Functional Metabolomics—A Useful Tool to Characterize Stress-Induced Metabolome Alterations Opening New Avenues towards Tailoring Food Crop Quality
Abstract
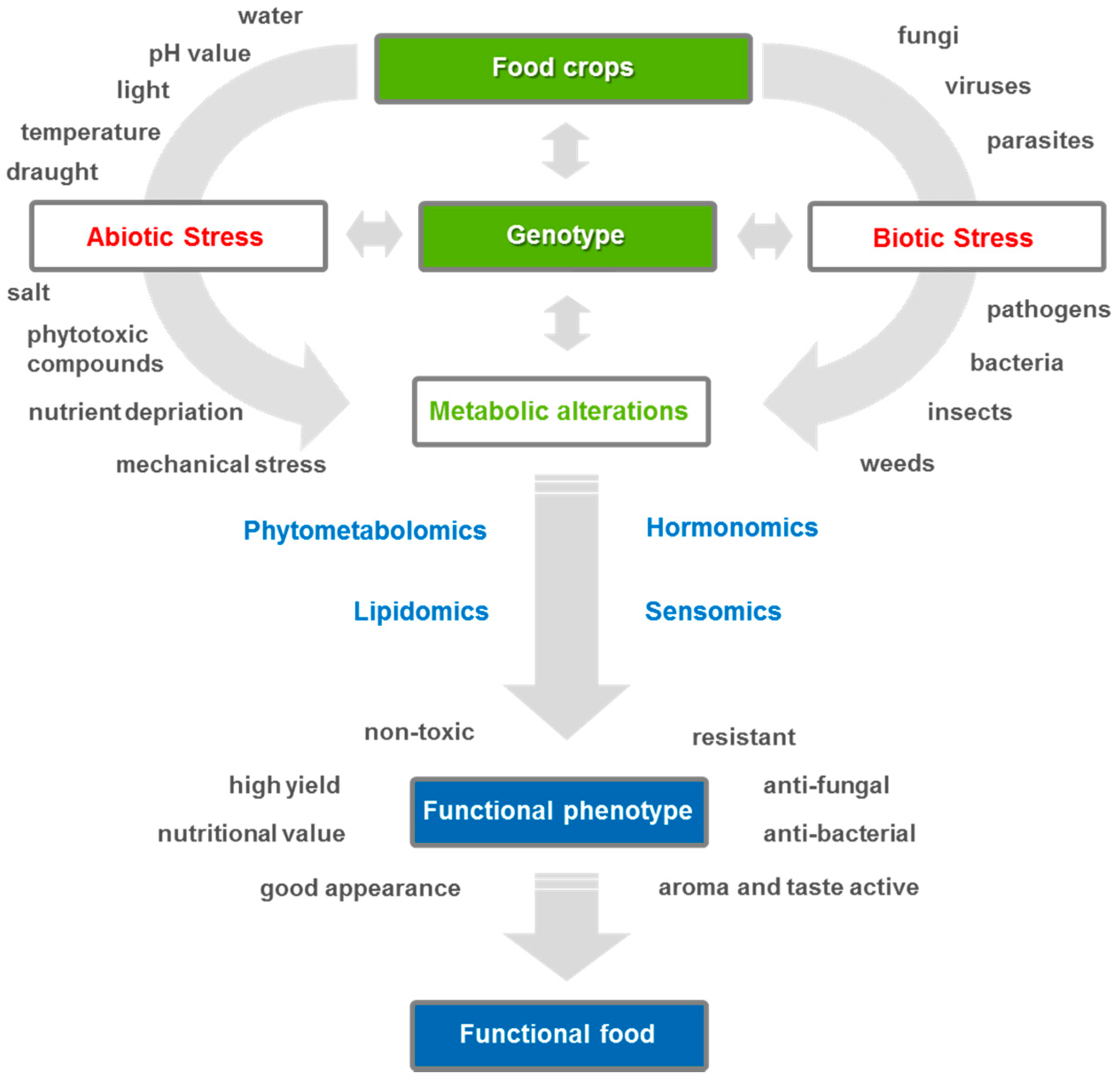
:1. Importance of Metabolomics for Agricultural Research
2. Analytical Techniques Used to Characterize Stress-Induced Metabolome Alterations in Plants
3. Phytometabolomics—From Plant Stress to Metabolic Response
3.1. From Abiotic Plant Stress to Metabolic Response
3.2. From Biotic Stress Metabolomics to Metabolic Response
3.3. Functional Phytometabolomics—Characterization Approach of Plant Stress Metabolites
4. Phytohormone Profiling by Means of Plant Hormonomics
5. Sensomics—A Phenotyping Tool to Characterize Crops Flavor Impression
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Börner, H. Pflanzenkrankheiten und Pflanzenschutz, 8th ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for Plant Improvement. Status and Prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Gang, D.R.; Charlton, A.J.; Fiehn, O.; Kuiper, H.A.; Reynolds, T.L.; Tjeerdema, R.S.; Jeffery, E.H.; German, J.B.; Ridley, W.P.; et al. Application of metabolomics in agriculture. J. Agric. Food Chem. 2006, 54, 8984–8994. [Google Scholar] [CrossRef] [PubMed]
- Arbona, V.; Manzi, M.; de Ollas, C.; Gómez-Cadenas, A. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Fernie, A. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Luca, V.; St Pierre, B. The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci. 2000, 5, 168–173. [Google Scholar] [CrossRef]
- D’Auria, J.C.; Greshenzon, J. The secondary metabolism of Arabidopsis thalina: Growing like a weed. Curr. Opin. Plant Biol. 2005, 8, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Davies, H. A role for “omics” technologies in food safety assessment. Food Control 2010, 21, 1601–1610. [Google Scholar] [CrossRef]
- Saito, K.; Matsuda, F. Metabolomics for funtional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 2010, 61, 463–489. [Google Scholar] [CrossRef] [PubMed]
- Cantu, D.; Govindarajulu, M.; Kozik, A.; Wang, M.; Chen, X.; Kojima, K.K.; Dubcovsky, J. Next generation sequencing provides rapid access to the genome of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS ONE 2011, 6, e24230. [Google Scholar]
- Schneeberger, K.; Weigel, D. Fast-forward genetics enabled by new sequencing technologies. Trends Plant Sci. 2011, 16, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Bino, R.J.; Hall, R.D.; Fiehn, O.; Kopka, J.; Saito, K.; Draper, J.; Nikolau, B.J.; Mendes, P.; Roessner-Tunali, U.; Beale, M.H.; et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004, 9, 1360–1385. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Lei, Z.; Nikolau, B.J.; Saito, K. Modern plant metabolomics. Advanced natural product gene discoveries, improved technologies, and future prospects. Nat. Prod. Rep. 2015, 32, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Keizer, L.C.; Bouwmeester, H.J.; Sun, Z.; Alvarez-Huerta, M.; Verhoeven, H.A.; Blaas, J.; van Houwelingen, A.M.; De Vos, R.C.; van der Voet, H.; et al. Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell. 2000, 12, 647–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carreno-Quintero, N.; Acharjee, A.; Maliepaard, C.; Bachem, C.W.B.; Mumm, R.; Bouwmeester, H.; Visser, R.G.F.; Keurentjes, J.J.B. Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol. 2012, 158, 1306–1318. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Long, Y.; Shi, L.; Shi, J.; Barker, G.; Meng, J. Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus. New Phytol. 2012, 193, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Keilwagen, J.; Lehnert, H.; Berner, T.; Budahn, H.; Mothnagel, T.; Ulrich, D.; Dunemann, F. The terpene synthase gene family of carrot (Daucus carota L.): Identification of candidate genes associated with terpenoid volatile compounds. Front. Plant Sci. 2017, 8, 1930. [Google Scholar] [CrossRef] [PubMed]
- Rambla, J.L.; Medina, A.; Fernández-Del-Carmen, A.; Barrantes, W.; Grandillo, S.; Cammareri, M.; López-Casado, G.; Rodrigo, G.; Alonso, A.; García-Martínez, S.; et al. Identification, introgression, and validation of fruit volatile QTLs from a red-fruited wild tomato species. J. Exp. Bot. 2017, 68, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Nakabayashi, R.; Yang, Z.; Okazaki, Y.; Yonemaru, J.; Ebana, K.; Yano, M.; Saito, K. Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant J. Cell Mol. Boil. 2015, 81, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for plant stress response. Physiol. Plant. 2008, 132, 199–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, F.; Kopka, J.; Haskell, D.W.; Zhao, W.; Schiller, K.C.; Gatzke, N.; Sung, D.Y.; Guy, C.L. Exploring the temperature-stress metabolom of Arabidopsis. Plant Physiol. 2004, 136, 4159–4168. [Google Scholar] [CrossRef] [PubMed]
- Urano, K.; Kurihara, Y.; Seki, M.; Shinozaki, K. ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr. Opin. Plant Biol. 2010, 13, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Urano, K.; Maruyama, K.; Ogata, Y.; Morishita, Y.; Takeda, M.; Sukarai, N.; Suzuki, H.; Saito, K.; Shibata, D.; Kobayashi, M.; et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009, 57, 1065–1078. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Metabolomics in Plant Stress Physiology. In Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Kushalappa, A.C.; Gunnaiah, R. Metabolo-proteomics to discover plant biotic stress resistance genes. Trends Plant Sci. 2013, 18, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.A.; Ricke, D.; Lan, T.H.; Presting, G.; Wang, R.L.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Hu, S.N.; Wang, J.; Wong, G.K.S.; Li, S.G.; Liu, B.; Deng, Y.J.; Dai, L.; Zhou, Y.; Zhang, X.Q.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [Green Version]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boue, S.M.; Cleveland, T.E.; Carter-Wientjes, C.; Shih, B.Y.; Bhatnagar, D.; McLachlan, J.M.; Burow, M.E. Phytoalexin-enriched functional foods. J. Agric. Food Chem. 2009, 57, 2614–2622. [Google Scholar] [CrossRef] [PubMed]
