Transferring a Biomass Enhancement Biotechnology from Glasshouse to Field: A Case Study on Wheat GWD RNAi
Abstract
:1. Introduction
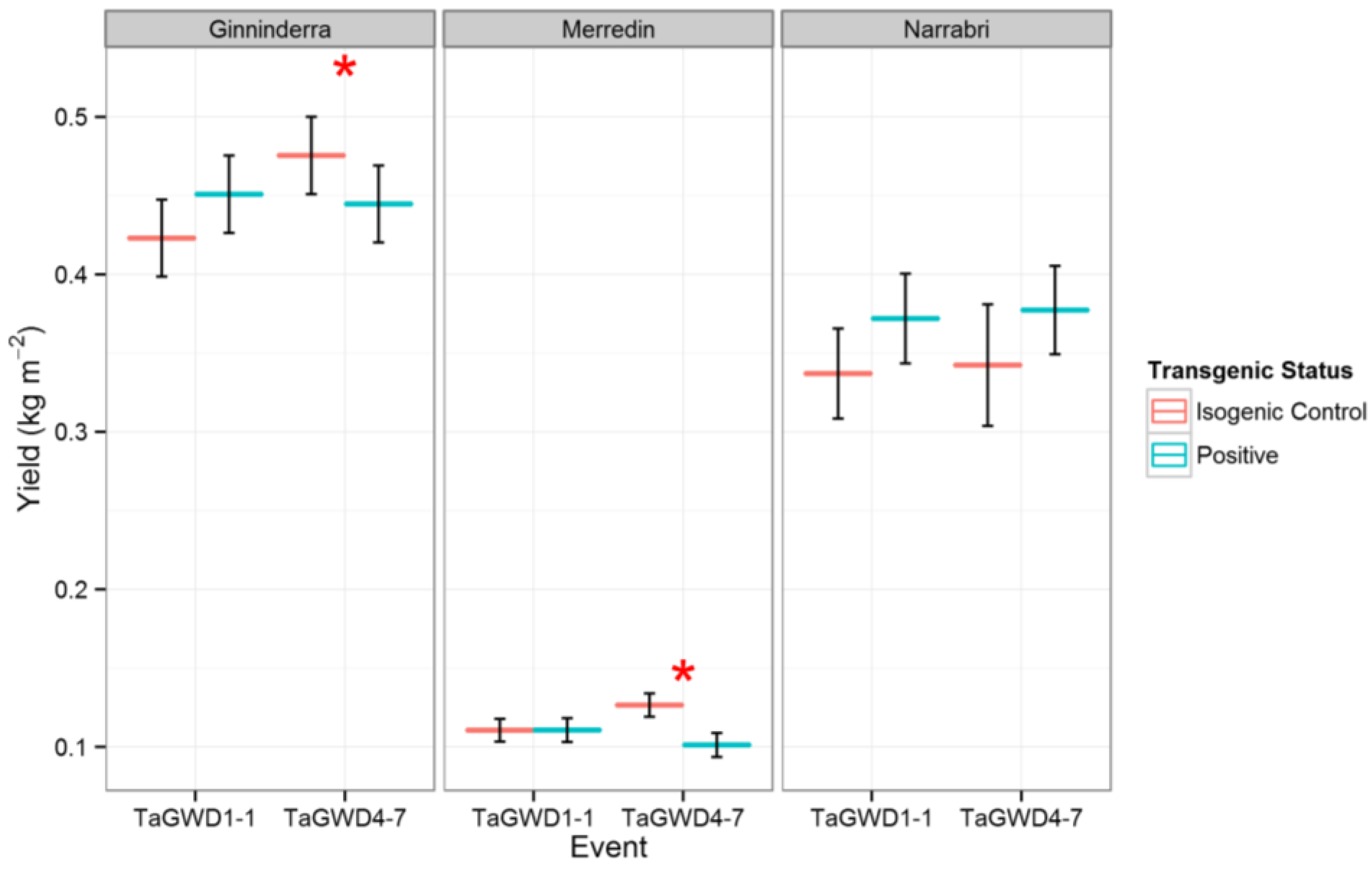
2. Results and Discussion
3. Experimental Section
3.1. Field Design
3.2. Measurements
3.3. Statistical Analysis
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
| Trait (Units) | Event | Site | Negative Segregant | GWD RNAi | ||
|---|---|---|---|---|---|---|
| Mean | SE | Mean | SE | |||
| TGW (g) | G1.1 | GES | 40.05 | 2.55 | 35.06 | 2.56 |
| TGW (g) | G4.7 | GES | 40.12 | 2.54 | 36.63 | 2.51 |
| TGW (g) | G1.1 | Merredin | 44.95 | 0.60 | 45.33 | 0.60 |
| TGW (g) * | G4.7 | Merredin | 43.20 | 0.60 | 45.36 | 0.60 |
| TGW (g) | G1.1 | Narrabri | 42.90 | 0.73 | 43.45 | 0.73 |
| TGW (g) | G4.7 | Narrabri | 42.71 | 0.73 | 43.48 | 0.73 |
| HC biomass (g) | G1.1 | GES | 236.38 | 20.04 | 212.43 | 19.99 |
| HC biomass (g) | G4.7 | GES | 248.63 | 20.07 | 224.68 | 26.96 |
| HC biomass (g) | G1.1 | Merredin | 319.75 | 21.03 | 278.55 | 21.03 |
| HC biomass (g) * | G4.7 | Merredin | 342.18 | 21.03 | 293.59 | 21.03 |
| HC biomass (g) | G1.1 | Narrabri | 303.77 | 25.27 | 320.78 | 40.72 |
| HC biomass (g) | G4.7 | Narrabri | 303.77 | 25.27 | 339.1 | 25.3 |
| Yield/spike (g) | G1.1 | GES | 1.59 | 0.06 | 1.54 | 0.06 |
| Yield/spike (g) | G4.7 | GES | 1.58 | 0.06 | 1.52 | 0.1 |
| Yield/spike (g) | G1.1 | Merredin | 1.42 | 0.06 | 1.38 | 0.06 |
| Yield/spike (g) | G4.7 | Merredin | 1.33 | 0.06 | 1.45 | 0.06 |
| Yield/spike (g) | G1.1 | Narrabri | 1.53 | 0.1 | 1.65 | 0.1 |
| Yield/spike (g) | G4.7 | Narrabri | 1.62 | 0.1 | 1.77 | 0.11 |
| Leaf area (cm2) * | G1.1 | Narrabri | 48.25 | 2.98 | 57.87 | 2.97 |
| Leaf area (cm2) * | G4.7 | Narrabri | 49.99 | 2.97 | 58.27 | 2.94 |
| Rachis nodes * | G1.1 | GES | 18.26 | 0.24 | 19.32 | 0.24 |
| Rachis nodes | G4.7 | GES | 18.33 | 0.24 | 18.67 | 0.24 |
| NDVI (Z14) | G1.1 | Narrabri | 0.38 | 0.06 | 0.42 | 0.06 |
| NDVI (Z14) | G4.7 | Narrabri | 0.44 | 0.06 | 0.37 | 0.06 |
| Field Trial Characteristics | Narrabri (NSW) | Ginnindera (NSW) | Merredin (WA) |
|---|---|---|---|
| Sowing density (seeds per m2) | 150 | 170 | 100 |
| Plot size (m2) | 10 | 10 | 8 |
| Nitrogen application (kg·Ha−1) | 40 (sowing) | 15 (sowing) | 7.5 (sowing) |
| 40 (anthesis) | 75 (anthesis) | 7.5 (anthesis) | |
| Planting date | 16/05/12 | 11/06/12 | 14/06/12 |
| Emergence | 26/05/12 | 26/06/12 | 07/07/12 |
| Anthesis | 11/10/12 | 10/11/12 | 07/09/12 |
| Harvest | 27/11/12 | 15/01/13 | 08/11/12 |
