Assessing Genotype-By-Environment Interactions in Aspergillus Ear Rot and Pre-Harvest Aflatoxin Accumulation in Maize Inbred Lines
Abstract
:1. Introduction
2. Results
2.1. Aspergillus Ear Rot Severity
2.2. Aspergillus Flavus Colonization
2.3. Aflatoxin Accumulation in Maize Inbred Lines
3. Discussion
4. Materials and Methods
4.1. Germplasm and Field Experimental Design
4.2. Field Locations
4.3. Inoculum Production and Field Inoculation
4.4. Assessment of Aspergillus Ear Rot (AER) Rating
4.5. Quantification of Aspergillus Flavus in Maize Grain
4.6. Toxin Analysis
4.7. Data Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Okoth, S. Improving the Evidence Base on Aflatoxin Contamination and Exposure in Africa; Agriculture and Nutrition Series, Working Paper 16/13; The Technical Centre for Agricultural and Rural Cooperation (CTA): Wageningen, The Netherlands, 2016; pp. 1–113. Available online: http://aflatoxinpartnership.org/uploads/CTA_PACA_Aflatoxin_Final_for%20publlication.pdf (accessed on 19 August 2017).
- Alim, M.; Iqbal, S.Z.; Selamat, J.; Ariño, A. Regulations for Food Toxins. In Food Safety; Selamat, J., Iqbal, S.Z., Eds.; Springer: Zurich, Switzerland, 2016; pp. 33–39. [Google Scholar]
- Atehnkeng, J.; Ojiambo, P.S.; Cotty, P.J.; Bandyopadhyay, R. Field efficacy of a mixture of atoxigenic Aspergillus flavus Link: Fr vegetative compatibility groups in preventing aflatoxin contamination in maize (Zea mays L.). Biol. Control 2014, 72, 62–70. [Google Scholar]
- Yu, J. Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 2012, 4, 1024–1057. [Google Scholar]