- Wüst, M. Smell of stress: Identification of induced biochemical pathways affecting the volatile composition and flavor quality of crops. J. Agric. Food Chem. 2018, 66, 3616–3618. [Google Scholar] [CrossRef] [PubMed]
- Blanksby, S.J.; Mitchell, T.W. Advances in mass spectrometry for lipidomics. Annu. Rev. Anal. Chem. 2010, 3, 433–465. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, R.; Frank, O.; Hofmann, T. Differential off-line LC-NMR (DOLC-NMR) metabolomics to monitor tyrosine-induced metabolome alterations in Saccharomyces cerevisiae. J. Agric. Food Chem. 2017, 65, 3230–3241. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted Metabolomics. Curr. Protoc. Mol. Biol. 2012, 30. [Google Scholar] [CrossRef] [PubMed]
- Desbrosses, G.; Steinhauser, D.; Kopka, J. Metabolom analysis using GC-MS. In Lotus Japonicus Handbook; Springer: Dordrecht, The Netherlands, 2005; pp. 165–174. [Google Scholar]
- Burton, L.; Ivosev, G.; Tate, S.; Impey, G.; Wingate, J.; Bonner, R. Instrumental and experimental effects in LC-MS-based metabolomics. J. Chromatogr. B 2008, 871, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Moco, S.; Bino, R.J.; Vorst, O.; Verhoeven, H.A.; de Groot, J.; van Beek, T.A.; Vervoort, J.; de Vos, C.H. A liquid-chromatography-mass spectrometry based metabolome database for tomato. Plant Physiol. 2006, 141, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- Frank, O.; Kreißl, J.K.; Daschner, A.; Hofmann, T. Accurate determination of reference materials and natural isolates by means of quantitative 1H NMR spectroscopy. J. Agric. Food Chem. 2014, 62, 2506–2515. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Mendes, P.; Dixon, R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics area. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef]
- Wang, D.; Bodowitz, S. Single cell analysis: The new fronzier in òmics. Trends Biotechnol. 2010, 28, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B.; Assmann, S.M.; Chen, S. Plant single-cell and single-cell-type metabolomics. Trends Plant Sci. 2014, 19, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Lange, B.M. Single cell genomics. Curr. Opin. Plant Biol. 2005, 8, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Lao, K.; Surani, M.A. Development and applications of single cell transcriptome analysis. Nat. Methods 2011, 8, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Chen, S. Single cell-type proteomics: Toward a holistic understanding of plant function. Mol. Cell. Proteom. 2012, 11, 1622–1630. [Google Scholar] [CrossRef] [PubMed]
- De Souza, L.P.; Naake, T.; Tohge, T.; Fernie, A.R. From chromatogram to analyte to metabolite. How to pick horses for courses from the massive web resources for mass spectral plant metabolomics. GigaScience 2017, 6, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mithöfer, A.; Boland, W. Do you speak chemistry. EMBO Rep. 2016, 17, 626–629. [Google Scholar] [CrossRef] [PubMed]
