Generation of Transgenic Rootstock Plum ((Prunus pumila L. × P. salicina Lindl.) × (P. cerasifera Ehrh.)) Using Hairpin-RNA Construct for Resistance to the Plum pox virus
Abstract
:1. Introduction
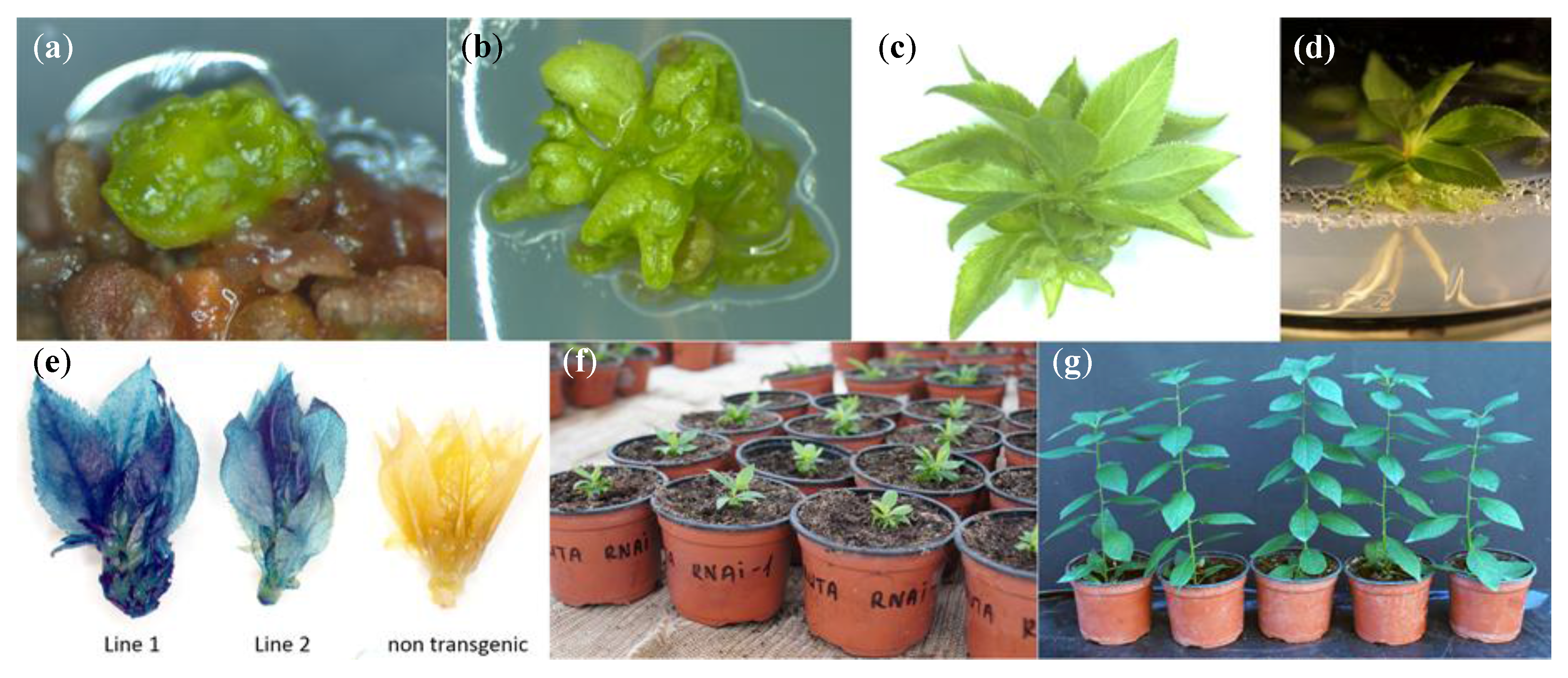
2. Results and Discussion
3. Materials and Methods
3.1. Plant Material
3.2. Vector
3.3. Agrobacterium-Mediated Transformation and Plant Regeneration
3.4. Histochemical GUS Assay
3.5. PCR Analysis and Southern Hybridization
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Scorza, R.; Callahan, A.; Dardick, C.; Ravelonandro, M.; Polak, J.; Malinowski, T.; Zagrai, I.; Cambra, M.; Kamenova, I. Genetic engineering of Plum pox virus resistance: ‘HoneySweet’ plum—From concept to product. Plant Cell Tissue Organ Cult. 2013, 115, 1–12. [Google Scholar] [CrossRef]
- Llacer, G.; Cambra., M. Hosts and symptoms of Plum pox virus: Fruiting Prunus species. EPPO Bull. 2006, 36, 219–221. [Google Scholar] [CrossRef]
- Cambra, M.; Capote, N.; Myrta, A.; Llacer, G. Plum pox virus and the estimated costs associated with sharka disease. Bull. OEPP/EPPO Bull. 2006, 36, 202–204. [Google Scholar] [CrossRef]
- Ilardi, V.; Tavazza, M. Biotechnological strategies and tools for Plum pox virusresistance: Trans-, intra-, cis-genesis, and beyond. Front. Plant Sci. 2015, 6, 379. [Google Scholar] [CrossRef]
- Rubio, M.; Pascal, T.; Bachellez, A.; Lambert, P. Quantitative trait loci analysis of Plum pox virus resistance in Prunus davidiana P1908: New insights on the organization of genomic resistance regions. Tree Genet. Genomes 2010, 6, 291–304. [Google Scholar] [CrossRef]
- Marandel, G.; Salava, J.; Abbott, A.; Candresse, T.; Decroocq, V. Quantitative trait loci meta-analysis of Plum pox virus resistance in apricot (Prunus armeniaca L.): New insights on the organization and the identification of genomic resistance factors. Mol. Plant Pathol. 2009, 10, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, W.; Neumuller, M. “Plum Breeding”, in Breeding Plantation Tree Crops: Temperate Species; Mohan, J.S., Priyadarshan, P.M., Eds.; Springer: New York, NY, USA, 2009; pp. 161–231. [Google Scholar]
- Scorza, R.; Ravelonandro, M.; Callahan, A.M.; Cordts, J.M.; Fuchs, M.; Dunez, J.; Gonsalves, D. Transgenic plum (Prunus domestica L.) express the Plum pox virus coat protein gene. Plant Cell Rep. 1994, 14, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Ravelonandro, M.; Scorza, R.; Bachelier, J.C.; Labonne, G.; Levy, L.; Damsteegt, V.; Callahan, A.M.; Dunez, J. Resistance of transgenic Prunus domestica to plum pox virus infection. Plant Dis. 1997, 81, 1231–1235. [Google Scholar] [CrossRef]
- Wang, A.; Tian, L.; Brown, D.C.W.; Svircev, A.M.; Stobbs, L.W.; Sanfaçon, H. Generation of efficient resistance to Plum pox virus (PPV) in Nicotiana benthamiana and Prunus domestica expressing triple-intron-spanned double-hairpin RNAs simultaneously targeting 5′ and 3′ conserved genomic regions of PPV. Acta Hortic. 2013, 1063, 77–84. [Google Scholar] [CrossRef]