- Mohammadi, R.; Amri, A. Genotype x Environment Interaction Implication: A Case Study of Durum Wheat Breeding in Iran. In Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits; Al-Khayri, J., Jain, S., Johnson, D., Eds.; Springer: Cham, Switzerland, 2016; pp. 515–558. [Google Scholar]
- Dhakal, R.; Windham, G.L.; Williams, W.P.; Subudhi, P.K. Quantitative trait loci (QTL) for reducing aflatoxin accumulation in corn. Mol. Breed. 2016, 36, 164. [Google Scholar]
- Mylroie, J.E.; Warburton, M.L.; Wilkinson, J.R. Development of a gene-based marker correlated to reduced aflatoxin accumulation in maize. Euphytica 2013, 194, 431–441. [Google Scholar]
- Rose, L.J.; Okoth, S.; Beukes, I.; Mouton, M.; Flett, B.C.; Makumbi, D.; Viljoen, A. Determining resistance to Fusarium verticillioides and fumonisin accumulation in maize inbred lines resistant to Aspergillus flavus and aflatoxins. Euphytica 2017, 213, 93. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists and Agronomists; CRC Press: Boca Raton, FL, USA, 2003; p. 271. [Google Scholar]
- Shu, X.; Livingston, D.P.; Franks, R.G.; Boston, R.S.; Woloshuk, C.P.; Payne, G.A. Tissue-specific gene expression in maize seeds during colonization by Aspergillus flavus and Fusarium verticillioides. Mol. Plant Pathol. 2015, 16, 662–674. [Google Scholar] [PubMed]
- Hintsa, G.H.; Fetien, A. AMMI and GGE biplot analysis of bread wheat genotypes in the Northern part of Ethiopia. J. Plant Breed. Genet. 2013, 1, 12–18. [Google Scholar]