- Mithöfer, A.; Boland, W. Plant defense against herbivores. Chemical aspects. Annu. Rev. Plant Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, F.; Guy, C.L. β-Amylse induction and the protective role of maltose during temperature shock. Plant Physiol. 2004, 135, 1674–1684. [Google Scholar] [CrossRef] [PubMed]
- Tenenboim, H.; Burgos, A.; Willmitzer, L.; Brotman, Y. Using lipidomics for expanding the knowledge on lipid metabolism in plants. Biochimie 2016, 130, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Frankel, E.N. Lipid Oxidation, 2nd ed.; Woodhead Publishing in Food Science, Technology and Nutrition: Philadelphia, PA, USA, 2012. [Google Scholar]
- El-Hafid, L.; Pham, T.A.; Zuily-Fodil, Y.; Vieira da Silva, J. Enzymatic Breakdown of Polar Lipids in Cotton Leaves under Water Stress. 1. Degradation of Monogalactosyl-Diacylglycerol. Plant Physiology Biochemistry 1989. Available online: http://agris.fao.org/agris-search/search.do?recordID=FR9001726 (accessed on 29 June 2018).
- Hubac, C.; Guerrier, D.; Ferran, J.; Tremolieres, A. Change of leaf lipid composition during water stress in two genotypes of lupinus albus resistant or susceptible to drought. Plant Physiol. Biochem. 1989, 27, 737–744. [Google Scholar]
- Pham, T.A.T.; Vieira da Silva, J.; Mazliak, P. The role of membrane lipids in drought resistance of plants. Bulletin de la Société Botanique de France. Actual. Bot. 1990, 137, 99–114. [Google Scholar]
- Quartacci, M.F.; Pinzino, C.; Sgherri, C.L.; Navari-Izzo, F. Lipid composition and protein dynamics in thylakoids of two wheat cultivars differently sensitive to drought. Plant Physiol. 1995, 108, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Kaoua, M.; Serraj, R.; Benichou, M.; Hsissou, D. Comparative sensitivity of two Moroccan wheat varieties to water stress: The relationship between fatty acids and proline accumulation. Bot. Stud. 2006, 47, 51–60. [Google Scholar]
- Moradi, P.; Mahdavi, A.; Khoshkam, M.; Iriti, M. Lipidomics unravels the role of leaf lipids in thyme plant response to drought stress. Int. J. Mol. Sci. 2017, 18, 2067. [Google Scholar] [CrossRef] [PubMed]
- De Paula, F.M.; Thi, A.P.; de Silva, J.V.; Justin, A.; Demandre, C.; Mazliak, P. Effects of water stress on the molecular species composition of polar lipids from Vigna unguiculata L. Leaves. Plant Sci. 1990, 66, 185–193. [Google Scholar] [CrossRef]
- Hamrouni, I.; Salah, H.B.; Marzouk, B. Effects of water-deficit on lipids of safflower aerial parts. Phytochemistry 2001, 58, 277–280. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, Q.; Shen, W.; Cram, D.; Fowler, D.B.; Wie, Y. Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant Cell 2015, 27, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Köhl, K. Metabolomics on Combined Abiotic Stress Effects in Crops. In Drought Stress Tolerance in Plants; Hossain, M., Wani, S., Bhattacharjee, S., Burritt, D., Tran, L.S., Eds.; Springer: Cham, Switzerland, 2016; Volume 2. [Google Scholar]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Boil. 2015, 24, 10–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okazaki, Y.; Saito, K. Integrated metabolomics and phytochemical genomics approaches for studies on rice. GigaScience 2016, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G. Genetics of plant disease. In Pant Pathology, 5th ed.; Elsvier Academic Press: Cambridge, MA, USA, 2005; pp. 125–174. [Google Scholar]
