Rep. 2016, 6, 28423. [Google Scholar] [CrossRef]
- DeBosch, B.J.; Heitmeier, M.R.; Mayer, A.L.; Higgins, C.B.; Crowley, J.R.; Kraft, T.E.; Chi, M.; Newberry, E.P.; Chen, Z.; Finck, B.N.; et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci. Signal. 2016, 9, ra21. [Google Scholar] [CrossRef]
- Sarkar, S.; Davies, J.E.; Huang, Z.; Tunnacliffe, A.; Rubinsztein, D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 2007, 282, 5641–5652. [Google Scholar] [CrossRef]
- Belzile, J.P.; Sabalza, M.; Craig, M.; Clark, E.; Morello, C.S.; Spector, D.H. Trehalose, an mTOR-Independent Inducer of Autophagy, Inhibits Human Cytomegalovirus Infection in Multiple Cell Types. J. Virol. 2016, 90, 1259–1277. [Google Scholar] [CrossRef]
- Wu, Q.; Jiang, D.; Huang, C.; van Dyk, L.F.; Li, L.; Chu, H.W. Trehalose-mediated autophagy impairs the anti-viral function of human primary airway epithelial cells. PLoS ONE 2015, 10, e0124524. [Google Scholar] [CrossRef]
- Stock, I. Human rhinovirus diseases–epidemiology, treatment and prevention. Med. Monatsschr. Pharm. 2014, 37, 44–53. [Google Scholar]
- Griffiths, P.; Baraniak, I.; Reeves, M. The pathogenesis of human cytomegalovirus. J. Pathol. 2015, 235, 288–297. [Google Scholar] [CrossRef]
- Berencsi, K.; Endresz, V.; Klurfeld, D.; Kari, L.; Kritchevsky, D.; Gonczol, E. Early atherosclerotic plaques in the aorta following cytomegalovirus infection of mice. Cell Adhes. Commun. 1998, 5, 39–47. [Google Scholar] [CrossRef]
- Rosenfeld, M.E.; Campbell, L.A. Pathogens and atherosclerosis: Update on the potential contribution of multiple infectious organisms to the pathogenesis of atherosclerosis. Thromb. Haemost. 2011, 106, 858–867. [Google Scholar] [CrossRef]
- Cobbs, C.S. Cytomegalovirus and brain tumor: Epidemiology, biology and therapeutic aspects. Curr. Opin. Oncol. 2013, 25, 682–688. [Google Scholar] [CrossRef]
- Pasquereau, S.; Al Moussawi, F.; Karam, W.; Diab Assaf, M.; Kumar, A.; Herbein, G. Cytomegalovirus, Macrophages and Breast Cancer. Open Virol. J. 2017, 11, 15–27. [Google Scholar] [CrossRef]
- Meier, J.L.; Grose, C. Variable Effects of Autophagy Induction by Trehalose on Herpesviruses Depending on Conditions of Infection. Yale J. Biol. Med. 2017, 90, 25–33. [Google Scholar]
- Macias-Ceja, D.C.; Cosin-Roger, J.; Ortiz-Masia, D.; Salvador, P.; Hernandez, C.; Esplugues, J.V.; Calatayud, S.; Barrachina, M.D. Stimulation of autophagy prevents intestinal mucosal inflammation and ameliorates murine colitis. Br. J. Pharmacol. 2017, 174, 2501–2511. [Google Scholar] [CrossRef]
- Antonucci, R.; Locci, C.; Clemente, M.G.; Chicconi, E.; Antonucci, L. Vitamin D deficiency in childhood: Old lessons and current challenges. J. Pediatr. Endocrinol. Metab. 2018, 31, 247–260. [Google Scholar] [CrossRef]
- Hoyer-Hansen, M.; Nordbrandt, S.P.S.; Jaattela, M. Autophagy as a basis for the health-promoting effects of vitamin D. Trends Mol. Med. 2010, 16, 295–302. [Google Scholar] [CrossRef]
- Wei, R.; Christakos, S. Mechanisms Underlying the Regulation of Innate and Adaptive Immunity by Vitamin D. Nutrients 2015, 7, 8251–8260. [Google Scholar] [CrossRef]
- Adams, J.S.; Hewison, M. Unexpected actions of vitamin D: New perspectives on the regulation of innate and adaptive immunity. Nat. Clin. Pract. Endocrinol. Metab. 2008, 4, 80–90. [Google Scholar] [CrossRef]
