Transfer of Synthetic Human Chromosome into Human Induced Pluripotent Stem Cells for Biomedical Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Lentivirus Preparations
2.2. Reprogramming Human Mesenchymal Stem Cells with OKSM/rt-TA
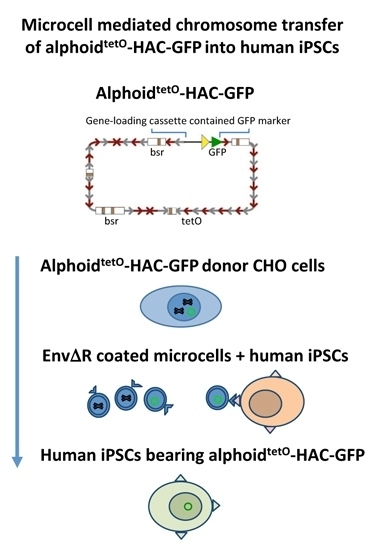
2.3. MMCT into hiPSCs
2.4. Preparation of Metaphase Spreads
2.5. Fluorescence In Situ Hybridization with the PNA Probes
2.6. FACS Sorting of hiPSCs
2.7. Karyotype Analysis
2.8. Cell Immunostaining
2.9. Southern-Blot Hybridization Analysis
2.10. Treatment of Cells with Inhibitors of Chromatin Modifiers
3. Results and Discussion
3.1. Reprogramming Human Endometrial MSCs in Lonza’s cGMP Culture Conditions
3.2. MLV Envelope Protein-Mediated Transfer of AlphoidtetO-HAC to Human iPSCs.
3.3. AlphoidtetO-HAC Maintenance in Human Induced-Pluripotent Stem Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Connor, T.P.; Crystal, R.G. Genetic medicines: Treatment strategies for hereditary disorders. Nat. Rev. Genet. 2006, 7, 261–276. [Google Scholar] [CrossRef]
- Costantini, F.; Radice, G.; Lee, J.L.; Chada, K.K.; Perry, W.; Son, H.J. Insertional mutations in transgenic mice. Prog. Nucleic Acid Res. Mol. Biol. 1989, 36, 159–169. [Google Scholar] [PubMed]
- Soriano, P.; Gridley, T.; Jaenisch, R. Retroviruses and insertional mutagenesis in mice: Proviral integration at the Mov 34 locus leads to early embryonic death. Genes Dev. 1987, 1, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Hotta, A.; Yamanaka, S. From genomics to gene therapy: Induced pluripotent stem cells meet genome editing. Annu. Rev. Genet. 2015, 49, 47–70. [Google Scholar] [CrossRef] [PubMed]
- Oshimura, M.; Katoh, M. Transfer of human artificial chromosome vectors into stem cells. Reprod. Biomed. Online 2008, 16, 57–69. [Google Scholar] [CrossRef]
- Kazuki, Y.; Hiratsuka, M.; Takiguchi, M.; Osaki, M.; Kajitani, N.; Hoshiya, H.; Hiramatsu, K.; Yoshino, T.; Kazuki, K.; Ishihara, C.; et al. Complete genetic correction of ips cells from Duchenne muscular dystrophy. Mol. Ther. 2010, 18, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Kouprina, N.; Tomilin, A.N.; Masumoto, H.; Earnshaw, W.C.; Larionov, V. Human artificial chromosome-based gene delivery vectors for biomedicine and biotechnology. Expert Opin. Drug Delivery 2014, 11, 517–535. [Google Scholar] [CrossRef]
- Oshimura, M.; Uno, N.; Kazuki, Y.; Katoh, M.; Inoue, T. A pathway from chromosome transfer to engineering resulting in human and mouse artificial chromosomes for a variety of applications to bio-medical challenges. Chromosome Res. 2015, 23, 111–133. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Kazuki, Y.; Nakayama, Y.; Nanba, E.; Oshimura, M.; Ohbayashi, T. A method for producing transgenic cells using a multi-integrase system on a human artificial chromosome vector. PLoS ONE 2011, 6, e17267. [Google Scholar] [CrossRef]
- Hoshiya, H.; Kazuki, Y.; Abe, S.; Takiguchi, M.; Kajitani, N.; Watanabe, Y.; Yoshino, T.; Shirayoshi, Y.; Higaki, K.; Messina, G.; et al. A highly stable and nonintegrated human artificial chromosome (HAC) containing the 2.4 Mb entire human dystrophin gene. Mol. Ther. 2009, 17, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, F.S.; Hoshiya, H.; D’Antona, G.; Gerli, M.F.; Messina, G.; Antonini, S.; Tonlorenzi, R.; Benedetti, S.; Berghella, L.; Torrente, Y.; et al. Stem cell-mediated transfer of a human artificial chromosome ameliorates muscular dystrophy. Sci. Transl. Med. 2011, 3, 96ra78. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, F.S.; Gerli, M.F.; Perani, L.; Benedetti, S.; Ungaro, F.; Cassano, M.; Antonini, S.; Tagliafico, E.; Artusi, V.; Longa, E.; et al. Transplantation of genetically corrected human iPSC-derived progenitors in mice with limb-girdle muscular dystrophy. Sci. Transl. Med. 2012, 4, 140ra189. [Google Scholar] [CrossRef] [PubMed]