- Aghnoum, R.; Marcel, T.C.; Johrde, A.; Pecchioni, N.; Schweizer, P.; Niks, R.E. Basal host resistance of barley to powdery mildew: Connecting quantitative trait loci and candidate genes. Mol. Plant Microbe Interact. 2010, 23, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Valdés-López, O.; Thibivilliers, S.; Qiu, J.; Xu, W.W.; Nguyen, T.H.; Libault, M.; Le, B.H.; Goldberg, R.B.; Hill, C.B.; Hartman, G.L.; et al. Identification of quantitative trait loci controlling gene expression during the innate immunity response of soybean. Plant Physiol. 2011, 157, 1975–1986. [Google Scholar] [CrossRef] [PubMed]
- Massman, J.; Cooper, B.; Horsley, R.; Neat, S.; Dill-Macky, R.; Chao, S.; Dong, Y.; Schwarz, P.; Muehlbauer, G.J.; Smith, K.P. Genome-wide association mapping of Fusarium head blight resistance in contemporary barley breeding germplasm. Mol. Breed. 2011, 27, 439–454. [Google Scholar] [CrossRef]
- Gunnaiah, R.; Kushalappa, A.C.; Duggavathi, R.; Fox, S.; Somers, D.J. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS ONE 2012, 7, e40695. [Google Scholar] [CrossRef] [PubMed]
- Bollina, V.; Kumaraswamy, G.K.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S.; Faubert, D.; Hamzehzarghani, H. Mass spectrometry-based metabolomics application to identify quantitative resistance-related metabolites in barley against Fusarium head blight. Mol. Plant Pathol. 2010, 11, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.R.; Lafuente, M.T.; de Vos, R.C.; Bovy, A.G.; González-Candelas, L. Citrus phenylpropanoids and defence against pathogens. Part I: Metabolic profiling in elicited fruits. Food Chem. 2013, 136, 178–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado, A.R.T.; Campos, V.A.C.; da Silva, W.J.R.; Campos, V.P.; de Mattos Zeri, A.C.; Olivera, D.F. Metabolic profiling in the roots of coffee plants exposed to the coffee root-knot nematode, Meloidogyne exigua. Eur. J. Plant Pathol. 2012, 134, 431–441. [Google Scholar] [CrossRef]
- Sana, T.R.; Fischer, S.; Wohlgemuth, G.; Katrekar, A.; Jung, K.H.; Ronald, P.C.; Fiehn, O. Metabolomic and transcriptomic analysis of the rice response to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae. Metabolomics 2010, 6, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Batovska, D.I.; Todorova, I.T.; Nedelcheva, D.V.; Parushev, S.P.; Atanassov, A.I.; Hvarleva, T.D.; Djakova, G.J.; Bankova, V.S.; Popov, S.S. Preliminary study on biomarkers for the fungal resistance in Vitis vinifera leaves. J. Plant Physiol. 2008, 165, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Suh, M.C.; Samuels, A.L.; Jetter, R.; Kunst, L.; Pollard, M.; Ohlrogge, J. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 2005, 139, 1649–1665. [Google Scholar] [CrossRef] [PubMed]