- Kamen, D.L.; Tangpricha, V. Vitamin D and molecular actions on the immune system: Modulation of innate and autoimmunity. J. Mol. Med. 2010, 88, 441–450. [Google Scholar] [CrossRef]
- Altieri, B.; Muscogiuri, G.; Barrea, L.; Mathieu, C.; Vallone, C.V.; Mascitelli, L.; Bizzaro, G.; Altieri, V.M.; Tirabassi, G.; Balercia, G.; et al. Does vitamin D play a role in autoimmune endocrine disorders? A proof of concept. Rev. Endocr. Metab. Dis. 2017, 18, 335–346. [Google Scholar] [CrossRef]
- Yuk, J.M.; Shin, D.M.; Lee, H.M.; Yang, C.S.; Jin, H.S.; Kim, K.K.; Lee, Z.W.; Lee, S.H.; Kim, J.M.; Jo, E.K. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe 2009, 6, 231–243. [Google Scholar] [CrossRef]
- Tang, J.; Yam, W.C.; Chen, Z. Mycobacterium tuberculosis infection and vaccine development. Tuberculosis (Edinb) 2016, 98, 30–41. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Ciric, B.; Ma, C.G.; Gran, B.; Rostami, A.; Zhang, G.X. Therapeutic effect of baicalin on experimental autoimmune encephalomyelitis is mediated by SOCS3 regulatory pathway. Sci. Rep. 2015, 5, 17407. [Google Scholar] [CrossRef]
- Liu, T.; Dai, W.; Li, C.; Liu, F.; Chen, Y.; Weng, D.; Chen, J. Baicalin Alleviates Silica-Induced Lung Inflammation and Fibrosis by Inhibiting the Th17 Response in C57BL/6 Mice. J. Nat. Prod. 2015, 78, 3049–3057. [Google Scholar] [CrossRef]
- Hung, C.H.; Wang, C.N.; Cheng, H.H.; Liao, J.W.; Chen, Y.T.; Chao, Y.W.; Jiang, J.L.; Lee, C.C. Baicalin Ameliorates Imiquimod-Induced Psoriasis-Like Inflammation in Mice. Planta Med. 2018, 84, 1110–1117. [Google Scholar] [CrossRef]
- Orzechowska, B.; Chaber, R.; Wisniewska, A.; Pajtasz-Piasecka, E.; Jatczak, B.; Siemieniec, I.; Gulanowski, B.; Chybicka, A.; Blach-Olszewska, Z. Baicalin from the extract of Scutellaria baicalensis affects the innate immunity and apoptosis in leukocytes of children with acute lymphocytic leukemia. Int. Immunopharmacol. 2014, 23, 558–567. [Google Scholar] [CrossRef]
- Yu, F.Y.; Huang, S.G.; Zhang, H.Y.; Ye, H.; Chi, H.G.; Zou, Y.; Lv, R.X.; Zheng, X.B. Effects of baicalin in CD4 + CD29 + T cell subsets of ulcerative colitis patients. World J. Gastroenterol. 2014, 20, 15299–15309. [Google Scholar] [CrossRef]
- Gao, X.; Guo, M.; Zhang, Z.; Shen, P.; Yang, Z.; Zhang, N. Baicalin promotes the bacteriostatic activity of lysozyme on S. aureus in mammary glands and neutrophilic granulocytes in mice. Oncotarget 2017, 8, 19894–19901. [Google Scholar] [CrossRef]
- Zhang, Q.; Sun, J.; Wang, Y.; He, W.; Wang, L.; Zheng, Y.; Wu, J.; Zhang, Y.; Jiang, X. Antimycobacterial and Anti-inflammatory Mechanisms of Baicalin via Induced Autophagy in Macrophages Infected with Mycobacterium tuberculosis. Front. Microbiol. 2017, 8, 2142. [Google Scholar] [CrossRef]
- Lee, G.Y.; Park, K.G.; Namgoong, S.; Han, S.K.; Jeong, S.H.; Dhong, E.S.; Kim, W.K. Effects of Panax ginseng extract on human dermal fibroblast proliferation and collagen synthesis. Int. Wound J. 2016, 13, 42–46. [Google Scholar] [CrossRef]
- Simu, S.Y.; Ahn, S.; Castro-Aceituno, V.; Singh, P.; Mathiyalagan, R.; Jimenez-Perez, Z.E.; Hurh, J.; Oi, L.Z.; Hun, N.J.; Kim, Y.J.; et al. Gold Nanoparticles Synthesized with Fresh Panax ginseng Leaf Extract Suppress Adipogenesis by Downregulating PPARgamma/CEBPalpha Signaling in 3T3-L1 Mature Adipocytes. J. Nanosci. Nanotechnol. 2019, 19, 701–708. [Google Scholar] [CrossRef]