- Kazuki, Y.; Kobayashi, K.; Aueviriyavit, S.; Oshima, T.; Kuroiwa, Y.; Tsukazaki, Y.; Senda, N.; Kawakami, H.; Ohtsuki, S.; Abe, S.; et al. Trans-chromosomic mice containing a human CYP3A cluster for prediction of xenobiotic metabolism in humans. Hum. Mol. Genet. 2013, 22, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Tomizuka, K.; Yoshida, H.; Uejima, H.; Kugoh, H.; Sato, K.; Ohguma, A.; Hayasaka, M.; Hanaoka, K.; Oshimura, M.; Ishida, I. Functional expression and germLine transmission of a human chromosome fragment in chimaeric mice. Nat. Genet. 1997, 16, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, Y.; Kasinathan, P.; Choi, Y.J.; Naeem, R.; Tomizuka, K.; Sullivan, E.J.; Knott, J.G.; Duteau, A.; Goldsby, R.A.; Osborne, B.A.; et al. Cloned transchromosomic calves producing human immunoglobulin. Nat. Biotechnol. 2002, 20, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Ebersole, T.; Okamoto, Y.; Noskov, V.N.; Kouprina, N.; Kim, J.H.; Leem, S.H.; Barrett, J.C.; Masumoto, H.; Larionov, V. Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucleic Acids Res. 2005, 33, e130. [Google Scholar] [CrossRef]
- Kim, J.H.; Kononenko, A.; Erliandri, I.; Kim, T.A.; Nakano, M.; Iida, Y.; Barrett, J.C.; Oshimura, M.; Masumoto, H.; Earnshaw, W.C.; et al. Human artificial chromosome (HAC) vector with a conditional centromere for correction of genetic deficiencies in human cells. Proc. Natl. Acad. Sci. USA 2011, 108, 20048–20053. [Google Scholar] [CrossRef] [Green Version]
- Nakano, M.; Cardinale, S.; Noskov, V.N.; Gassmann, R.; Vagnarelli, P.; Kandels-Lewis, S.; Larionov, V.; Earnshaw, W.C.; Masumoto, H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell 2008, 14, 507–522. [Google Scholar] [CrossRef]
- Kouprina, N.; Samoshkin, A.; Erliandri, I.; Nakano, M.; Lee, H.S.; Fu, H.; Iida, Y.; Aladjem, M.; Oshimura, M.; Masumoto, H.; et al. Organization of synthetic alphoid DNA array in human artificial chromosome (HAC) with a conditional centromere. ACS Synth. Biol. 2012, 1, 590–601. [Google Scholar] [CrossRef]
- Kouprina, N.; Earnshaw, W.C.; Masumoto, H.; Larionov, V. A new generation of human artificial chromosomes for functional genomics and gene therapy. Cell. Mol. Life Sci. 2013, 70, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.C.; Kononenko, A.V.; Lee, H.S.; Tolkunova, E.N.; Liskovykh, M.A.; Masumoto, H.; Earnshaw, W.C.; Tomilin, A.N.; Larionov, V.; Kouprina, N. Protecting a transgene expression from the HAC-based vector by different chromatin insulators. Cell. Mol. Life Sci. 2013, 70, 3723–3737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liskovykh, M.; Ponomartsev, S.; Popova, E.; Bader, M.; Kouprina, N.; Larionov, V.; Alenina, N.; Tomilin, A. Stable maintenance of de novo assembled human artificial chromosomes in embryonic stem cells and their differentiated progeny in mice. Cell Cycle 2015, 14, 1268–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohzeki, J.; Bergmann, J.H.; Kouprina, N.; Noskov, V.N.; Nakano, M.; Kimura, H.; Earnshaw, W.C.; Larionov, V.; Masumoto, H. Breaking the HAC Barrier: Histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012, 31, 2391–2402. [Google Scholar] [CrossRef] [Green Version]