- Keymer, A.; Pimprikar, P.; Wewer, V.; Huber, C.; Brands, M.; Bucerius, S.L.; Delaux, P.M.; Klingl, V.; Röpenack-Lahaye, E.V.; Wang, T.L.; et al. Lipid transfer from plants to arbuscular mycorrhiza fungi. Elife 2017, 6, e29107. [Google Scholar] [CrossRef] [PubMed]
- Cellini, F.; Chesson, A.; Colquhoun, I.; Constable, A.; Davies, H.V.; Engel, K.H.; Gatehouse, A.M.R.; Kärenlami, S.; Kok, E.J.; Leguay, J.-J.; et al. Unintended effects and their detection in genetically modified crops. Food Chem. Toxicol. 2004, 24, 1089–1125. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R. Metabolomics—The new frontier in food safety and quality research. Food Res. Int. 2015, 72, 80–81. [Google Scholar] [CrossRef]
- Shephard, G.S. Current status of mycotoxin analysis: A critical review. J. AOAC Int. 2016, 99, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Taiz, L.; Zeiger, E. Plant Physiology, 5th ed.; Sinauer Associates: Sunderland, MA, USA, 2010. [Google Scholar]
- Novák, O.; Napier, R.; Ljung, K. Zooming in on plant hormone analysis: Tissue- and cell-specific approaches. Annu. Rev. Plant Biol. 2017, 68, 323–348. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.J. Plant Hormones: Biosynthesis, Signal Transduction, Action! 3rd ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Estelle, M. The ubiquitin—Proteasome system regulates plant hormone signaling. Plant J. 2010, 61, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Porfírio, S.; Gomes da Silva, M.D.R.; Peixe, A.; Cabrita, M.J.; Azadi, P. Current analytical methods for plant auxin quantification—A review. Anal. Chim. Acta 2016, 902, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowska, D.; Strnad, M.; Ljung, K.; Novak, O. Plant hormonomics: Multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Rhy, H.; Cho, Y. Plant hormons in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar]
- Ramos, L. Critical overview of selected contemporary sample preparation techniques. J. Chromatogr. A 2012, 1221, 84–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prinsen, E.; Van Dongen, W.; Esmans, E.L.; Van Onckelen, H.A. HPLC linked electrospray tandem mass spectrometry: A rapid and reliable method to analyse indole-3-acetic acid metabolism in bacteria. J. Mass Spectrom. 1997, 32, 12–22. [Google Scholar] [CrossRef]
- Waterval, J.; Lingeman, H.; Bult, A.; Underberg, W.J. Derivatization trends in capillary electrophoresis. Electrophoresis 2000, 21, 4029–4045. [Google Scholar] [CrossRef]
- Egging, M.; Wijtmans, M.; Ekkebus, R.; Lingeman, H.; de Esch, I.J.; Kool, J.; Niessen, W.M.A.; Irth, H. Development of a selective ESI-MS derivatization reagent: Synthesis and optimization for the analysis of aldehydes in biological mixtures. Anal. Chem. 2008, 80, 9042–9051. [Google Scholar] [CrossRef] [PubMed]
- International Food Information Council Foundation, Washington, D.C. 2016 Food and Health Survey. Available online: http://www.foodinsight.org/articles/2016-food-and-health-survey-food-decision-2016-impact-growing-national-food-dialogue (accessed on 3 December 2016).