- Yun, S.N.; Moon, S.J.; Ko, S.K.; Im, B.O.; Chung, S.H. Wild ginseng prevents the onset of high-fat diet induced hyperglycemia and obesity in ICR mice. Arch. Pharm. Res. 2004, 27, 790–796. [Google Scholar] [CrossRef]
- Lee, J.H.; Min, D.S.; Lee, C.W.; Song, K.H.; Kim, Y.S.; Kim, H.P. Ginsenosides from Korean Red Ginseng ameliorate lung inflammatory responses: Inhibition of the MAPKs/NF-kappaB/c-Fos pathways. J. Ginseng. Res. 2018, 42, 476–484. [Google Scholar] [CrossRef]
- Hou, Y.L.; Tsai, Y.H.; Lin, Y.H.; Chao, J.C. Ginseng extract and ginsenoside Rb1 attenuate carbon tetrachloride-induced liver fibrosis in rats. BMC Complement. Altern. Med. 2014, 14, 415. [Google Scholar] [CrossRef]
- Chen, Y.; Bi, L.; Luo, H.; Jiang, Y.; Chen, F.; Wang, Y.; Wei, G.; Chen, W. Water extract of ginseng and astragalus regulates macrophage polarization and synergistically enhances DDP’s anticancer effect. J. Ethnopharmacol. 2018, 232, 11–20. [Google Scholar] [CrossRef]
- Yu, J.S.; Roh, H.S.; Baek, K.H.; Lee, S.; Kim, S.; So, H.M.; Moon, E.; Pang, C.; Jang, T.S.; Kim, K.H. Bioactivity-guided isolation of ginsenosides from Korean Red Ginseng with cytotoxic activity against human lung adenocarcinoma cells. J. Ginseng. Res. 2018, 42, 562–570. [Google Scholar] [CrossRef]
- Kim, C.; Lee, J.H.; Baek, S.H.; Ko, J.H.; Nam, D.; Ahn, K.S. Korean Red Ginseng Extract Enhances the Anticancer Effects of Sorafenib through Abrogation of CREB and c-Jun Activation in Renal Cell Carcinoma. Phytother. Res. 2017, 31, 1078–1089. [Google Scholar] [CrossRef]
- Yang, P.; Ling, L.; Sun, W.; Yang, J.; Zhang, L.; Chang, G.; Guo, J.; Sun, J.; Sun, L.; Lu, D. Ginsenoside Rg1 inhibits apoptosis by increasing autophagy via the AMPK/mTOR signaling in serum deprivation macrophages. Acta Biochim. Biophys. Sin. (Shanghai) 2018, 50, 144–155. [Google Scholar] [CrossRef]
- Qiao, L.; Zhang, X.; Liu, M.; Liu, X.; Dong, M.; Cheng, J.; Zhang, X.; Zhai, C.; Song, Y.; Lu, H.; et al. Ginsenoside Rb1 Enhances Atherosclerotic Plaque Stability by Improving Autophagy and Lipid Metabolism in Macrophage Foam Cells. Front. Pharmacol. 2017, 8, 727. [Google Scholar] [CrossRef]
- Zhou, P.; Xie, W.; Luo, Y.; Lu, S.; Dai, Z.; Wang, R.; Zhang, X.; Li, G.; Sun, G.; Sun, X. Inhibitory Effects of Ginsenoside Rb1 on Early Atherosclerosis in ApoE−/− Mice via Inhibition of Apoptosis and Enhancing Autophagy. Molecules 2018, 23, 2912. [Google Scholar] [CrossRef]
- Li, M.; Li, J.; Zeng, R.; Yang, J.; Liu, J.; Zhang, Z.; Song, X.; Yao, Z.; Ma, C.; Li, W.; et al. Respiratory Syncytial Virus Replication Is Promoted by Autophagy-Mediated Inhibition of Apoptosis. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Eskenazi, B.; Warner, M.L. Epidemiology of endometriosis. Obstet Gynecol Clin. North. Am. 1997, 24, 235–258. [Google Scholar] [CrossRef]
- Pritts, E.A.; Taylor, R.N. An evidence-based evaluation of endometriosis-associated infertility. Endocrinol. Metab. Clin. North. Am. 2003, 32, 653–667. [Google Scholar] [CrossRef]