- Ayahlneh, T.; Tesfaye, L.; Mohammed, A. Assessment of stability, adaptability and yield Performance of bread wheat (Triticum aestivum L.) Cultivars in South Eastern Ethiopia. Am.-Eurasian J. Agric. Environ. Sci. 2013, 13, 885–890. [Google Scholar]
- Tamene, T.T.; Gemechu, K.; Tadesse, S.; Musa, J.; Yeneneh, B. GXE interaction and performance stability for grain yield in field pea (Pisum sativum L.) genotypes. Int. J. Plant Breed. 2013, 7, 116–123. [Google Scholar]
- Mideros, S.X.; Warburton, M.L.; Jamann, T.M.; Windham, G.L.; Williams, W.P.; Nelson, R.J. Quantitative trait loci for resistance to Aspergillus ear rot: Analysis by linkage mapping, characterization of near-isogenic lines and meta-analysis. Crop Sci. 2014, 54, 127–142. [Google Scholar]
- Yan, W. GGE biplot—A windows application for graphical analysis of multi-environment trial data and other types of two-way data. Agron. J. 2001, 93, 1111–1118. [Google Scholar] [CrossRef]
- Brown, R.L.; Williams, W.P.; Windham, G.L.; Menkir, A.; Chen, Z.Y. Evaluation of African-bred maize germplasm lines for resistance to aflatoxin accumulation. Agronomy 2016, 6, 24. [Google Scholar] [CrossRef]
- Henry, W.B.; Blanco, M.H.; Rowe, D.E.; Windham, G.L.; Murray, S.C.; Williams, W.P. Diallel analysis of diverse maize germplasm lines for agronomic characteristics. Crop Sci. 2014, 54, 2547–2556. [Google Scholar] [CrossRef]
- Williams, W.P.; Windham, G.L. Aflatoxin accumulation in a maize diallel cross. Agriculture 2015, 5, 344–352. [Google Scholar] [CrossRef]