- Dunkel, A.; Steinhaus, M.; Kotthoff, M.; Nowak, B.; Krautwurst, D.; Schieberle, P.; Hofmann, T. Nature’s chemical signatures in human olfaction: A foodborne perspective for future biotechnology. Angew. Chem. Int. Ed. Engl. 2014, 53, 7124–7143. [Google Scholar] [CrossRef] [PubMed]
- Schieberle, P.; Hofmann, T. Mapping the combinatorial code of food flavors by means of molecular sensory science approach. In Food Flavors—Chemical, Sensory and Technological Properties; Jelen, H., Ed.; CRC Press: Boca Raton, FL, USA, 2012; pp. 411–437. [Google Scholar]
- Li, J.-X.; Schieberle, P.; Steinhaus, M. Insights into the key compounds of durian (Durio zibethinus L. ‘Monthong’) pulp odor by odorant quantitation and aroma simulation experiments. J. Agric. Food Chem. 2016, 65, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Munafo, J.P.; Didzbalis, J.; Schnell, R.J.; Steinhaus, M. Insights into the key aroma compounds in mango (Mangifera indica L. ‘Haden’) fruits by stable isotope dilution quantitation and aroma simulation experiments. J. Agric. Food Chem. 2016, 64, 4312–4318. [Google Scholar] [CrossRef] [PubMed]
- Czepa, A.; Hofmann, T. Quantitative studies and sensory analyses on the influence of cultivar, spatial tissue distribution, and industrial processing on the bitter Off-taste of carrots (Daucus carota L.) and carrot products. J. Agric. Food Chem. 2004, 52, 4508–4514. [Google Scholar] [CrossRef] [PubMed]
- Schmiech, L.; Uemra, D.; Hofmann, T. Reinvestigation of the bitter compounds in carrots (Daucus carota L.) by using a molecular sensory science approach. J. Agric. Food Chem. 2008, 56, 10252–10260. [Google Scholar] [CrossRef] [PubMed]
- Stark, T.; Hofmann, T. Isolation, structure determination, synthesis, and sensory activity of N-phenylpropenoyl-l-amino acids from cocoa (Theobroma cacao). J. Agric. Food Chem. 2005, 53, 5419–5428. [Google Scholar] [CrossRef] [PubMed]
- Dawid, C.; Hofmann, T. Structural and sensory characterization of bitter tasting steroidal saponins from asparagus spears (Asparagus officinalis L.). J. Agric. Food Chem. 2012, 60, 11889–11900. [Google Scholar] [CrossRef] [PubMed]
- Dawid, C.; Hofmann, T. Identification of sensory-active phytochemicals in asparagus (Asparagus officinalis L.). J. Agric. Food Chem. 2012, 60, 11877–11888. [Google Scholar] [CrossRef] [PubMed]
- Dawid, C.; Hofmann, T. Quantitation and bitter taste contribution of saponins in fresh and cooked white asparagus (Asparagus officinalis L.). Food Chem. 2013, 145, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Dawid, C.; Henze, A.; Frank, O.; Glabasnia, A.; Rupp, M.; Buening, K.; Orlikowski, D.; Bader, M.; Hofmann, T. Structural and sensory characterization of key pungent and tingling compounds from black pepper (Piper nigrum L.). J. Agric. Food Chem. 2012, 60, 2884–2895. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, B.; Hofmann, T. Sensory-Guided Decomposition of Red Current Juice (Ribes rubrum) and Structure Determination of Key Astringent Compounds. J. Agric. Food Chem. 2007, 55, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Scharbert, S.; Holzmann, N.; Hofmann, T. Identification of the astringent taste compounds in black tea infusions by combining instrumental analysis and human bioresponse. J. Agric. Food Chem. 2004, 52, 3498–3508. [Google Scholar] [CrossRef] [PubMed]
- Hellfritsch, C.; Brockhoff, A.; Stähler, F.; Meyerhof, W.; Hofmann, T. Human psychometric and taste receptor responses to steviol glycosides. J. Agric. Food Chem. 2012, 60, 6782–6793. [Google Scholar] [CrossRef] [PubMed]