- Mei, J.; Zhu, X.Y.; Jin, L.P.; Duan, Z.L.; Li, D.J.; Li, M.Q. Estrogen promotes the survival of human secretory phase endometrial stromal cells via CXCL12/CXCR4 up-regulation-mediated autophagy inhibition. Hum. Reprod. 2015, 30, 1677–1689. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Yuan, Q.; Cui, Q.; Liu, C.; Zhou, Z.; Zhao, H.; Dun, Y.; Wang, T.; Zhang, C. Vaccine adjuvant ginsenoside Rg1 enhances immune responses against hepatitis B surface antigen in mice. Can. J. Physiol. Pharmacol. 2016, 94, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Ko, E.; Lee, J.; Rho, S.; Ko, S.; Shin, M.K.; Min, B.I.; Hong, M.C.; Kim, S.Y.; Bae, H. Ginsenoside Rg1 enhances CD4(+) T-cell activities and modulates Th1/Th2 differentiation. Int. Immunopharmacol. 2004, 4, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xiang, Z.; Wang, Y.; Li, X.; Fang, C.; Song, S.; Li, C.; Yu, H.; Wang, H.; Yan, L.; et al. (-)-Epigallocatechin Gallate Targets Notch to Attenuate the Inflammatory Response in the Immediate Early Stage in Human Macrophages. Front. Immunol. 2017, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Gao, C.; Yan, P.; Zhang, M.; Wang, Y.H.; Hu, Y.; Wu, X.Y.; Wang, X.J.; Sheng, J. EGCG Reduces Obesity and White Adipose Tissue Gain Partly Through AMPK Activation in Mice. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef]
- Luo, K.W.; Wei, C.; Lung, W.Y.; Wei, X.Y.; Cheng, B.H.; Cai, Z.M.; Huang, W.R. EGCG inhibited bladder cancer SW780 cell proliferation and migration both in vitro and in vivo via down-regulation of NF-kappaB and MMP-9. J. Nutr. Biochem. 2017, 41, 56–64. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Lu, J.L.; Liang, Y.R.; Li, Q.S. Suppressive Effects of EGCG on Cervical Cancer. Molecules 2018, 23, 2334. [Google Scholar] [CrossRef]
- Zhou, J.; Farah, B.L.; Sinha, R.A.; Wu, Y.; Singh, B.K.; Bay, B.H.; Yang, C.S.; Yen, P.M. Epigallocatechin-3-gallate (EGCG), a green tea polyphenol, stimulates hepatic autophagy and lipid clearance. PLoS ONE 2014, 9, e87161. [Google Scholar] [CrossRef]
- Tsai, C.Y.; Chen, C.Y.; Chiou, Y.H.; Shyu, H.W.; Lin, K.H.; Chou, M.C.; Huang, M.H.; Wang, Y.F. Epigallocatechin-3-Gallate Suppresses Human Herpesvirus 8 Replication and Induces ROS Leading to Apoptosis and Autophagy in Primary Effusion Lymphoma Cells. Int. J. Mol. Sci. 2017, 19, 16. [Google Scholar] [CrossRef]
- Li, S.; Xia, Y.; Chen, K.; Li, J.; Liu, T.; Wang, F.; Lu, J.; Zhou, Y.; Guo, C. Epigallocatechin-3-gallate attenuates apoptosis and autophagy in concanavalin A-induced hepatitis by inhibiting BNIP3. Drug Des. Devel. Ther. 2016, 10, 631–647. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xia, Y.; Liu, T.; Wang, J.; Dai, W.; Wang, F.; Zheng, Y.; Chen, K.; Li, S.; Abudumijiti, H.; et al. Protective effects of astaxanthin on ConA-induced autoimmune hepatitis by the JNK/p-JNK pathway-mediated inhibition of autophagy and apoptosis. PLoS ONE 2015, 10, e0120440. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yu, H.; Gu, W.; Luo, X.; Li, R.; Zhang, J.; Xu, Y.; Yang, L.; Shen, N.; Feng, L.; et al. Autophagy Negatively Regulates Transmissible Gastroenteritis Virus Replication. Sci. Rep. 2016, 6, 23864. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Gu, M.J.; Kim, C.G.; Kye, Y.C.; Lim, Y.; Lee, J.E.; Park, B.C.; Chu, H.; Han, S.H.; Yun, C.H. Rapamycin-induced autophagy restricts porcine epidemic diarrhea virus infectivity in porcine intestinal epithelial cells. Antiviral Res. 2017, 146, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Chiramel, A.I.; Brady, N.R.; Bartenschlager, R. Divergent roles of autophagy in virus infection. Cells 2013, 2, 83–104. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.C.; Zhao, L.L.; Li, S.Q.; Niu, Y.J.; Jiang, X.J.; Xu, L.J.; Lu, T.F.; Han, L.X.; Liu, S.W.; Chen, H.Y. Autophagy activated by duck enteritis virus infection positively affects its replication. J. Gen. Virol. 2017, 98, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Stylianou, K.; Petrakis, I.; Mavroeidi, V.; Stratakis, S.; Vardaki, E.; Perakis, K.; Stratigis, S.; Passam, A.; Papadogiorgaki, E.; Giannakakis, K.; et al. The PI3K/Akt/mTOR pathway is activated in murine lupus nephritis and downregulated by rapamycin. Nephrol. Dial. Transplant. 2011, 26, 498–508. [Google Scholar] [CrossRef]
- Reddy, P.S.; Legault, H.M.; Sypek, J.P.; Collins, M.J.; Goad, E.; Goldman, S.J.; Liu, W.; Murray, S.; Dorner, A.J.; O’Toole, M. Mapping similarities in mTOR pathway perturbations in mouse lupus nephritis models and human lupus nephritis. Arthritis Res. Ther. 2008, 10, R127. [Google Scholar] [CrossRef]
- Qi, Y.Y.; Zhou, X.J.; Cheng, F.J.; Hou, P.; Ren, Y.L.; Wang, S.X.; Zhao, M.H.; Yang, L.; Martinez, J.; Zhang, H. Increased autophagy is cytoprotective against podocyte injury induced by antibody and interferon-alpha in lupus nephritis. Ann. Rheum. Dis. 2018, 77, 1799–1809. [Google Scholar] [CrossRef]
- Jia, X.; Cao, B.; An, Y.; Zhang, X.; Wang, C. Rapamycin ameliorates lipopolysaccharide-induced acute lung injury by inhibiting IL-1beta and IL-18 production. Int. Immunopharmacol. 2018, 67, 211–219. [Google Scholar] [CrossRef]
- Shoji-Kawata, S.; Sumpter, R.; Leveno, M.; Campbell, G.R.; Zou, Z.; Kinch, L.; Wilkins, A.D.; Sun, Q.; Pallauf, K.; MacDuff, D.; et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 2013, 494, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Mei, Y.; Sanishvili, R.; Levine, B.; Colbert, C.L.; Sinha, S. Targeting gamma-herpesvirus 68 Bcl-2-mediated down-regulation of autophagy. J. Biol. Chem. 2014, 289, 8029–8040. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Mani, J.; Kumar, P.; Haridas, S.; Upadhyay, P.; Bhaskar, S. Activation of anti-tumor immune response and reduction of regulatory T cells with Mycobacterium indicus pranii (MIP) therapy in tumor bearing mice. PLoS ONE 2011, 6, e25424. [Google Scholar] [CrossRef] [PubMed]
- Halder, K.; Banerjee, S.; Ghosh, S.; Bose, A.; Das, S.; Chowdhury, B.P.; Majumdar, S. Mycobacterium indicus pranii (Mw) inhibits invasion by reducing matrix metalloproteinase (MMP-9) via AKT/ERK-1/2 and PKCalpha signaling: A potential candidate in melanoma cancer therapy. Cancer Biol. Ther. 2017, 18, 850–862. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, M.; In, L.L.; Kumar, A.; Ahmed, N.; Nagoor, N.H. Cytotoxic and apoptotic effects of heat killed Mycobacterium indicus pranii (MIP) on various human cancer cell lines. Sci. Rep. 2016, 6, 19833. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Ahmad, F.J.; Ahmad, F.; Gupta, U.D.; Natarajan, M.; Katoch, V.; Bhaskar, S. Efficacy of Mycobacterium indicus pranii immunotherapy as an adjunct to chemotherapy for tuberculosis and underlying immune responses in the lung. PLoS ONE 2012, 7, e39215. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Chowdhury, B.P.; Goswami, A.; Parveen, S.; Jawed, J.; Pal, N.; Majumdar, S. Mycobacterium indicus pranii (MIP) mediated host protective intracellular mechanisms against tuberculosis infection: Involvement of TLR-4 mediated signaling. Tuberculosis (Edinb) 2016, 101, 201–209. [Google Scholar] [CrossRef]