- Lauren, L.; Onsongo, M.; Njapau, H.; Schurz-Rogers, H.; Luber, G.; Kieszak, S.; Nyamongo, J.; Backer, L.; Dahiye, M.; Misore, A.; et al. Aflatoxin Contamination of Commercial Maize Products during an Outbreak of Acute Aflatoxicosis in Eastern and Central Kenya. Environ. Health Perspect. 2005, 113, 1763–1767. [Google Scholar]
- Okoth, S.; Nyongesa, B.; Ayugi, V.; Kang’ethe, E.; Korhonen, H.; Joutsjoki, V. Toxigenic potential of Aspergillus species occurring on maize kernels from two agro-ecological zones in Kenya. Toxins 2012, 4, 991–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okoth, S.; Rose, L.J.; Ouko, A.; Beukes, I.; Mouton, M.; Flett, B.C.; Makumbi, D.; Viljoen, A. Field evaluation of resitance to aflatoxin accumulation inmaize inbred lines in Kenya and South Africa. J. Crop Improv. 2017, 31, 862–878. [Google Scholar] [CrossRef]
- Zummo, N.; Scott, G.E. Evaluation of field inoculation techniques for screening maize genotypes against kernel infection by Aspergillus flavus in Mississippi. Plant Dis. 1989, 73, 313–316. [Google Scholar] [CrossRef]
- Henry, W.B.; Williams, W.P.; Windham, G.L.; Hawkins, L.K. Evaluation of maize inbred lines for resistance to Aspergillus and Fusarium ear rot and mycotoxin accumulation. Agron. J. 2009, 101, 1219–1226. [Google Scholar] [CrossRef]
- Boutigny, A.-L.; Beukes, I.; Small, I.; Zühlke, S.; Spiteller, M.; Van Rensburg, B.J.; Flett, B.; Viljoen, A. Quantitative detection of Fusarium pathogens and their mycotoxins in South Africa. Plant Pathol. 2012, 61, 522–531. [Google Scholar] [CrossRef]
- Rose, L.J.; Mouton, M.; Beukes, I.; Flett, B.C.; van der Vyver, C.; Viljoen, A. Multi-environment evaluation of maize inbred lines for resistance to Fusarium ear rot and fumonisins. Plant Dis. 2016, 100, 2134–2144. [Google Scholar] [CrossRef]