References
- Tanno, K.-I.; Willcox, G. How fast was wild wheat domesticated? Science 2006, 311, 1886. [Google Scholar] [CrossRef] [PubMed]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Millet, E.; Rong, J.K.; Qualset, C.O.; Mcguire, P.E.; Bernard, M.; Sourdille, P.; Feldman, M. Grain yield and grain protein percentage of common wheat lines with wild emmer chromosome-arm substitutions. Euphytica 2014, 195, 69–81. [Google Scholar] [CrossRef]
- Yang, Z.; Bai, Z.; Li, X.; Wang, P.; Wu, Q.; Yang, L.; Li, L.; Li, X. SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor. Appl. Genet. 2012, 125, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C. Wheat genomics comes of age. Curr. Opin. Plant Biol. 2017, 36, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Whan, A.; Dielen, A.S.; Mieog, J.; Bowerman, A.F.; Robinson, H.M.; Byrne, K.; Colgrave, M.; Larkin, P.J.; Howitt, C.A.; Morell, M.K.; et al. Engineering alpha-amylase levels in wheat grain suggests a highly sophisticated level of carbohydrate regulation during development. J. Exp. Bot. 2014, 65, 5443–5457. [Google Scholar] [CrossRef] [PubMed]
- Bowerman, A.F.; Newberry, M.; Dielen, A.; Whan, A.; Larroque, O.; Pritchard, J.; Gubler, F.; Howitt, C.A.; Pogson, B.J.; Morell1, M.K.; et al. Suppression of glucan, water dikinase in the endosperm alters wheat grain properties, germination and coleoptile growth. Plant Biotechnol. J. 2016, 14, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Smidansky, E.D.; Clancy, M.; Meyer, F.D.; Lanning, S.P.; Blake, N.K.; Talbert, L.E.; Giroux, M.J. Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc. Natl. Acad. Sci. USA 2002, 99, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- Smidansky, E.D.; Meyer, F.D.; Blakeslee, B.; Weglarz, T.E.; Greene, T.W.; Giroux, M.J. Expression of a modified ADP-glucose pyrophosphorylase large subunit in wheat seeds stimulates photosynthesis and carbon metabolism. Planta 2007, 225, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Tuncel, A.; Okita, T.W. Improving starch yield in cereals by over-expression of ADPglucose pyrophosphorylase: Expectations and unanticipated outcomes. Plant Sci. Int. J. Exp. Plant Biol. 2013, 211, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Liu, G.; Peng, X.; Wei, L.; Wang, C.; Zhu, Y.; Ma, Y.; Jiang, Y.; Guo, T. Increasing the starch content and grain weight of common wheat by overexpression of the cytosolic AGPase large subunit gene. Plant Physiol. Biochem. PPB 2013, 73, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Ral, J.