- Doherty, A.M.; Fisher, E.M. Microcell-mediated chromosome transfer (MMCT): Small cells with huge potential. Mammalian Genome 2003, 14, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Fournier, R.E.; Ruddle, F.H. Microcell-mediated transfer of murine chromosomes into mouse, Chinese hamster, and human somatic cells. Proc. Natl. Acad. Sci. USA 1977, 74, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Liskovykh, M.; Lee, N.C.; Larionov, V.; Kouprina, N. Moving toward a higher efficiency of microcell-mediated chromosome transfer. Mol. Ther. 2016, 3, 16043. [Google Scholar] [CrossRef] [PubMed]
- Paulis, M.; Bensi, M.; Orioli, D.; Mondello, C.; Mazzini, G.; D’Incalci, M.; Falcioni, C.; Radaelli, E.; Erba, E.; Raimondi, E.; et al. Transfer of a human chromosomal vector from a hamster cell line to a mouse embryonic stem cell line. Stem Cells 2007, 25, 2543–2550. [Google Scholar] [CrossRef] [PubMed]
- Paulis, M. Chromosome transfer via cell fusion. Methods Mol. Biol. 2011, 738, 57–67. [Google Scholar] [CrossRef]
- Suzuki, N.; Itou, T.; Hasegawa, Y.; Okazaki, T.; Ikeno, M. Cell to cell transfer of the chromatin-packaged human β-globin gene cluster. Nucleic Acids Res. 2010, 38, e33. [Google Scholar] [CrossRef]
- Suzuki, T.; Kazuki, Y.; Oshimura, M.; Hara, T. Highly efficient transfer of chromosomes to a broad range of target cells using Chinese hamster ovary cells expressing murine leukemia virus-derived envelope proteins. PLoS ONE 2016, 11, e0157187. [Google Scholar] [CrossRef]
- Tang, Y.; Garson, K.; Li, L.; Vanderhyden, B.C. Optimization of lentiviral vector production using polyethylenimine-mediated transfection. Oncol. Lett. 2015, 9, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Skvortsova, E.V.; Sinenko, S.A.; Tomilin, A. Immortalized murine fibroblast cell lines are refractory to reprogramming to pluripotent state. Oncotarget 2018, 9, 35241–35250. [Google Scholar] [CrossRef]
- Liskovykh, M.; Chuykin, I.; Ranjan, A.; Safina, D.; Popova, E.; Tolkunova, E.; Mosienko, V.; Minina, J.M.; Zhdanova, N.S.; Mullins, J.J.; et al. Derivation, characterization, and stable transfection of induced pluripotent stem cells from Fischer344 rats. PLoS ONE 2011, 6, e27345. [Google Scholar] [CrossRef]
- Kostina, A.; Bjork, H.; Ignatieva, E.; Irtyuga, O.; Uspensky, V.; Semenova, D.; Maleki, S.; Tomilin, A.; Moiseeva, O.; Franco-Cereceda, A.; et al. Notch, BMP and WNT/β-catenin network is impaired in endothelial cells of the patients with thoracic aortic aneurysm. Atheroscler. Suppl. 2018, 35, e6–e13. [Google Scholar] [CrossRef] [PubMed]
- Malashicheva, A.; Kanzler, B.; Tolkunova, E.; Trono, D.; Tomilin, A. Lentivirus as a tool for lineage-specific gene manipulations. Genesis 2007, 45, 456–459. [Google Scholar] [CrossRef] [PubMed]
- Shilina, M.A.; Grinchuk, T.M.; Anatskaya, O.V.; Vinogradov, A.E.; Alekseenko, L.L.; Elmuratov, A.U.; Nikolsky, N.N. Cytogenetic and transcriptomic analysis of human endometrial MSC retaining proliferative activity after sublethal heat shock. Cells 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Malashicheva, A.; Bogdanova, M.; Zabirnyk, A.; Smolina, N.; Ignatieva, E.; Freilikhman, O.; Fedorov, A.; Dmitrieva, R.; Sjoberg, G.; Sejersen, T.; et al. Various lamin A/C mutations alter expression profile of mesenchymal stem cells in mutation specific manner. Mol. Genet. Metab. 2015, 115, 118–127. [Google Scholar] [CrossRef]
- Carey, B.W.; Markoulaki, S.; Hanna, J.; Saha, K.; Gao, Q.; Mitalipova, M.; Jaenisch, R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA 2009, 106, 157–162. [Google Scholar] [CrossRef]
- Somers, A.; Jean, J.C.; Sommer, C.A.; Omari, A.; Ford, C.C.; Mills, J.A.; Ying, L.; Sommer, A.G.; Jean, J.M.; Smith, B.W.; et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells 2010, 28, 1728–1740. [Google Scholar] [CrossRef]