- Gauche, H.G.; Zobel, R.W. AMMI analysis of yield trials. In Genotype by Environment Interaction; Kang, M.S., Gauche, H.G., Eds.; CRC Press: Boca Raton, FL, USA, 1996; pp. 85–122. [Google Scholar]
- Payne, R.; Murray, D.A.; Harding, S.A.; Baird, D.B.; Soutar, D.M. An Introduction to GenStat for Windows, 15th ed.; VSN International: Hemel Hempstead, UK, 2012; p. 262. [Google Scholar]
- Yan, W.; Hunt, L.A.; Sheng, Q.; Szlavnics, Z. Cultivar evaluation and mega-environment investigation based on the GGE Biplot. Crop Sci. 2000, 40, 597–605. [Google Scholar] [CrossRef]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef]
- Yan, W.; Rajcan, I. Biplot analysis of test sites and trait relations of soybean in Ontario. Crop Sci. 2002, 42, 11–20. [Google Scholar] [CrossRef] [PubMed]
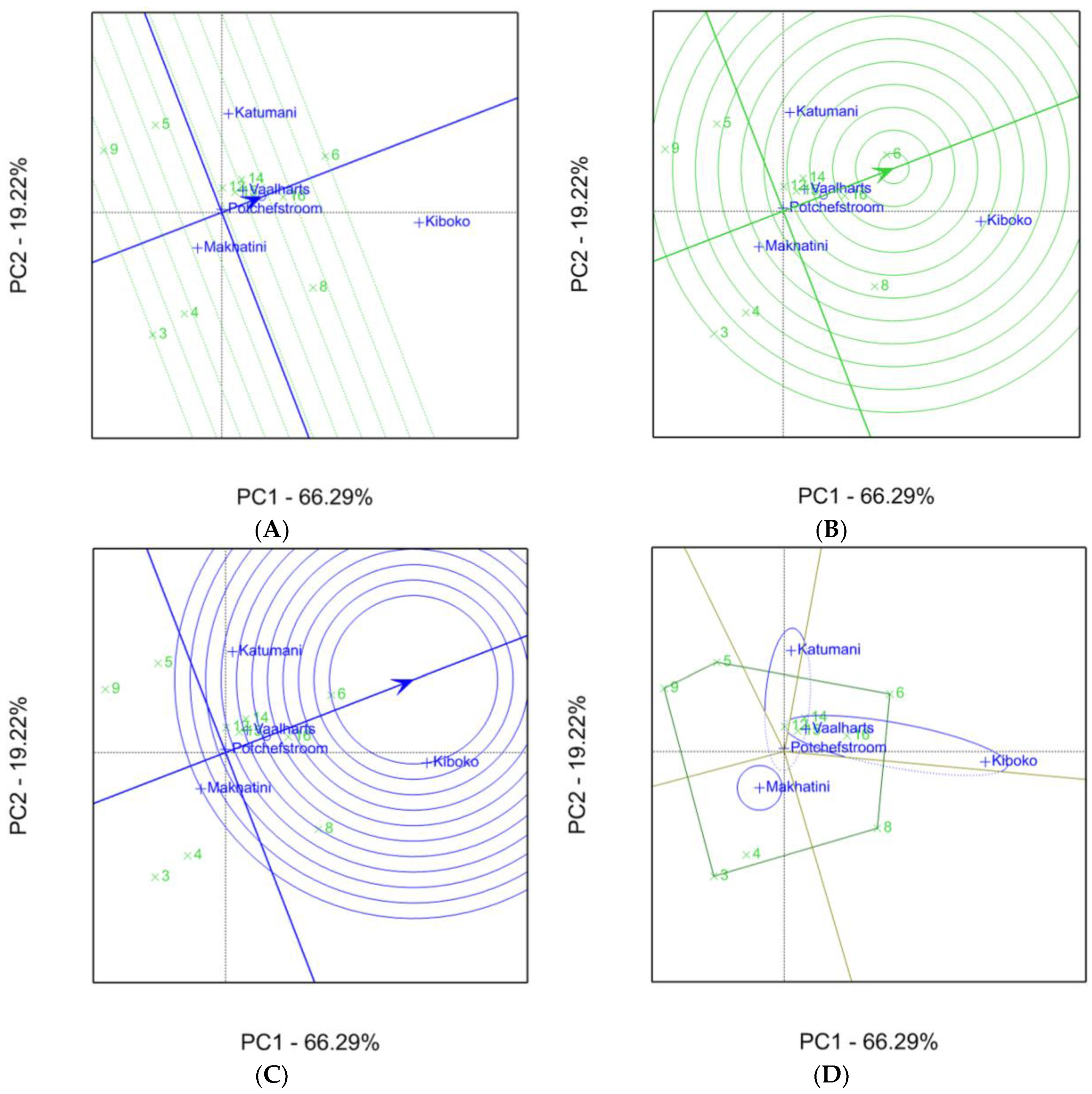
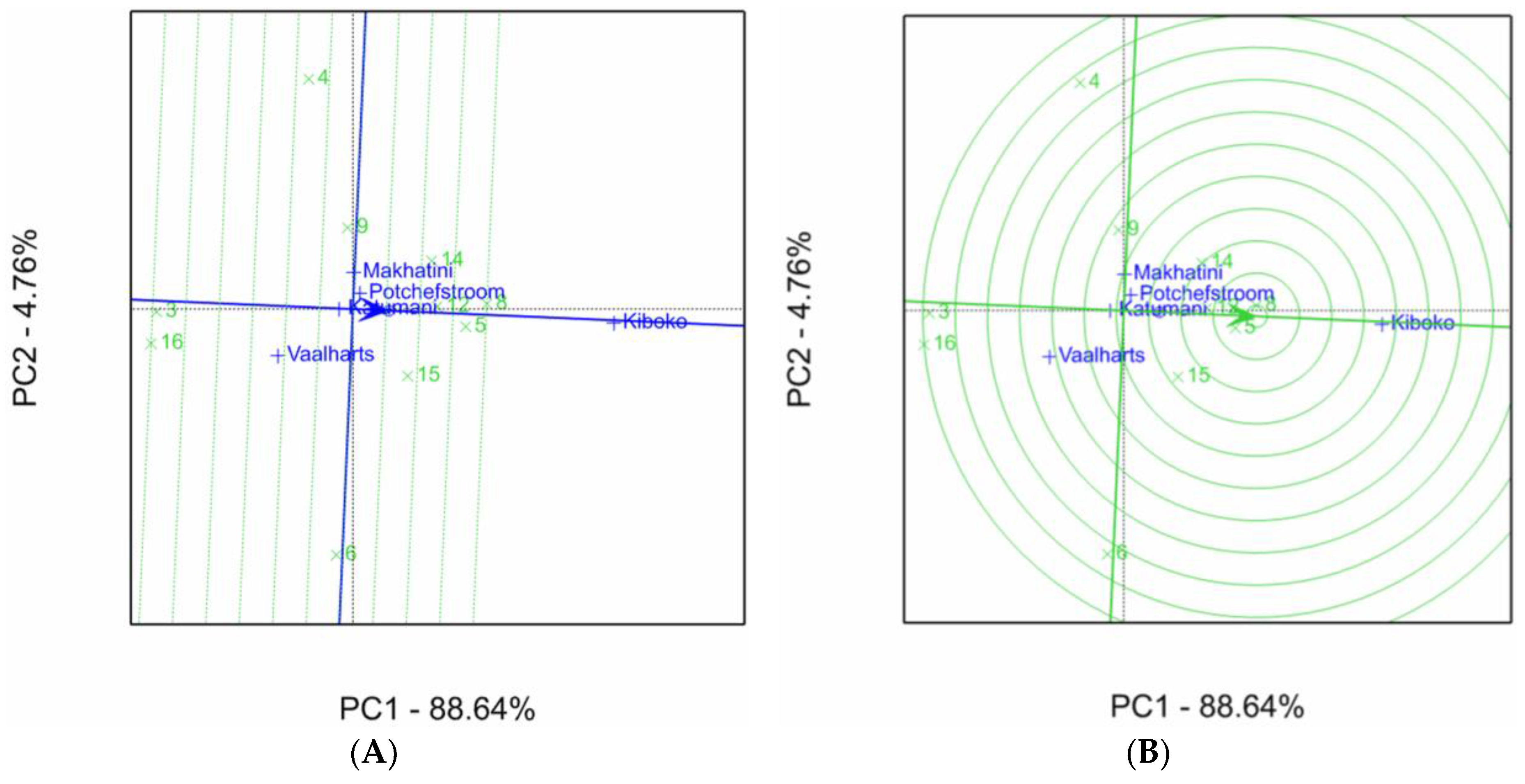
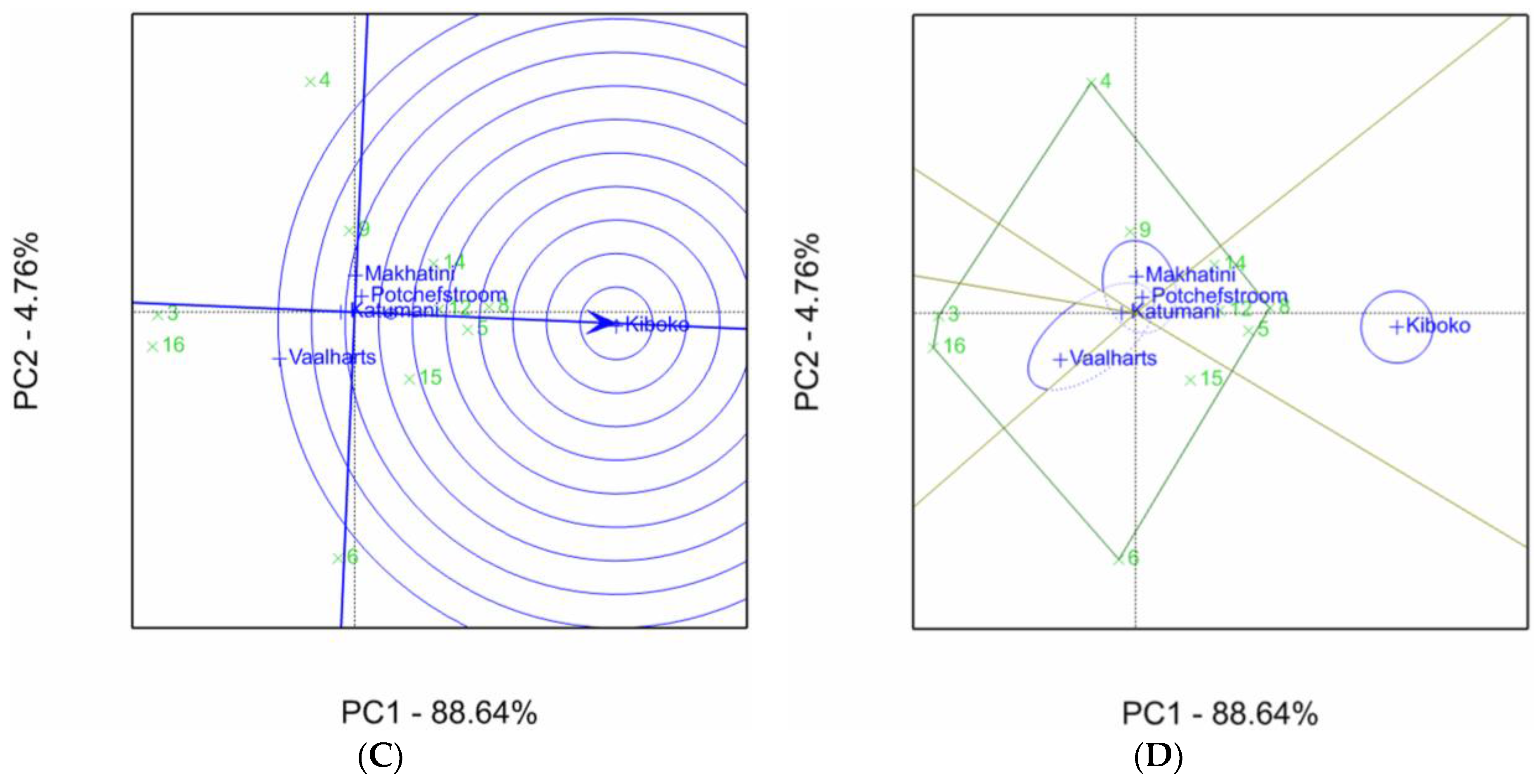
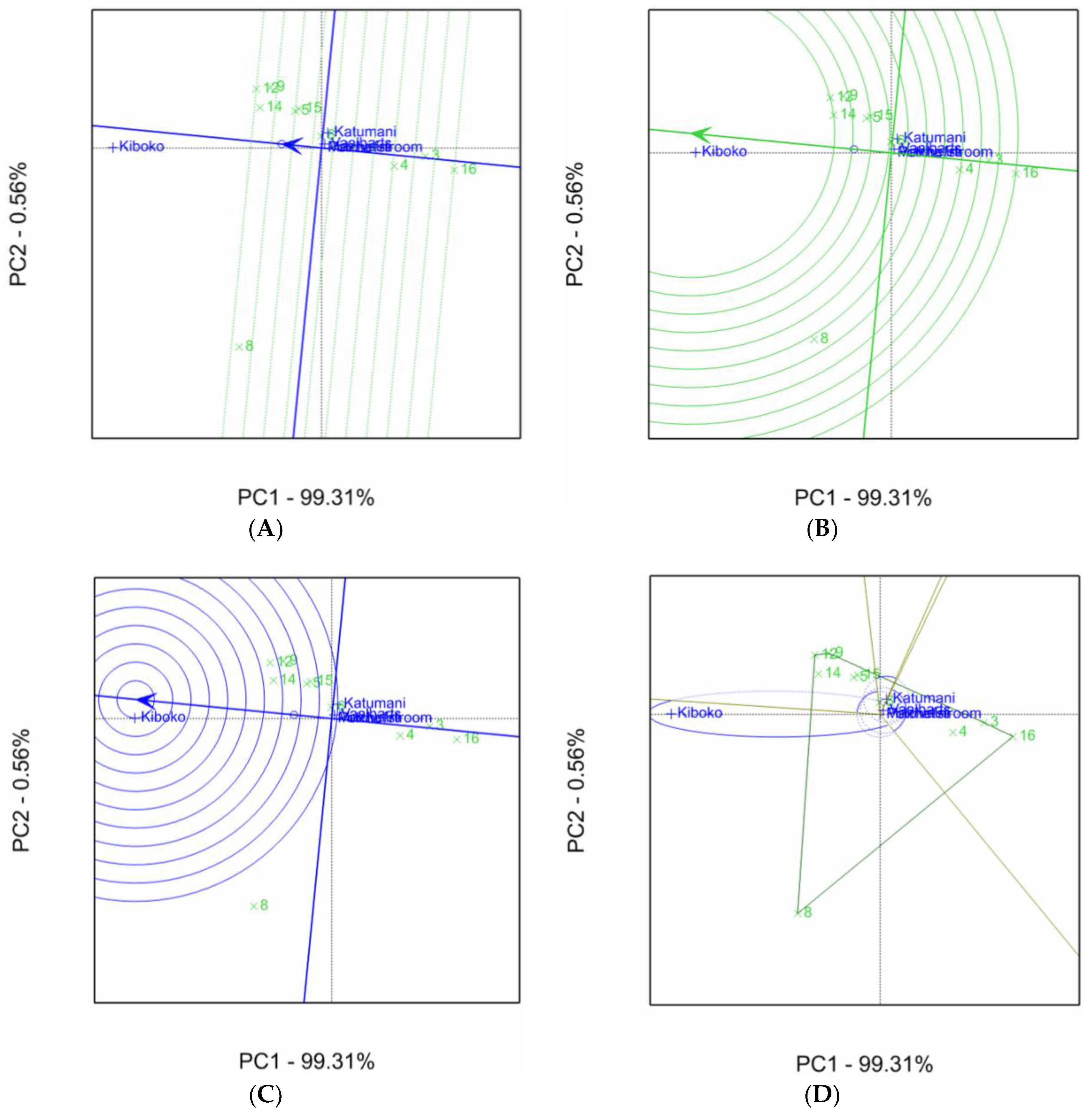
| Inbred Line Name | Aspergillus Ear Rot Severity (%) 1,2 | |||||
|---|---|---|---|---|---|---|
| Katumani | Kiboko | Makhatini | Potchefstroom | Vaalharts | Across Environment | |
| CKL05003 | 4.9 a | 18.9 a–c | 1.5 ab | 0.1 a | 1.4 a | 0.2 a |
| CKL05015 | 6.2 ab | 8.5 a–d | 0.1 c | 0.1 a | 0.6 a–c | 0.1 a–c |
| CKL05019 | 0.0 c | 16.7 ab | 1.6 ab | 0.0 a | 0.1 bc | 0.0 bc |
| CKL05022 | 0.3 bc | 0.0 e | 4.7 a | 0.0 a | 0.2 bc | 0.1 a–c |
| CML247 | 1.9 a–c | 0.0 e | 0.0 c | 0.0 a | 0.0 c | 0.0 c |
| CML264 | 0.0 c | 35.6 a | 0.2 bc | 0.1 a | 0.2 bc | 0.1 a–c |