- Singh, B.; Saqib, M.; Gupta, A.; Kumar, P.; Bhaskar, S. Autophagy induction by Mycobacterium indicus pranii promotes Mycobacterium tuberculosis clearance from RAW 264.7 macrophages. PLoS ONE 2017, 12, e0189606. [Google Scholar] [CrossRef]

| Compounds | Mechanism of Inducing Autophagy (Activation/Inhibition) | Related Immunity Model | References |
|---|---|---|---|
| Spermine | Increase ATG5 expression (Autophagy activation) | Liver inflammation | [68] |
| Spermidine | Unknown (Autophagy activation) | CD8(+) T cell response | [72] |
| Resveratrol | Increase SIRT3 expression & phosphorylation of AMPK (Autophagy activation) | Macrophage reactive oxygen species | [87] |
| Increase SIRT3 expression & increase Beclin-1 expression (Autophagy activation) | Splenocyte viability | [88] | |
| Increase AMPK phosphorylation (Autophagy activation) | Peritoneal inflammatory injury | [89] | |
| Activation of cAMP-PRKA-AMPK-SIRT1 pathway (Autophagy activation) | Atherosclerosis | [90] | |
| Artesunate | Inhibit TRAF6-Beclin 1-PI3KC3 (Autophagy inhibition) | Inflammation in peritoneal macrophages | [103] |
| Trehalose | Increase ATG5 expression (Autophagy activation) | Human rhinovirus infection (increase) | [114] |
| Unknown (Autophagy activation) | Human cytomegalovirus infection (decrease) | [113] | |
| Unknown (Autophagy activation) | Colitis | [122] | |
| Vitamin D3 (1a,25-dihydroxyvitamin D3) | Increase Beclin 1 & ATG5 expression (Autophagy activation) | Mycobacterium tuberculosis infection | [129] |
| Baicalin | Suppress PI3K-Akt-mTOR pathway (Autophagy activation) | Mycobacterium tuberculosis infection | [137] |
| Ginsenosides Rb1 | Increase Beclin 1 expression (Autophagy activation) | Atherosclerosis | [147] |
| Protopanaxadiol | Unknown (Autophagy activation) | NK cell cytotoxicity in eESCs | [149] |
| Epigallocatechin-3-gallate (EGCG) | Increase phosphorylation of AMPK (Autophagy activation) | Hepatosteastosis | [159] |
| Increase Beclin 1 (Autophagy activation) | Protection against Human herpesvirus 8 | [160] | |
| Inhibit IL-6/JAKs/STAT3/BNIP3 pathway (Autophagy inhibition) | Liver injury | [161] | |
| Rapamycin | Inhibit mTORC1 (Autophagy activation) | Coronavirus infection (decrease) | [163,164] |
| Inhibit mTORC1 (Autophagy activation) | Respiratory syncytial virus & duck enteritis virus infection (increase) | [149,166] | |
| Inhibit mTORC1 (Autophagy activation) | Lupus nephritis | [167,169] | |
| Inhibit mTORC1 (Autophagy activation) | Acute lung injury | [170] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, Y.J.; Kim, J.H.; Byun, S. Modulation of Autophagy for Controlling Immunity. Cells 2019, 8, 138. https://doi.org/10.3390/cells8020138
Jang YJ, Kim JH, Byun S. Modulation of Autophagy for Controlling Immunity. Cells. 2019; 8(2):138. https://doi.org/10.3390/cells8020138
Chicago/Turabian StyleJang, Young Jin, Jae Hwan Kim, and Sanguine Byun. 2019. "Modulation of Autophagy for Controlling Immunity" Cells 8, no. 2: 138. https://doi.org/10.3390/cells8020138