- Brock, A.; Hofmann, T. Identificatioion of the Key Astringent Compounds in Spinach (Spinacia oleracea) by Means of the Taste Dilution Analysis. Chem. Percept. 2008, 1, 268–281. [Google Scholar] [CrossRef]
- Hofmann, T.; Krautwurst, D.; Schieberle, P. Current status and future perspectives in flavor research: Highlights of the 11th Wartburg Symposium on flavor chemistry & biology. J. Agric. Food Chem. 2018, 66, 2197–2203. [Google Scholar] [PubMed]
- Harding, V.K.; Heale, J.B. The accumulation of inhibitory compounds in the induced resistance response of carrot root slices to Botrytis cinerea. Physiol. Plant Pathol. 1981, 18, 7–15. [Google Scholar] [CrossRef]
- Lund, E.D.; White, J.M. Polyacetylenes in Normal and waterstressed ‘Orlando Gold’ carrots (Daucus carota). J. Sci. Food Agric. 1990, 51, 507–516. [Google Scholar] [CrossRef]
- Olsson, K.; Svensson, R. The influence of polyacetylenes on the susceptibility of carrots to storage diseases. J. Phytopathol. Phytopathol. Z. 1996, 144, 441–447. [Google Scholar] [CrossRef]
- Kreutzmann, S.; Christensen, L.P.; Edelenbos, M. Investigation of bitterness in carrots (Daucus carota L.) based on quantitative chemical and sensory analyses. LWT Food Sci. Technol. 2008, 41, 193–205. [Google Scholar] [CrossRef]
- Kreutzmann, S.; Svensson, V.T.; Thybo, A.K.; Bro, R.; Petersen, M.A. Prediction of sensory quality in raw carrots (Daucus carota L.) using multi-block LS-ParPLS. Food Qual. Preference 2008, 19, 609–617. [Google Scholar] [CrossRef]
- Kidmose, U.; Hansen, S.L.; Christensen, L.P.; Edelenbos, M.; Larsen, E.; Nørbæk, R. Effects of genotype, root size, storage, and processing on bioactive compounds in organically grown carrots (Daucus carota L.). J. Food Sci. 2004, 69, S388–S394. [Google Scholar] [CrossRef]
- Lund, E.D.; Bruemmer, J.H. Acetylenic compounds in stored packaged carrots. J. Sci. Food Agric. 1991, 54, 287–294. [Google Scholar] [CrossRef]
- Singldinger, B.; Dunkel, A.; Bahmann, D.; Bahmann, C.; Kadow, D.; Bisping, B.; Hofmann, T. New taste-active 3-(O-β-d-glucosyl)-2-oxoindole-3-acetic acids and diarylheptanoids in Cimiciato-infected hazelnuts. J. Agric. Food Chem. 2018, 66, 4660–4673. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Liu, X.; Zhou, Y.; Wang, X.; Zeng, L.; Fu, X.; Li, J.; Tang, J.; Dong, F.; Yang, Z. Formation and emission of linalool in tea (Camellia sinensis) leaves infested by tea green leafhopper (Empoasca (Matsumurasca) onukii Matsuda). Food Chem. 2017, 237, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zeng, L.; Liu, X.; Gui, J.; Mei, X.; Fu, X.; Dong, F.; Tang, J.; Zhang, L.; Yang, Z. Formation of (E)-nerolidol in tea (Camellia sinensis) leaves exposed to multiple stresses during tea manufacturing. Food Chem. 2017, 231, 78–86. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dawid, C.; Hille, K. Functional Metabolomics—A Useful Tool to Characterize Stress-Induced Metabolome Alterations Opening New Avenues towards Tailoring Food Crop Quality. Agronomy 2018, 8, 138. https://doi.org/10.3390/agronomy8080138
Dawid C, Hille K. Functional Metabolomics—A Useful Tool to Characterize Stress-Induced Metabolome Alterations Opening New Avenues towards Tailoring Food Crop Quality. Agronomy. 2018; 8(8):138. https://doi.org/10.3390/agronomy8080138
Chicago/Turabian StyleDawid, Corinna, and Karina Hille. 2018. "Functional Metabolomics—A Useful Tool to Characterize Stress-Induced Metabolome Alterations Opening New Avenues towards Tailoring Food Crop Quality" Agronomy 8, no. 8: 138. https://doi.org/10.3390/agronomy8080138