| CML495 | 0.2 bc | 6.0 b–d | 0.3 bc | 0.0 a | 0.4 bc | 0.0 bc |
| La Posta | 0.2 bc | 3.4 c–e | 2.2 ab | 0.0 a | 0.0 c | 0.0 c |
| MIRTC5 | 1.3 bc | 4.6 b–e | 1.1 a–c | 0.0 a | 0.7 ab | 0.1 a–c |
| P502c2 | 1.0 bc | 1.8 de | 2.3 ab | 0.3 a | 0.0 c | 0.3 ab |
| Mean | 1.6 bc | 9.6 a | 1.4 b | 0.1 d | 0.4 cd | - |
| Inbred Line Name | A. flavus Target DNA (ng/µL) 1,2 | |||||
|---|---|---|---|---|---|---|
| Katumani | Kiboko | Makhatini | Potchefstroom | Vaalharts | Across Localities | |
| CKL05003 | 0.26 ab | 53.4 a | 0.01 b | 0.44 ab | 2.36 a | 0.26 ab |
| CKL05015 | 0.06 b | 14.87 ab | 0.02 b | 0.03 c | 5.70 a | 0.03 bc |
| CKL05019 | 0.11 b | 4.67 b | 0.00 b | 0.08 bc | 16.20 a | 0.06 bc |
| CKL05022 | 0.08 b | 12.07 ab | 0.78 a | 0.37 a–c | 10.08 a | 0.46 a |
| CML247 | 0.97 a | 4.27 b | 0.02 b | 0.07 bc | 6.21 a | 0.21 a–c |
| CML264 | 0.07 b | 11.10 ab | 0.00 b | 0.08 bc | 7.60 a | 0.05 bc |
| CML495 | 0.06 b | 5.77 b | 0.00 b | 0.03 c | 26.35 a | 0.03 c |
| La Posta | 0.15 b | 10.53 b | 0.00 b | 0.05 c | 2.23 a | 0.05 bc |
| MIRTC5 | 0.08 b | 6.03 b | 0.01 b | 0.46 a | 10.41 a | 0.23 a–c |
| P502c2 | 0.07 b | 43.5 a | 0.09 b | 0.12 a–c | 4.81 a | 0.10 bc |
| Mean | 0.19 c | 16.62 a | 0.09 c | 0.17 c | 9.19 b | - |
| Inbred Line Name | Total Aflatoxins (μg/kg) 1,2 | |||||
|---|---|---|---|---|---|---|
| Katumani | Kiboko | Makhatini | Potchefstroom | Vaalharts | Across localities | |
| CKL05003 | 0.00 b | 4.72 ab | 0.00 a | 0.00 ab | 0.00 a | 0.001 a |
| CKL05015 | 0.00 b | 3.86 a–c | 0.00 a | 0.00 ab | 0.06 a | 0.000 ab |