- Ravelonandro, M.; Scorza, R.; Hily, J.M.; Briard, P. The efficiency of RNA interference for conferring stable resistance to Plum pox virus. Plant Cell Tissue Organ Cult. 2014, 118, 347–356. [Google Scholar] [CrossRef]
- Mikhailov, R.V.; Dolgov, S.V. Agrobacterium-mediated transformation of plum cv. Startovaya by selfcomplementary hairpin RNA of PPV-CP gene. Acta Hortic. 2011, 941, 85–90. [Google Scholar] [CrossRef]
- Sidorova, T.N.; Vagapova, T.I.; Dolgov, S.V. Intron-hairpin-RNA construct provides stable resistance to Plum pox virus in plum cultivar ‘Startovaja’. Acta Hortic. 2016, 1110, 197–202. [Google Scholar] [CrossRef]
- Lemgo, G.N.; Sabbadini, S.; Pandolfini, T.; Mezzetti, B. Biosafety considerations of RNAi-mediated virus resistance in fruit-tree cultivars and in rootstock. Transgenic Res. 2013, 22, 1073–1088. [Google Scholar] [CrossRef] [PubMed]
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 2017, 8, 1418. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Song, G.Q. Rootstock-to-scion transfer of transgene-derived small interfering RNAs and their effect on virus resistance in nontransgenic sweet cherry. Plant Biotechnol. J. 2014, 12, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Flachowsky, H.; Trankner, C.; Szankowski, I.; Waidmann, S.; Hanke, M.; Treutter, D.; Fischer, C.T. RNA-mediated gene silencing signals are not graft transmissible from the rootstock to the scion in greenhouse-grown apple plants malus spp. Int. J. Mol. Sci. 2012, 13, 9992–10009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padilla, I.M.G.; Burgos, L. Aminoglycoside antibiotics: Structure, functions and effects on in vitro plant culture and genetic transformation protocols. Plant Cell Rep. 2010, 29, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Sidorova, T.; Mikhailov, R.; Pushin, A.; Miroshnichenko, D.; Dolgov, S. A non-antibiotic selection strategy uses the phosphomannose-isomerase (PMI) gene and green fluorescent protein (GFP) gene for Agrobacterium-mediated transformation of Prunus domestica L. leaf explants. Plant Cell Tissue Organ Cult. 2017, 128, 197–209. [Google Scholar] [CrossRef]
- Gambino, G.; Gribaudo, I. Genetic transformation of fruit trees: Current status and remaining challenges. Transgenic Res. 2012, 21, 1163–1181. [Google Scholar] [CrossRef] [PubMed]
- Dolgov, S.; Mikhaylov, R.; Serova, T.; Shulga, O.; Firsov, A. Pathogen-derived methods for improving resistance of transgenic plums (Prunus domestica L.) for Plum pox virus infection. Julius Kühn Arch. 2010, 427, 133–140. [Google Scholar]
- Padilla, I.M.; Golis, A.; Gentile, A.; Damiano, C.; Scorza, R. Evaluation of transformation in peach Prunus persica explants using green fluorescent protein (GFP) and beta-glucuronidase (GUS) reporter genes. Plant Cell Tissue Organ Cult. 2006, 84, 309–314. [Google Scholar] [CrossRef]
- Ramesh, S.A.; Kaiser, B.N.; Franks, T.; Collins, G.; Sedgley, M. Improved methods in Agrobacterium-mediated transformation of almond using positive (mannose/pmi) or negative (kanamycin resistance) selection-based protocols. Plant Cell Rep. 2006, 25, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Magnusson, V.; Johnson, C. Agrobacterium-Mediated Transformation of Chokecherry (Prunus virginiana L.). HortScience 2007, 42, 140–142. [Google Scholar]