- Uno, N.; Uno, K.; Zatti, S.; Ueda, K.; Hiratsuka, M.; Katoh, M.; Oshimura, M. The transfer of human artificial chromosomes via cryopreserved microcells. Cytotechnology 2013, 65, 803–809. [Google Scholar] [CrossRef] [Green Version]
- Baghbaderani, B.A.; Syama, A.; Sivapatham, R.; Pei, Y.; Mukherjee, O.; Fellner, T.; Zeng, X.; Rao, M.S. Detailed characterization of human induced pluripotent stem cells manufactured for therapeutic applications. Stem Cell Rev. 2016, 12, 394–420. [Google Scholar] [CrossRef] [PubMed]
- Baghbaderani, B.A.; Tian, X.; Neo, B.H.; Burkall, A.; Dimezzo, T.; Sierra, G.; Zeng, X.; Warren, K.; Kovarcik, D.P.; Fellner, T.; et al. cGMP-manufactured human induced pluripotent stem cells are available for pre-clinical and clinical applications. Stem Cell Rep. 2015, 5, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Zhao, R.; West, J.A.; Yabuuchi, A.; Huo, H.; Ince, T.A.; Lerou, P.H.; Lensch, M.W.; Daley, G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008, 451, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Dance, A. Core concept: Human artificial chromosomes offer insights, therapeutic possibilities, and challenges. Proc. Natl. Acad. Sci. USA 2017, 114, 9752–9754. [Google Scholar] [CrossRef] [PubMed]
- Kononenko, A.V.; Lee, N.C.; Earnshaw, W.C.; Kouprina, N.; Larionov, V. Re-engineering an alphoid(tetO)-HAC-based vector to enable high-throughput analyses of gene function. Nucleic Acids Res. 2013, 41, e107. [Google Scholar] [CrossRef] [PubMed]
- Kouprina, N.; Larionov, V. Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma 2016, 125, 621–632. [Google Scholar] [CrossRef] [Green Version]
- Hiratsuka, M.; Uno, N.; Ueda, K.; Kurosaki, H.; Imaoka, N.; Kazuki, K.; Ueno, E.; Akakura, Y.; Katoh, M.; Osaki, M.; et al. Integration-free iPS cells engineered using human artificial chromosome vectors. PLoS ONE 2011, 6, e25961. [Google Scholar] [CrossRef]
- Park, I.H. DYS-HAC-iPS cells: The combination of gene and cell therapy to treat duchenne muscular dystrophy. Mol. Ther. 2010, 18, 238–240. [Google Scholar] [CrossRef]
- Yakura, Y.; Ishihara, C.; Kurosaki, H.; Kazuki, Y.; Komatsu, N.; Okada, Y.; Doi, T.; Takeya, H.; Oshimura, M. An induced pluripotent stem cell-mediated and integration-free factor VIII expression system. Biochem. Biophys. Res. Commun. 2013, 431, 336–341. [Google Scholar] [CrossRef]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinenko, S.A.; Skvortsova, E.V.; Liskovykh, M.A.; Ponomartsev, S.V.; Kuzmin, A.A.; Khudiakov, A.A.; Malashicheva, A.B.; Alenina, N.; Larionov, V.; Kouprina, N.; et al. Transfer of Synthetic Human Chromosome into Human Induced Pluripotent Stem Cells for Biomedical Applications. Cells 2018, 7, 261. https://doi.org/10.3390/cells7120261
Sinenko SA, Skvortsova EV, Liskovykh MA, Ponomartsev SV, Kuzmin AA, Khudiakov AA, Malashicheva AB, Alenina N, Larionov V, Kouprina N, et al. Transfer of Synthetic Human Chromosome into Human Induced Pluripotent Stem Cells for Biomedical Applications. Cells. 2018; 7(12):261. https://doi.org/10.3390/cells7120261
Chicago/Turabian StyleSinenko, Sergey A., Elena V. Skvortsova, Mikhail A. Liskovykh, Sergey V. Ponomartsev, Andrey A. Kuzmin, Aleksandr A. Khudiakov, Anna B. Malashicheva, Natalia Alenina, Vladimir Larionov, Natalay Kouprina, and et al. 2018. "Transfer of Synthetic Human Chromosome into Human Induced Pluripotent Stem Cells for Biomedical Applications" Cells 7, no. 12: 261. https://doi.org/10.3390/cells7120261