| CKL05019 | 0.00 b | 1.15 cd | 0.00 a | 0.00 b | 0.03 a | 0.000 ab |
| CKL05022 | 0.00 b | 2.12bd | 0.00 a | 0.00 b | 0.07 a | 0.001 ab |
| CML247 | 0.19 a | 0.30 d | 0.00 a | 0.00 b | 0.07 a | 0.000 ab |
| CML264 | 0.00 b | 0.67 d | 0.00 a | 0.00 b | 0.02 a | 0.000 ab |
| CML495 | 0.00 b | 0.50 d | 0.00 a | 0.00 b | 0.00 a | 0.000 b |
| La Posta | 0.00 b | 0.55 d | 0.00 a | 0.00 b | 0.00 a | 0.001 ab |
| MIRTC5 | 0.00 b | 1.10 cd | 0.00 a | 0.01 a | 0.05 a | 0.000 ab |
| P502c2 | 0.00 b | 7.09 a | 0.00 a | 0.00 b | 0.02 a | 0.000 ab |
| Mean | 0.02 b | 2.20 a | 0.00 b | 0.00 b | 0.03 b | - |
| Locality | Country | AER Severity (%) versus Total Aflatoxin (μg/kg) | AER Severity (%) versus A. flavus DNA Content (ng/µL) | A. flavus DNA Content (ng/µL) versus Total Aflatoxin (μg/kg) |
|---|---|---|---|---|
| Makhatini | South Africa | 0.200 NS (p = 0.289) | 0.149 NS (p = 0.433) | 0.321 NS (p = 0.084) |
| Potchefstroom | South Africa | 0.055 NS (p = 0.774) | −0.33 NS (p = 0.864) | 0.328 NS (p = 0.077) |
| Vaalharts | South Africa | 0.094 NS (p = 0.623) | 0.290 NS (p = 0.120) | 0.237 NS (p = 0.207) |
| Katumani | Kenya | 0.027 NS (p = 0.887) | 0.066 NS (p = 0.073) | 0.951 ** (p = 0.000) |
| Kiboko | Kenya | 0.010 NS (p = 0.957) | 0.243 NS (p = 0.196) | 0.772 ** (p = 0.000) |
| Combined | - | 0.257 ** (p = 0.002) | 0.176 * (p = 0.031) | 0.761 ** (p = 0.000) |
| Name/Pedigree | Line Code | Characteristics |
|---|---|---|
| CKL05003 | 3 | MA adaptation, turcicum leaf blight (TLB) and gray leaf spot (GLS) tolerant, semi-dent, white grain |
| CKL05015 | 4 | MA adaptation, TLB and maize streak virus (MSV) tolerant, semi-flint |