-P.; Bowerman, A.F.; Li, Z.; Sirault, X.; Furbank, R.; Pritchard, J.R.; Bloemsma, M.; Cavanagh, C.R.; Howitt, C.A.; Morell, M.K. Down-regulation of Glucan, Water-Dikinase activity in wheat endosperm increases vegetative biomass and yield. Plant Biotechnol. J. 2012, 10, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Ritte, G.; Heydenreich, M.; Mahlow, S.; Haebel, S.; Kotting, O.; Steup, M. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006, 580, 4872–4876. [Google Scholar] [CrossRef] [PubMed]
- Ritte, G.; Scharf, A.; Eckermann, N.; Haebel, S.; Steup, M. Phosphorylation of transitory starch is increased during degradation. Plant Physiol. 2004, 135, 2068–2077. [Google Scholar] [CrossRef] [PubMed]
- Streb, S.; Eicke, S.; Zeeman, S.C. The simultaneous abolition of three starch hydrolases blocks transient starch breakdown in Arabidopsis. J. Biol. Chem. 2012, 287, 41745–41756. [Google Scholar] [CrossRef] [PubMed]
- Blennow, A.; Engelsen, S.B. Helix-breaking news: Fighting crystalline starch energy deposits in the cell. Trends Plant Sci. 2010, 15, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Zeeman, S.C.; Smith, S.M. Starch degradation. Annu. Rev. Plant Biol. 2005, 56, 73–98. [Google Scholar] [CrossRef] [PubMed]
- MacNeill, G.J.; Mehrpouyan, S.; Minow, M.A.A.; Patterson, J.A.; Tetlow, I.J.; Emes, M.J. Starch as a source, starch as a sink: The bifunctional role of starch in carbon allocation. J. Exp. Bot. 2047, 68, 4433–4453. [Google Scholar] [CrossRef] [PubMed]
- Zeller, S.L.; Kalinina, O.; Brunner, S.; Keller, B.; Schmid, B. Transgene × Environment Interactions in Genetically Modified Wheat. PLoS ONE 2010, 5, e11405. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Whan, A.P.; Verbyla, A.P.; Mieog, J.C.; Howitt, C.A.; Ral, J.-P. Transferring a Biomass Enhancement Biotechnology from Glasshouse to Field: A Case Study on Wheat GWD RNAi. Agronomy 2017, 7, 82. https://doi.org/10.3390/agronomy7040082
Whan AP, Verbyla AP, Mieog JC, Howitt CA, Ral J-P. Transferring a Biomass Enhancement Biotechnology from Glasshouse to Field: A Case Study on Wheat GWD RNAi. Agronomy. 2017; 7(4):82. https://doi.org/10.3390/agronomy7040082
Chicago/Turabian StyleWhan, Alex P., Arunas P. Verbyla, Jos C. Mieog, Crispin A. Howitt, and Jean-Philippe Ral. 2017. "Transferring a Biomass Enhancement Biotechnology from Glasshouse to Field: A Case Study on Wheat GWD RNAi" Agronomy 7, no. 4: 82. https://doi.org/10.3390/agronomy7040082