- Petri, C.; Wang, H.; Alburquerque, N.; Faize, M.; Burgos, L. Agrobacterium-mediated transformation of apricot (Prunus armeniaca L.) leaf explants. Plant Cell Rep. 2008, 27, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Pijut, P.M. Agrobacterium-mediated transformation of mature Prunus serotina (black cherry) and regeneration of transgenic shoots. Plant Cell Tissue Organ Cult. 2010, 101, 49–57. [Google Scholar] [CrossRef]
- Urtubia, C.; Devia, J.; Castro, Á.; Zamora, P.; Aguirre, C.; Tapia, E.; Barba, P.; Dell’Orto, P.; Moynihan, M.R.; Petri, C.; et al. Agrobacterium-mediated genetic transformation of Prunus salicina. Plant Cell Rep. 2008, 27, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Canli, F.A.; Wang, X.; Sibbald, S. Genetic transformation of Prunus domestica L. using the hpt gene coding for hygromycin resistance as the selectable marker. Sci. Hortic. 2009, 119, 339–343. [Google Scholar] [CrossRef]
- Lazo, G.R.; Stein, P.A.; Ludwig, R.A. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Nat. Biotechnol. 1991, 9, 963–967. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth bio assays with tobacco tissue culture. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Jacoboni, A.; Standardi, A. La moltiplicazione “in vitro” del melo cv. Wellspur. Riv. Ortoflorofruttic. Ital. 1982, 66, 217–229. [Google Scholar]
- Jefferson, R.A. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 1987, 5, 387–405. [Google Scholar] [CrossRef]
- Rogers, S.; Bendich, A. Extraction of total cellular DNA from plants, algae and fungi. In Plant Molecular Biology Manual; Gelvin, S., Schiperoort, R., Eds.; Springer: Dordrecht, The Netherlands, 1994; Section 7-1; pp. 183–190. [Google Scholar]


| Experiment Event | Number of Explants | Number of Explants Produced Shoot Clusters | Number of PCR Positive Lines | Transformation Rate, % |
|---|---|---|---|---|
| 1 | 113 | 1 | 0 | 0.0 |
| 2 | 148 | 2 | 1 | 0.7 |
| 3 | 125 | 2 | 1 | 0.8 |
| total | 386 | 5 | 2 | 0.5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sidorova, T.; Pushin, A.; Miroshnichenko, D.; Dolgov, S. Generation of Transgenic Rootstock Plum ((Prunus pumila L. × P. salicina Lindl.) × (P. cerasifera Ehrh.)) Using Hairpin-RNA Construct for Resistance to the Plum pox virus. Agronomy 2018, 8, 2. https://doi.org/10.3390/agronomy8010002
Sidorova T, Pushin A, Miroshnichenko D, Dolgov S. Generation of Transgenic Rootstock Plum ((Prunus pumila L. × P. salicina Lindl.) × (P. cerasifera Ehrh.)) Using Hairpin-RNA Construct for Resistance to the Plum pox virus. Agronomy. 2018; 8(1):2. https://doi.org/10.3390/agronomy8010002
Chicago/Turabian StyleSidorova, Tatiana, Alexander Pushin, Dmitry Miroshnichenko, and Sergey Dolgov. 2018. "Generation of Transgenic Rootstock Plum ((Prunus pumila L. × P. salicina Lindl.) × (P. cerasifera Ehrh.)) Using Hairpin-RNA Construct for Resistance to the Plum pox virus" Agronomy 8, no. 1: 2. https://doi.org/10.3390/agronomy8010002