| CKL05019 | 5 | MA adaptation, intermediate maturity, TLB and GLS tolerant, flint, white |
| CKL05022 | 6 | MA adaptation, TLB and GLS tolerant, flint, white |
| CML247 | 8 | Lowland tropical (LT) adaptation, GLS tolerant, semi-dent, white |
| CML264 | 9 | LT adaptation, flint, white |
| CML495 | 12 | LT adaptation, flint, white |
| La Posta Seq C7-F103-2-1-1-1xMIRTC5 Bco F80-4-2-1-1-1-3-1-B-B-B | 14 | LT adaptation, drought tolerant background |
| MIRTC5 Bco F78-2-2-1-1-1xDERRc2 15-3-7-1-1-B-B-B-B | 15 | LT adaptation, semi-flint, white |
| P502c2-185-3-4-2-3-B-2-B-B-B-B-B-B | 16 | LT adaptation, semi-dent, white |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okoth, S.; Rose, L.J.; Ouko, A.; Netshifhefhe, N.E.I.; Sila, H.; Viljoen, A. Assessing Genotype-By-Environment Interactions in Aspergillus Ear Rot and Pre-Harvest Aflatoxin Accumulation in Maize Inbred Lines. Agronomy 2017, 7, 86. https://doi.org/10.3390/agronomy7040086
Okoth S, Rose LJ, Ouko A, Netshifhefhe NEI, Sila H, Viljoen A. Assessing Genotype-By-Environment Interactions in Aspergillus Ear Rot and Pre-Harvest Aflatoxin Accumulation in Maize Inbred Lines. Agronomy. 2017; 7(4):86. https://doi.org/10.3390/agronomy7040086
Chicago/Turabian StyleOkoth, Sheila, Lindy J. Rose, Abigael Ouko, Nakisani E. I. Netshifhefhe, Henry Sila, and Altus Viljoen. 2017. "Assessing Genotype-By-Environment Interactions in Aspergillus Ear Rot and Pre-Harvest Aflatoxin Accumulation in Maize Inbred Lines" Agronomy 7, no. 4: 86. https://doi.org/10.3390/agronomy7040086





